1. Introduction
Coronary artery disease is a cardiovascular disease that has been found to be the leading cause of death in both developed and developing countries [
1]. Coronary artery calcium (CAC) scoring has been used to predict the risk of coronary heart disease [
2]. Determination of the CAC score (CACS) by computed tomography (CT) is based on axial slices, with a thickness of 3 mm, without overlapping or gaps and limited to the cardiac region. Calcification is identified as areas of hyper-attenuation in the CT images by using the Agatston method [
3]. The total calcium scores were calculated based on the number, area, and peak CT numbers of the calcific lesions detected. Previously, CAC scoring and its validation were performed manually.
Many methods for automatic CAC scoring are based on classical machine learning and digital signal processing techniques [
4,
5]. Classical machine learning methods are initially required to define specific data features and then use them as inputs to train the models. In these methods, feature engineering to transform the raw data into a suitable representation is necessary, and the used features significantly affect the performance of the classifier such as support vector machine.
Deep-learning models have overcome this limitation by extracting relevant features directly from the inputs without prior domain knowledge. Recently, deep-learning techniques have been successfully used in many fields, including image recognition, speech recognition, and medical diagnosis applications [
6,
7,
8,
9,
10].
Several deep-learning techniques for automatic CAC scoring and coronary artery plaque detection have been developed [
11,
12,
13,
14]. These methods mainly dealt with two tasks in CT examination: calcified region segmentation and the corresponding volume measurement.
Wang et al. introduced a neural network model with the ResNet architecture for quantization of CACS from the CT data of 530 patients [
11]. First, the CT scans are converted into three-dimensional (3-D) volume data. All the voxel points with high radiodensity value are segmented as the candidate calcified regions, which are fed as input to a neural network for automated analysis. The neural network classified each suspected calcified region into five categories according to the degree of calcification.
Shadmi et al. introduced an automatic method based on fully convolutional networks (U-Net [
15] and FCDenseNet) to segment coronary calcium and predict the Agaston score from 1054 non-contrast chest CTs [
12]. The dataset reflects a variety of originating institutions, acquisition devices, and manufacturers. They applied a set of heuristics to predict a bounding box around the heart, which is divided into consecutive slices. After feeding all the cropped axial slices through the network, the predicted volume is assembled where each voxel intensity represents its probability of being a CAC voxel. Then, two-dimensional (2-D) candidate blobs on each axial slice are identified as coronary artery calcification via a thresholding value, and a connected components analysis is performed. The cardiovascular-disease risk is categorized into five sub-groups based on the Agaston score. It was difficult to differentiate the 1–10 category from other sub-groups because it was highly sensitive to small prediction mistakes. Specifically, the precision, recall, and F-1 scores in the category are 60.0%, 15.0%, and 24.0%, respectively.
Zreik et al. proposed a multi-task recurrent convolutional neural network (CNN) for automatic detection and classification of coronary artery plaque and stenosis in coronary CT angiography [
13]. The features obtained by 3-D CNN are aggregated by a recurrent neural network that performs two simultaneous multi-class classification tasks: the type of coronary artery plaque detection and the anatomical significance of the coronary artery detection. Santini et al. introduced a CNN-based automatic calcium scoring system to identify true coronary calcifications and discard other lesions [
14]. The Agatston-based risk assessment of 56 patients was calculated and compared with manual annotations provided by an expert operator, considered as the ground truth reference. This method achieved 91.1% risk categorization accuracy. However, CT images were employed, and the number of CT image datasets was limited: 45 CT volumes for training, 18 volumes for validation, and 56 volumes for testing. In contrast, our model can predict CAC scores directly using a large-scaled electrocardiogram (ECG) dataset.
Cardiac rhythm abnormalities differ from the normal rhythm of the heartbeat cycle and are known as heart arrhythmia. The heart’s rhythmic irregularities can be detected using electrocardiogram (ECG). ECG is a one-dimensional (1-D) signal representing a time series, widely used as a basic diagnostic tool for the analysis of the cardiac condition of patients. ECG signals represent the recording of the bioelectrical activities of the heart. Millions of ECGs are recorded annually, with the majority being automatically analyzed, followed by an intermediate interpretation. Digital ECG programs providing diagnostic interpretation have been actively proposed [
16]. However, ECG interpretation is a mixture of both subjective and objective aspects, where even experienced cardiologists or experts can disagree [
17]. This significant interobserver variability makes ECG interpretation difficult; consequently, digital ECG analysis may lead to erroneous diagnosis, which may lead to unnecessary actions (such as a surgery or an operation) taken on the patient.
Several methods to diagnose patients’ heart condition from ECG signals have been proposed. Sengupta et al. introduced a method to predict the presence of abnormal cardiac muscle relaxation in reference [
18]. Using continuous wavelet transform, an ECG signal is converted into a normalized energy distribution in the frequency domain, which is used to calculate multiple indices, such as T-wave peak. Small changes on the surface ECG frequency spectrum, which are associated with the development of myocardial relaxation abnormalities, are magnified. Then, a random forest ensemble classifier with a Monte Carlo cross-validation procedure is used to identify patients at risk for left ventricular diastolic dysfunction.
Recently, deep-learning networks have been widely applied to ECG data, for screening hyperkalemia [
19], cardiac contractile dysfunction [
20], and cardiac arrhythmias [
21]. Ullah et al. introduced a 2-D CNN model for the classification of ECG signals into eight classes, including all major types of arrhythmia—from normal beat to ventricular escape beat arrhythmias [
22]. By applying a short-time Fourier transform to the 1-D ECG time series signals, they obtained 2-D spectrograms that encapsulate the time and frequency information within a single matrix. The proposed CNN model works on 2-D images of ECG signals as input data, in which the augmentation method, changing the image size with operations such as cropping, is employed.
Ebrahimi et al. reviewed 75 deep-learning-based studies reported in 2017 and 2018 for arrhythmia classification using ECG signals. They concluded that CNN-based methods have shown excellent performance in classifying different types of arrhythmia [
23]. Peimanker et al. presented a deep-learning model for real-time segmentation of heartbeats using ECG as inputs [
24]. They combined CNN and a long short-term memory (LSTM) model to detect knowledge about the location and morphology of different segment waveforms (P-wave, QRS complex, and T-wave) in ECG records, which can be used for arrhythmia classification. QRS complex, which is a combination of the Q wave, R wave and S wave, represents ventricular depolarization. Ribeiro et al. introduced a residual neural network to recognize six ECG abnormalities with the 12-lead ECG [
25].
Since ECG signals represent heart activation conditions, most studies on ECG mainly focus on cardiac arrhythmias. Sabour et al. discussed the relation of left ventricular hypertrophy and ECG abnormalities with CAC among 566 postmenopausal women selected from a population-based cohort study [
26]. In their statistical-analysis-based studies, many women with ECG abnormalities reflecting subclinical ischemia have CAC. In other words, authors showed only the relevance of repolarization abnormalities (T-axis and QRS-T angle) in ECG and CAC. However, they did not propose any quantitative method to compute CAC from the patient’s ECG data.
In this study, we introduce a deep-learning neural network model to predict CACS using a large scaled ECG dataset and the participant’s demographic information. Here, only the participant’s gender and age in the demographic information are used. To the best of our knowledge, no studies have been conducted to predict CACS directly using ECG data. Additionally, since CACS is measured using CT scans, the patient is exposed to radiation while the scan is being captured. The increased use of CT scans in a patient’s life has raised concerns regarding potential cancer risks [
27]. Furthermore, a CT scan is troublesome for the patient as it is time-consuming and expensive. Since a CT scan is not used for our CACS regression, the proposed method is safer, simpler, and less expensive than previous methods. In contrast, our study adopts a deep-learning model to predict the CACSs with ECG datasets.
The remainder of this paper is organized as follows. In
Section 2, we describe the pre-processing step to normalize the characteristics of the ECGs and introduce our neural network architecture. Then, we explain the experimental results in
Section 3 and conclude the paper in
Section 4.
2. Deep-Learning Model Design
In ECG diagnostics, data from 12 ECG leads (I, II, V0–5, III, aVR, aVL, aVF) are acquired simultaneously. In a 12-lead ECG, ten electrodes are placed on the patient’s limbs and chest surface. An electrode is a conductive pad that is attached to the skin and enables the recording of electrical currents. An ECG lead is a graphical description of the electrical activity of the heart, and it is created by analyzing several electrodes. In other words, each ECG lead is computed by analyzing the electrical currents detected by several electrodes [
28]. ECG signals are captured over a period of time (typically, 10 s).
Specifically, leads I, II, and III compare the electrical potential differences between two electrodes. Lead I compares the electrode on the left arm (exploring electrode) with the electrode on the right arm. Lead II compares the left leg with the right arm, and lead III compares the left leg with the left arm. The spatial organization of these leads forms a triangle in the chest (Einthoven’s triangle). According to Kirchhoff’s law, the sum of all currents in a closed circuit must be zero. As Einthoven’s triangle can be viewed as a circuit, Kirchhoff’s law can be applied to it. Einthoven’s law indicates that the sum of potentials in leads I and II is equal to the potential in lead III. Additionally, aVR, aVL, and aVF leads are a linear combination of leads I, II, and III according to the Goldberger equation. Leads aVR, aVL, and aVF can be calculated using leads I, II, and III. For example, the ECG wave in lead aVF is the average of the ECG deflection in leads II and III. Therefore, leads III, aVR, aVL, and aVF do not provide any new information; however, they provide new angles to view the same information. There are six electrodes on the chest wall and thus six chest leads (V0–5). Each chest lead offers unique information that cannot be derived from other leads. Based on the ECG lead characteristics, we trained a deep-learning neural network model on eight-lead combinations of the ECGs (I, II, V0–5) among the 12-lead ECGs [
28].
Figure 1a depicts the changes in the duration, amplitude, and interval of the data from the eight-lead ECG. In
Figure 1a, the x and y axes represent the time (seconds) and millivoltage (mV), respectively.
ECG signal properties vary from person to person and depends on various factors, such as age, gender, physical conditions, and lifestyle. Specifically, the characteristics of the sub-waves (P-waves, QRS complexes, and T-waves) of the ECG, including their duration, amplitude, and R–R interval, vary according to the subject. In ECG waveforms, the first deflection (wave) is the P-wave, which represents the activation (depolarization) of the atria. Ventricular depolarization is visible as the QRS complex. The T-wave represents the repolarization of the ventricles [
29]. ECG signals have very small amplitude (mV) and duration. The noise added in the capturing process usually degrades the performance of the classifier. Additionally, ECG signal detection has limitations regarding inter-patient and intra-patient variability, which means that two different beats can be of the same morphology among different patients. Such morphological variation can be seen within the same patient as well [
30]. This suggests that the characteristics of a single ECG lead can change when recording an ECG.
To improve the performance of the deep-learning model, we introduce a pre-processing step to normalize the characteristics of the ECGs. First, each lead of the ECG is segmented into several waves. The intervals of the segmented waves are determined as the period from the starting point of a P-wave to the end point of a T-wave. The number of segmented waves, H, is equal to the number of heartbeats of the subject per 10 s. Second, bidirectional weight interpolation, median pooling, and smoothing filtering are applied to the segmented waves. Here, interpolation and median pooling are implemented using library functions “scipy.interpolate.interp1d” with “cubic” argument and “skimage.measure.block_reduce” with “numpy.mean” parameter. The smoothing filtering is implemented using “numpy.convolve” function, in which a hanning widow of size 17 × 1 is used.
Figure 1b illustrates the pre-processed eight-lead ECG waves of one cycle, which are the eight-lead ECG waves in
Figure 1a from 1–2 s. In
Figure 1b, the x axis represents the sample points, and the y axis represents the voltage (mV). By choosing a sample with a maximum value between two samples, which is similar to max pooling with a 2 × 1 receptive field in deep learning, the number of sample points (400) of the pre-processed ECG waves is reduced to 200. Then, amplitude normalization is applied to the segmented waves. Through this pre-processing, the segmented waves of one cycle are transformed to normalized waves, which consist of 200 sample points with an amplitude of 0–1. In this work, we term the normalized wave of an interval as a unit wave for the deep-learning model.
As the unit waves of the 200 sample points are obtained from the one-lead ECG and we employ eight-lead ECGs, the input ECG dataset has two dimensions (200 × 8). The amplitude and interval of the unit waves are normalized, while maintaining the characteristics of the original ECGs. The unit waves of eight leads are employed as the training data. The total size of the training dataset is determined as the product of the number of times the ECGs are recorded, corresponding to the number of visits of a subject, by the heart rate, H.
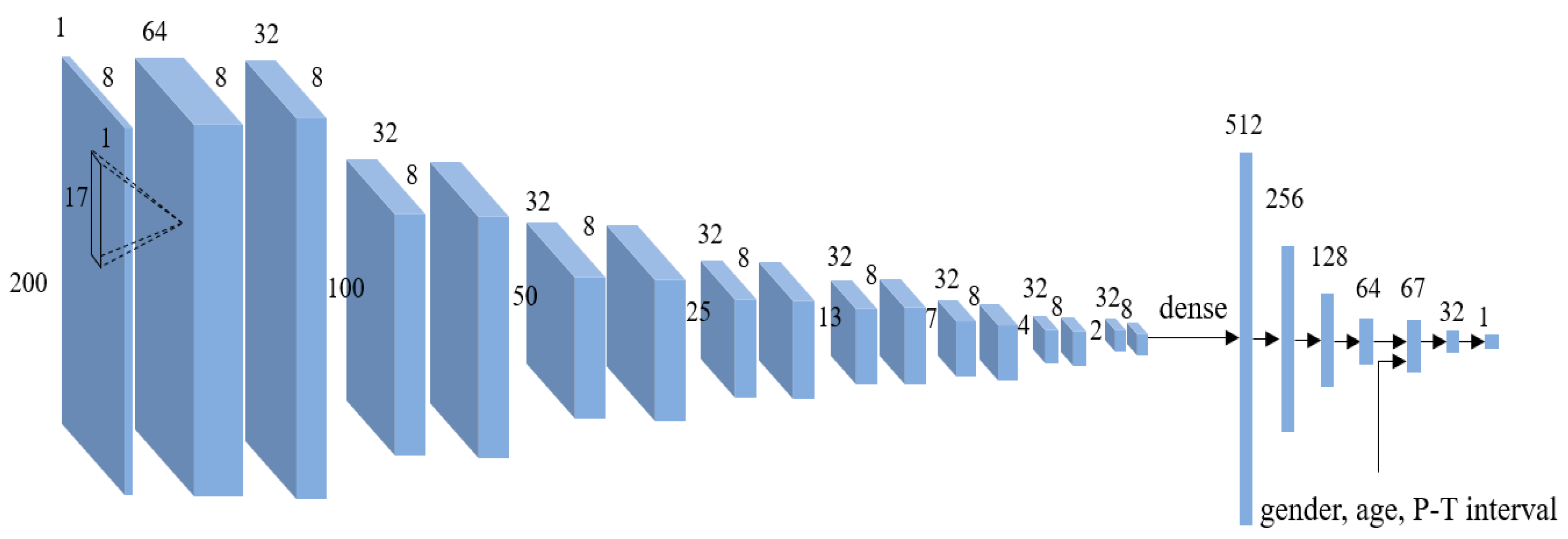
The neural network model architecture is a CNN with 16 layers and 5 fully connected layers (
Figure 2). We employ a 1-D filter to examine the unit wave of one lead, rather than using a 2-D filter to examine the coherence among the unit waves of eight leads. This is because, by using a 1-D filter, we can effectively derive features from the unit waves in the deep-learning model.
Table 1 presents the CNN layer configuration. Our deep-learning model comprises 16 convolutional layers, some of which are followed by max pooling layers. We employ max pooling with a 2 × 1 receptive field and a stride of 1.
Along with the output of the CNN, the P–T interval of the segmented waves of the ECGs before pre-processing is input to the dense layer. The normalized waves obtained by the pre-processing procedure consist of 200 sample points. This means that an absolute duration of the ECG wave is lost. To further consider the participant’s ECG characteristic, the P–T interval (duration) information of the ECG wave is also input to the dense layers. Here, the participant’s gender and age in the demographic information are also considered, as shown in
Figure 2. In this paper, the gender information of the male participant is coded to 0 and that of the female participant is coded to 1. The age of 100 years is scaled to 1.0. For example, the age of 45 years is scaled to 0.45. We use a rectified linear unit (ReLU) function as the activation function in the construction of our neural network architecture and use an adaptive moment estimation (Adam) optimizer for the adaptive computation of empirical gradients in the course of training process in which the gradient of the loss function with respect to the model parameters is computed following the stochastic process using a random subset of training data. Thus, the robustness of the proposed model is associated with the randomness in utilizing training data and it is naturally taken into consideration during the training phase.
As our goal is the early detection of heart disease possibility in the near future, the ECG dataset is collected from a regular medical screening at a healthcare center. Therefore, in general, the majority of participants have CACSs of zero or near zero, which means the dataset has an unequal distribution. Instead of computing the sum of the squared errors between the ground truth CACSs and the predicted values, a cosine similarity considering a relative magnitude (disease seriousness) of the ground truth CACSs in the batch dataset is used as a cost function of our neural network. Here, both the ground truth CACSs and the predicted values are represented as L2 normalized unit vectors, whose dimension number is the same as the batch size during the training stage. The L2 norm calculates the vector coordinate distance from the origin of the vector space. The degree of closeness between the ground truth CACSs and the predicted values is computed with an inner product of two normalized vectors as follows:
where
G and
P represent the ground truth CACSs and predicted values, respectively, and
Gi and
Pi their corresponding vector components,
n is the batch size. In our experiment, the batch size is 256, therefore, we define a 256-dimensional space as the cost domain. Our model makes the estimated vector closer to the ground truth vector in a learning process, which means we can estimate precise CACSs in a batch learning process.
In the pre-processing, the ECG of each lead is segmented into H unit waves. Each unit wave of each lead is used as part of the training dataset; therefore, the number of training data points is increased by H. This approach implies that the training datasets are considerably larger than the number of ECG readings collected from the study participants. As a single unit wave is used per lead, the proposed model is applicable to situations in which ECG data are acquired only for a short time.
3. Experimental Results
The ECG signals are captured using an electrocardiograph (supplied by Fukuda Denshi Co. Ltd.) with a duration of 10 s. As the ECGs are recorded on the paper with the fixed grid, the electrocardiograph machines generally provide the displaying and recording sensitivity function. The sensitivity of the ECG machine by Fukuda Denshi Co. Ltd. (Tokyo, Japan) is able to set at 1/4, 1/2, 1.0, and 2.0 cm/mV. The amplitude scale of ECG is dependent on the sensitivity configuration. However, in our pre-processing procedure, the amplitude value of ECGs is normalized to 0–1, and the ECG of each lead is segmented based on the heartbeats of the subject. Here, the number of segmented waves is equal to the number of heartbeats of the subject per 10 s. That means our deep learning model did not learn the absolute amplitude values of ECGs. The relative amplitude variations within one ECG and those among eight ECGs are used in CASC regression. Therefore, the sensitivity configuration of the ECG machine does not affect the performance of our model. In our CNN model, 1-D filters (17 × 1) are employed.
The ECG dataset has been collected from a regular medical screening for eight years (March 2010~November 2018) at the Total Healthcare Center of Kangbuk Samsung Hospital, Seoul, South Korea. CT scan is one of major examination courses in a regular medical screening at this healthcare center. The CT images of participants have been captured, and the medical professionals measured the CACS using the participant’s CT images. In our experiment, the CACS is used as the ground truth dataset. By comparing the CACS by our deep-learning neural network model with the ground truth dataset, the performance of our model can be validated quantitatively. The number of participants was 134,058. This means that our ECG dataset has been collected from a single hospital, despite large numbers of participants. Of the total number of participants, 74.24% (99,521 participants) were men and 25.76% (34,537 participants) were women. Some participants underwent the regular medical screening several times during that period. In our experiment, 177,547 ECG readings were used. The ECG characteristics of even the same participant changed over time. If the training dataset is set to a smaller percentage of the class (women), the total number of datasets decreases. Instead of balancing with the gender ratio in the dataset, our deep-learning model was trained on a large-scaled dataset. In other words, we employed a gender imbalanced dataset. The network model is trained using 142,037 ECG readings, and the test dataset consists of readings from 35,510 ECGs. The ratio between the training data and the test data is set to 8:2.
As depicted in
Figure 2, the unit waves (200 × 8) are input into our deep-learning model. The heart rate,
H, of the subjects was found to be 9–11 beats in 10 s on average, and thus the total number of datasets for training and testing increased to 1,792,919. Our model was trained for 100 epochs with a batch size of 256.
We used the CACS with the neural network model to determine whether a participant had a coronary artery disease. Traditionally, people with positive CAC, with scores in the ranges of 1–100, 100–400, and >400, are considered to be at low, intermediate, and high risk of both ischemia and cardiovascular disease, respectively [
5].
In
Table 2, the proposed network model is evaluated at an operating point that is selected such that the sensitivity and specificity are equal [
18]. This means that both sensitivity and specificity are the same as accuracy measure. In other words, in
Table 2, sensitivity, specificity, and accuracy have the same value. This point (Q* index) has been suggested as a possible global parameter to summarize the test accuracy of cognitive screening instruments and as a definition for the optimal test cut-off [
31]. A threshold is applied to the test datasets to evaluate the network model performance. The training and testing processes are repeated 10 times, and the obtained performances are averaged. The dataset was randomly assigned to the training and test subsets. Here, the Python NumPy library function “numpy.random.permutation” is employed to randomly permute a dataset. To compare the performances of our model under the same input (eight-lead ECG waves) condition, we implemented two network models: the deep neural network (DNN) and recurrent neural network (RNN). The first is based on the DNN model with six dense layers, in which ReLUs are used as active functions. The unit waves (200 × 8), by our pre-processing procedure, are flattened into 1600 × 1. The number of nodes of hidden layers are 1600, 256, 128, 66, 32, and 16. The gender and age information are also input to the fourth hidden layer. The second model is based on the RNN model with two hidden layers—the one with 256 and the other with 64 states—in which the number of output states is 32. To transform ECG waves to sequence data, the unit waves (200 × 8) are transposed into 8 × 200. Along with the output of the RNN, the gender and age information are input to the dense layers. The number of nodes of dense layers are 34, 32, and 16. In
Table 2, our method and two models were compared with respect to the accuracy and area under the receiver operating characteristic curve (AUC) measures. Here, the minimum (Min), maximum (Max), and average (Avg.) of the results were presented. Our model can predict the CACS using ECG data and demographic information (gender and age). As our main goal is to generate information about the heart-disease possibility, the CACS is used to generate a clinical interpretation of the heart disease. To more clearly identify the heart-disease possibility, the cardiovascular-disease risk has been categorized into seven cases: CACS ≥ 1, 25, 50, 100, 150, 200, and 400. As presented in
Table 2, our network model achieves an average AUC of 0.801–0.890, and the average accuracy of the proposed network model is in the range of 72.9–80.6%.
To validate the performance of our model, we also use metrics such as precision, recall rate, and F-1 score. Specifically, we focus on two cases (CACS ≥ 1 and 25) in the cardiovascular-disease-risk categorization based on the CACS. The first reason is that the previous method encountered a difficulty to differentiate the category (CACS = 1–10) from other categories [
12]. In Reference [
12], Shadmi et al. mentioned that it was highly sensitive to small prediction mistakes. As described in the Introduction, the precision, recall, and F-1 score in the category were 60.0%, 15.0%, and 24.0%, respectively. The second reason is that our goal is the early detection of heart-disease possibility. This means that it is important to generate a precise clinical interpretation of the coronary artery disease in the lower category. In the first case (CACS ≥ 1) and the second case (CACS ≥ 25), the percentages of the positive ECG data in the test dataset are 22.1% and 12.2%, respectively. This means that the dataset is imbalanced. In the first category, the precision, recall rate, and F-1 score of our model are 43.5%, 73.1%, and 54.6%, respectively. In the second category, the precision, recall rate, and F-1 score are 28.7%, 74.4%, and 41.1%, respectively. The experimental results indicate that our model outperformed the previous method [
12] in identifying patients at risk to a coronary artery disease, with respect to the recall rate and F-1 measures.
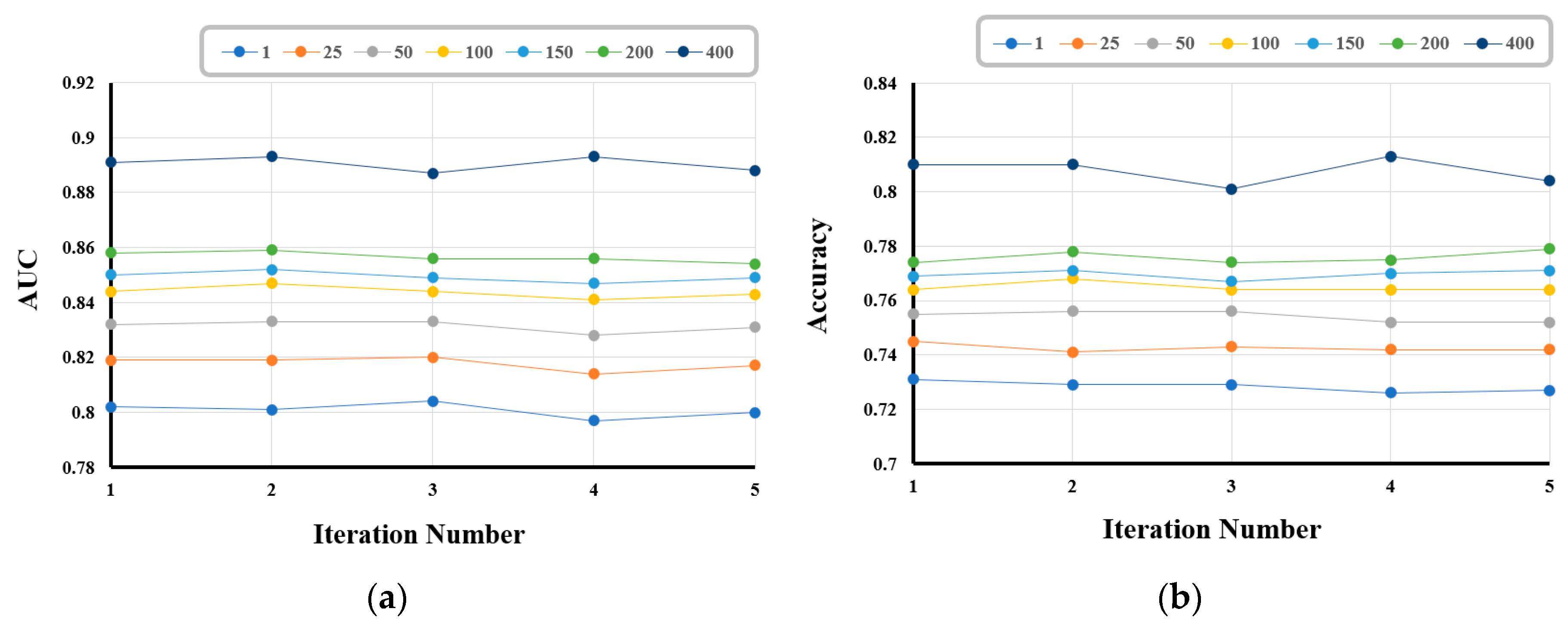
In the quantitative evaluation of the generalization induced by our model, we employ 5-fold cross validation, wherein all the samples in the dataset are randomly split into 5 smaller sets (folds). In the first iteration, the first fold is used to test the model, and the rest are used to train the model. In the second iteration, the second fold is used as the testing set, whereas the rest serve as the training set. This process is repeated until each of the 5 folds has been used as the testing set.
Figure 3 shows the performances of our model over five iterations with respect to AUC and accuracy; the performances do not change significantly during the iterations.
Table 3 shows that the mean and standard deviation (std. dev.) of the Matthews correlation coefficient, accuracy, and AUC in the 5-fold cross- validation framework. The experimental results demonstrate that our model is well generalized.
Figure 4a,b depict the ROC curves of the model performance when the CACS is greater than 1 and 400, respectively. The experimental results indicate that the proposed network model can effectively screen patients for the risk of coronary artery disease.
To further validate the robustness of our model, we included the experimental results according to the number of ECG leads.
Table 2 presents the performance when eight-lead (I, II, and V0–5) ECG waves are used as the input to our model. In the first case, two-lead (I and II) ECG waves are input to our model. The two leads compare the electrodes of the two arms and the left leg. In the second case, we use six-lead (V0–5) ECG waves, in which the exploring electrode is on the chest overlying the heart or its vicinity. The performances obtained in the two cases are presented in
Table 4. The number of ECG waves affect the performance of our model to some extent. In
Table 4, even if two-lead ECG waves are used, there is no significant difference in performance, compared with that in
Table 2.