Abstract
Cell nuclei segmentation is a challenging task, especially in real applications, when the target images significantly differ between them. This task is also challenging for methods based on convolutional neural networks (CNNs), which have recently boosted the performance of cell nuclei segmentation systems. However, when training data are scarce or not representative of deployment scenarios, they may suffer from overfitting to a different extent, and may hardly generalise to images that differ from the ones used for training. In this work, we focus on real-world, challenging application scenarios when no annotated images from a given dataset are available, or when few images (even unlabelled) of the same domain are available to perform domain adaptation. To simulate this scenario, we performed extensive cross-dataset experiments on several CNN-based state-of-the-art cell nuclei segmentation methods. Our results show that some of the existing CNN-based approaches are capable of generalising to target images which resemble the ones used for training. In contrast, their effectiveness considerably degrades when target and source significantly differ in colours and scale.
1. Introduction
Cell nuclei segmentation from stained tissue specimens is a useful computer vision functionality in histopathological image analysis [1,2]. Unlike cell nuclei detection, it provides whole nuclei binary masks that it can be used not only to extract the number and distribution of the cells but also to extract detailed information of each nucleus, such as colour, shape and texture features [3,4,5]. This information is helpful since it allows to count normal or cancer cells and recognise tissue structures, and because it is critical in cancer identification, grading, and prognosis [6,7,8,9].
The analysis of tissue specimens is performed after a staining procedure that highlights the interesting components of tissues [10]. The most used stain materials are hematoxylin & eosin (H&E) [11,12], which impart the blue-purple and pink colour to the nuclei and cytoplasm, respectively. For more than a century and still today, the interpretation of tissue specimens is performed under a microscope by human experts [10,13,14]. Nevertheless, this task requires a substantial manual effort for pathologists, whose results are subjective, as they are influenced by the level of expertise and tiredness, even though accuracy and efficiency are crucial in this field.
The growing need for objective and efficient automatic techniques has attracted the attention of many researchers in this field; these researchers have employed computer vision techniques to create computer-aided diagnosis (CAD) tools [4,15] for improving work efficiency and reducing error rates for pathologists [16,17]. In this paper, we focus on the nuclei segmentation task—in particular, on convolutional-neural-networks (CNNs)-based methods, that have recently demonstrated state-of-the-art performance [18,19]. Indeed, more straightforward approaches, such as thresholding, are efficient, but they fail with noisy images or colour variations [2,20], while more advanced image segmentation methods such as deformable models are complicated and computationally expensive [2,21].
However, despite the considerable effort spent so far by the research community, and the performance improvements achieved by current methods based on CNNs on benchmark datasets [22,23], cell nuclei segmentation remains a challenging task [24,25,26] due to chromatic stain variability, non-homogenous background, nuclear overlap and occlusion and differences in cell morphology and stain density [2,19,27]. This task is even more challenging in real applications where the specimens could belong to different tissue types. A further variability in optical image quality comes from the use of various staining protocols and digital microscopes or scanners [1,28].
We focus on this challenging application scenario which was inspired by our work in the recent HistoDSSP (Histopathology Decision Support Systems for Pathologists, based on whole slide imaging analysis) project, aimed at the design and implementation of a prototype of decision support system for pathologists (DSSP) to be integrated into a laboratory information system (LIS).
In particular, we consider fully unsupervised application scenarios in which a system has to be deployed on a target image database for which it is not possible to collect annotations for training or fine-tuning. Additionally, the target image database is updated daily with images belonging to different laboratories. Although considerable progress has been achieved so far and some solutions based on domain adaptation (DA) or transfer learning have already been proposed [29,30], cell nuclei segmentation remains a challenging task in the setting mentioned above. In particular, only limited cross-dataset evaluations of existing methods have been provided in the respective papers, partly due to the small number and tiny size of publicly available datasets, but mainly because the authors focused their attention on DA problems similar to other fields of applications, neglecting the real DA problems that can be present in this field of application.
Therefore, the literature is still lacking a thorough evaluation and analysis of the performance of existing methods under realistic cross-dataset settings. These are, however, necessary steps towards further development of nuclei segmentation methods that can be effectively deployed in real application scenarios. Accordingly, this work aims to evaluate the performance gap of state-of-the-art CNN-based cell nuclei segmentation methods between same-dataset and cross-dataset scenarios, i.e., where manually annotated images of the target dataset are or are not available for training, respectively. To this aim, we simulate cross-dataset settings using the available benchmark datasets and consider several state-of-the-art CNN-based methods, for which the code was made available by the authors.
The remainder of our paper is organised as follows. In Section 2, we review some related works dealing with cell nuclei segmentation. In Section 3, we discuss the challenges and the open issues in this field in detail, focusing on the application scenario mentioned above. The used datasets, the tested methods and experimental settings are shown in Section 4. In Section 5, we present the obtained results. Finally, the conclusion and perspectives are drawn in Section 6.
2. Related Works
A variety of standard image analysis methods are based on H&E image analysis in order to extract cell nuclei [31,32], with some based on detection [33,34] and other based on segmentation [35,36]. For the reasons mentioned above, segmentation-based methods, including segmentation methods that exploit thresholding, clustering, watershed algorithms, active contours and CNNs, have been preferred over detection-based ones. A variety of pre- and post-processing techniques have also been extensively used—in particular, in combination with simpler segmentation techniques [4].
Automatic-thresholding-based methods were the most used cell nuclei segmentation techniques in the early stage of this research area [2], including single Otsu thresholding [37] or multi-level Otsu thresholding [38]. In most cases, the segmentation results, obtained by thresholding, were refined through morphological operations such as morphological reconstruction [20] or multiple morphological operations [39]. Even the watershed transformation [40] was used for nuclei segmentation, but mostly as a post-processing method for overlapping objects segmentation. It was used both in its classical version, coupled with the distance transform [41], and in its marker-controlled version, where the markers are extracted with a conditional erosion [23,42].
However, the segmentation results of thresholding-based methods could be heavily affected by chromatic stain variability and a non-homogenous background. For these reasons, more advanced image segmentation methods have been proposed over the years. For instance, the active contour model has been combined with the expectation–maximisation (EM) algorithm [43]. Different level set methods have been used to delineate the cell boundaries, both by using a multi-phase approach [44] and exploiting the mean shift clustering as the initialisation points [1]. Even the graph-cut method was used to segment the cell nuclei coupled with an improved Laplacian-of-Gaussian filter to detect the seed points [24].
Nevertheless, the methods mentioned above could be complex and too sensitive to the initialisation and setting parameters. Various machine-learning-based nuclear segmentation approaches have been investigated to better tackle this issue and the variation over cell morphology and colour. The former type of machine-learning-based method exploits handcrafted features, such as filter bank responses, geometric and texture descriptors, that are forwarded to standard classification algorithms [45,46]. Recently, the great success of deep learning methods gave birth to a new type of machine-learning-based methods for nuclei segmentation, achieving outstanding performances [2,19,22,28,47]. In these methods, the images are directly used to feed CNN to label pixels as nuclear (foreground) or non-nuclear (background). Even with CNN-based methods, some additional post-processing operations have been used to separate the overlapping or touching nuclei in the foreground regions by using the shape deformation method [19] or the watershed transformation [23,28,36]. Other approaches instead added a third class for the nuclei boundary to define a precise separation between background and foreground regions, especially between touching nuclei, avoiding the post-processing procedures. The boundary region can be directly used to separate the touching nuclei into individual ones by removing the boundary from the foreground [47], or it can be used as a constraint for a growing region that starts from the foreground seed points [22].
Unfortunately, CNN-based methods require massive amounts of annotated and representative training data. The amount of the required training data is even larger for a network deployed in a real application for the segmentation of nuclei from various organs. As a consequence, when training data is scarce or not representative of deployment scenarios, CNNs may also suffer from overfitting to a different extent, and may hardly generalise to images coming from various tissues. Moreover, the performances are bounded by the quality of the annotated images. To solve the problems mentioned above, CycleGANs [48] have been used to create some additional synthetic training images of different tissue types [49] and even coupled with a specific loss function to take into account to the quality of synthesised images [29].
3. Open Issues and Goal of this Work
As previously reported, cell nuclei segmentation in histopathology images has been extensively studied using a variety of methods. Most work has focused on developing nuclei segmentation methods for single tissues and specific applications, while others focused on more generic application by developing nuclei segmentation methods for multiple tissues. However, there is still a lack of training datasets for certain organs, and even the single-tissue and multi-tissue datasets do not provide an exhaustive overview of the possible morphological variations of the cells caused by a greater disease grade. Furthermore, even images belonging to the same organ could present different appearances due to chromatic stain variability, non-homogenous background and variability in optical image quality caused by various digital microscopes or scanners [1,28]. An example of such a chromatic issue is shown in Figure 1.
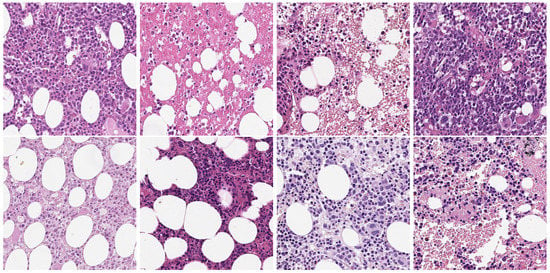
Figure 1.
Example of chromatic issues on images belonging to the same organ.
Recently, some authors tried to incorporate the benefits of the unsupervised domain adaptation (UDA) methods to tackle these issues. UDA methods allow adapting a model trained on a source domain to another target domain by exploiting some target images without annotations [50,51,52,53,54]. Such methods achieve state-of-the-art performance on UDA on object detection and classification. However, they are not specifically designed for semantic segmentation; thus, they ignore the domain shift at the semantic level. Indeed, several researchers exploiting UDA have, in their respective studies, proposed for semantic segmentation on many medical images using domain-invariant intermediate features [55] or both pixel- and feature-level adaptations with [29,30,54,56]. Among them, only two methods are designed for cell nuclei segmentation, providing an adaptation only at a pixel level [29] or at both the pixel and feature levels [30].
Although some solutions based on domain adaptation (DA) have already been proposed, they are mainly inspired by other fields of applications, neglecting most of the issues that can be present in this field, especially in real applications. In particular, they all require representative, although unlabelled, images of the target domain, which is not feasible in the real application scenario considered in this work. In this scenario we are dealing with in the HistoDSSP project, a system has to be deployed on a target image database for which it is not possible to collect annotations for training or fine-tuning; additionally, the target image database is updated daily with images belonging to various laboratories. In other words, just a few images of the same domain will be available to perform the adaptation.
Furthermore, only limited cross-dataset evaluations were provided in the respective papers, and most of the experiments focused on the adaptation from fluorescence microscopy images to H&E stained histopathology images [30], which is quite far from a real application. A realistic cross-dataset evaluation for cell nuclei segmentation is, therefore, still lacking in the literature. This is, however, a fundamental issue which must be addressed in order to be able to develop nuclei segmentation methods in real applications. Accordingly, this work aims to provide a comprehensive evaluation on the performance gap of state-of-the-art cell nuclei segmentation methods between same-dataset and cross-dataset scenarios, i.e., where manually annotated images of the target dataset are or are not available for training, respectively.
4. Experimental Evaluation
In this section, we evaluate and compare the cell nuclei segmentation performances of state-of-the-art methods on benchmark datasets on a real application scenario. The goal of our experiments is to evaluate the effectiveness of existing CNN-based methods for nuclei segmentation on a real application scenario. To this aim, we carried out the experiments using a representative selection of five cell nuclei segmentation methods on two benchmark datasets. In the following, we first describe the methods and datasets we used; then, we describe the experimental set-up.
4.1. Nuclei Segmentation Methods
In order to make a fair comparison, the CNN-based methods tested in this work were selected from among the ones whose source code was publicly available. These constraints greatly facilitated the research and allowed us to select five representative methods based on different CNN architectures. The first one exploits a fully convolutional network (FCN) for semantic segmentation [57] (implemented in the Python programming language https://github.com/appiek/Nuclei_Segmentation_Experiments_Demo) in order to deal with the large appearance variations of the nuclei. Indeed, in this architecture, the fully connected layers are replaced with convolutional ones that are able to produce a pixel-wise prediction on arbitrary-sized images. The second one is based on the holistically-nested edge detector (HED) [58] (implemented in the Python programming language https://github.com/appiek/Nuclei_Segmentation_Experiments_Demo), which is a learning-based end-to-end edge detection system that uses a common CNN to predict a rich hierarchical edge map to image at multiple scales by fusing the output of all convolutional layers. Thus, the HED model is naturally multi-scale. The third method is based on a UNet architecture [59], which is one of the most used for cell nuclei segmentation, even combined with other architectures or other UNets; however, in this case, we used a single UNet (implemented in the Python programming language https://github.com/limingwu8/UNet-pytorch). The UNet is a particular type of FCN that owes its name to its symmetric shape, which is different from other FCN variants. The fourth approach is based on the mask-regional CNN (Mask R-CNN) [60] that, differently from the previously mentioned approaches, as it is an extension of object detection CNNS, performs an instance segmentation. Thus, Mask R-CNNs should be able to individuate and separate single cells [61] (implemented in the Python programming language https://github.com/hwejin23/histopathology_segmentation). The last methods exploit a specific architecture for nuclei segmentation called Deep Interval-Marker-Aware Network (DIMAN) [23] (implemented in the Python programming language https://github.com/appiek/Nuclei_Segmentation_Experiments_Demo), which learns multi-scale feature maps that are stacked as a feature pyramid by skip connections. Unlike the other methods that predict just the foreground regions, this architecture predicts the foreground, the interval regions and the marker of nuclei simultaneously. Thus, it has been designed to facilitate and simplify further post-processing operations.
4.2. Datasets
In our experiments, we evaluate the performance of nuclei segmentation methods on two public histopathology image datasets that are two of the most used for cell nuclei detection and segmentation. MIC17 (available at http://miccai.cloudapp.net/competitions/) is the dataset provided for the MICCAI 2017 Digital Pathology Challenge. It includes 32 annotated squared image patches of sizes or that were manually cropped from H&E-stained histopathology Whole Slide Images (WSIs) of four various types of cancer, namely glioblastoma (GBM), lower grade glioma (LGG), head and neck squamous cell carcinoma (HNSC) and lung squamous cell carcinoma (LUSC). The first row of Figure 2 shows an image example for each type of cancer present in the dataset. During our experiments, we used the same partitions already used in [23] that consist of a training set composed by 24 randomly selected images (six for each cancer type) and a testing set composed by the remaining images (two for each cancer type). BNS (available at https://peterjacknaylor.github.io/PeterJackNaylor.github.io/2017/01/15/Isbi/), presented by Naylor et al. [28], contains 33 manually annotated images that were obtained by randomly cropping the patches with a fixed size of 512 × 512 from the original H&E-stained histopathology WSIs. The third row of Figure 2 shows some images present in the dataset. Even in this case, we used the same partitions proposed in [23] that consist of a training set composed of 23 randomly selected samples and a testing set composed by the remaining images.
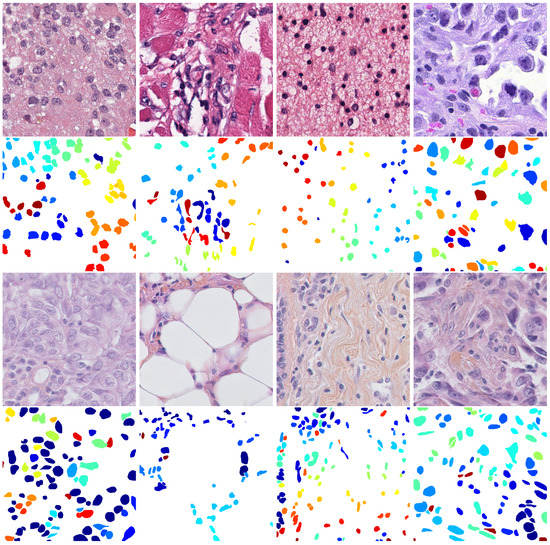
Figure 2.
Example of the images present in the used datasets: on top, the images belonging to MIC17 (GBM, LGG, HNSC and LUSC, respectively), and on bottom, the images belonging to BNS.
Both datasets present H&E images in PNG format, and each image is associated with a file containing the ground truth of the manual segmentation performed by expert pathologists. As can be observed in the second and fourth lines of Figure 2, the ground truth images contain different labels for each cell, meaning that they can be used to evaluate both the segmentation and counting performances.
In addition, we also used images that were expressly collected for the HistoDSSP project, and therefore cannot yet be shared publicly. The original WSIs were acquired with an Aperio AT2 scanner with a magnification of ×20, which was lower than the magnification used to acquire the public image datasets, which was 40×. Then, from the H&E-stained histopathology WSIs, a series of semiautomated crops with a fixed size of 512 × 512 were selected. Some images present in the dataset are shown in Figure 1. Unfortunately, for matters of time and resources, we could not get the manual annotations. Still, this set of images was constructive to visually evaluate the performances of the segmentation methods on images with a different scale from the training images.
4.3. Experimental Set-Up
All experiments were conducted under the same conditions. Considering that CNN-based methods need a large number of image data and that the number of training data were minimal for both datasets, to avoid overfitting, data augmentation methods were used to expand the training data. To this aim, we used random cropping, vertical and horizontal flipping (already implemented in the MATLAB programming language and available at https://github.com/appiek/Nuclei_Segmentation_Experiments_Demo). The random cropping procedure extracted 20 random crops of size 224 that were flipped vertically and horizontally with a probability of 50%. This procedure increased the original training sets by twenty times, so the total numbers of training images for MIC17 and BNS were 480 and 460, respectively. The training processes were carried out for 300 epochs with a learning rate of 0.0002, decreased by a factor of 0.8 per epoch. For the DIMAN loss function, we used the parameters suggested by the same authors [23]. For fairness, we did not exploited the post-processing procedures provided in the respective papers, since they could have significantly influenced the result by benefitting specific methods and not benefitting others. We conducted all the experiments on a single machine with the following hardware configuration: Intel(R) Core(TM) i9-8950HK @ 2.90 GHz CPU with 32 GB RAM and NVIDIA GTX1050 Ti 4 GB GPU.
To evaluate the performances on nuclei segmentation, we used the same evaluation metrics provided by the gland segmentation challenge contest (already implemented in the MATLAB programming language and available at https://warwick.ac.uk/fac/sci/dcs/research/tia/glascontest/evaluation/); they were F1-score (F1), precision (P), recall (R), object-level Dice index (ODI) and object-level Hausdorff distance (OHD). The first three metrics were used to evaluate the counting and object-level nuclei segmentation performances. is the average harmonic mean between the P and R for each object, where P and R gives information on the number of correctly segmented cells; in particular, P penalises oversegmentation (the lower the P value, the higher the oversegmentation) and R penalises undersegmentation (the lower the R value, the higher the undersegmentation). , P and R values range over the interval and are computed following Equations (1)–(3):
where (true positive) indicates the number of segmented cells that are present in the ground truth image, (false positive) provides the number of segmented cells that are not present in the ground truth image and (false negative) gives the number of ground truth cells that are not present in the segmented image.
The last two metrics instead were used to evaluate not only the object-level but also the pixel-level nuclei segmentation performances. The usual Dice index measures the similarity between the set of pixels belonging to the ground truth image G and the set of pixels belonging to the segmented image S as follows:
Instead, the ODI also takes into account the information of individual cells. Considering and , the number of cells in the ground truth and the segmented images, respectively, the ODI is defined as
where
Equation (5) present two summation terms that are weighted by the relative area of the cell, the first summation term reflects how well each ground truth cell overlaps its segmented cell, and the second summation term reflects how well each segmented cell overlaps its ground truth cell. The ODI value ranges over the interval , where the value of 1 means perfect segmentation. The OHD measures the boundary-based segmentation accuracy between the segmented cells and the ground truth cells. Differently from the other metrics that compute a score or a similarity value, OHD computes a distance value; the lower the OHD value, the better the segmentation accuracy, thus OHD = 0 means perfect segmentation. The usual definition of a Hausdorff distance between ground truth object G and segmented object S is
where denotes the Euclidean distance. Similarly to ODI, the OHD takes into account also the information of individual cells, and is defined as:
Finally, to simulate a real application scenario, we performed several cross-dataset experiments using training images taken from one source dataset and testing the resulting model on images taken from a different (target) dataset. As a baseline to evaluate the performance gap between a laboratory scenario and real application scenario, for each model, we also include the same-dataset results, obtained using training and testing samples from the same dataset.
5. Results
Table 1 show the results of the cross-dataset and same-dataset experiments on the above-mentioned datasets with the selected nuclei segmentation methods. For ease of comparison, for each target dataset, the same-dataset performance is highlighted in grey.

Table 1.
Same-dataset (highlighted in grey) and cross-dataset performances of FCN, HED, UNet, Mask R-CNN and DIMAN nuclei segmentation methods. The best cross-dataset result for each target (test) dataset is reported in bold.
As can be seen, the same-dataset performances of the various methods are quite aligned with each other, with a slight advantage for the HED method in both datasets and on almost all metrics. Obviously, these results represent ideal performances since both the training and test images come from the same dataset. The most interesting results instead came from the cross-dataset comparison, which simulated the behaviour of the segmentation methods in a real application. As expected, the performances decreased significantly when the target dataset was different from the source dataset. The most evident drop in performance was observed when the target dataset was BNS. In this case, the only method with satisfactory results was DIMAN, while when the target dataset was MIC17, the drop in performance was less evident and both DIMAN and HED performed well. In general, in cross-dataset experiments, the most remarkable drop occurred in the R value, when the target dataset was BNS, and in the P value, when the target dataset was MIC17. In other words, it means that in the first case, the models tended to underestimate the number of cell nuclei (many false negatives), while in the second case they tended to overestimate the number of cell nuclei (many false positives). Both errors could have a significant influence in a real application that relies on the cellularity value to identify or grade the presence of pathology. To test the significance of our results, we used the Wilcoxon test [62], which is the statistical test recommended for this scenario [63]. This test demonstrated that the drop in performances between same-dataset and cross-dataset is statistically significant (p-value ) in almost all the used metrics except the OHD. To better highlight the statistical differences between the same-dataset and cross-dataset performances, we reported in Figure 3 the bar charts of the p-value obtained with the Wilcoxon test. We evaluated the null hypothesis for precision and recall obtained with the same-dataset and cross-dataset for all the nuclei segmentation methods trained with the MIC17 dataset (in red) and the BNS dataset (in blue).
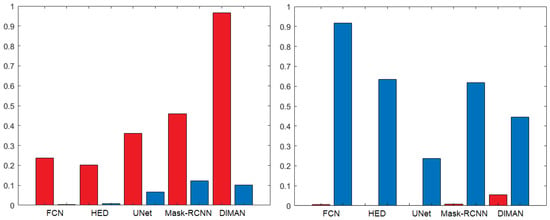
Figure 3.
Bar charts showing the p-value for the Wilcoxon test on precision (left) and recall (right) obtained with the same-dataset and cross-dataset for all the nuclei segmentation methods trained with the MIC17 dataset (in red) and the BNS dataset (in blue).
As can be observed, the two charts were complementary; indeed, for all the methods and the BNS dataset, the statistical difference was evident (low p value) in precision, while for the MIC17 dataset the statistical difference was evident in recall, as we already pointed out previously. In order to carry out a more detailed analysis and understand the generalisation power reached by the CNN-based methods, we performed further experiments involving the models previously trained with the MIC17 and BNS datasets, but using as target sets the individual subsets of the different types of cancer included in the MIC17 dataset. The results of these experiments are reported in Table 2.

Table 2.
Generalisation ability of FCN, HED, UNet, Mask R-CNN and DIMAN nuclei segmentation methods trained on MIC17 and BNS datasets and tested using the single MIC17 subsets, which were GBM, LGG, HNSC and LUSC.
As expected, the results were much better when the source dataset was MIC17, since images of the same domain were used both for training and testing the models. Even for this reason, it was not expected to see such a low generalisation capacity of these models that were instead overfitted on a single image subset. All the nuclei segmentation methods performed well with GBM target images. At the same time, they had a noticeable drop for the other target images—in particular, for LCC, where both Mask R-CNN and DIMAN showed the most critical drops. The HED method was the one that showed a more constant trend with the MIC17 source dataset, and the best performances with the BNS source dataset, even if the performance gap between the various subsets was not negligible. Indeed, even when the source dataset was BNS, all the nuclei segmentation methods performed well with GBM target images. In some cases, their performances were even better (for HED and UNet) than the corresponding performances when the source dataset was MIC17. Even in this case, we tested the significance of our results with the Wilcoxon test; that demonstrated that the gap in performances inside the individuals subsets was statistically significant (p value ) in almost all the used metrics. This gap in the performances obtained with the individual subsets was mainly due to the visual differences that their images present. To highlight these differences, we have reported in Table 3 some chromatic statistics both for the whole images and for the nuclei regions only.

Table 3.
Colour statistics extracted from the used datasets and subsets. From the left: average and standard deviation for the whole image, red (R), green (G), blue (B) and average and standard deviation for the nuclei only, R nuclei, G nuclei and B nuclei.
As can be observed, the datasets and subsets have different chromatic features and it is not a coincidence that the subset which differ the most from the others is precisely the LCC subset. Certainly, the set of features reported in Table 3 is not exhaustive, but it is representative of the issues present in histopathology images.
A further issue is represented by the difference in the shape and size of the cells. Indeed, most nuclei segmentation methods perform well for cells with standard shape and size, otherwise they could fail. To confirm what has just been said, we visually evaluated the performances of the nuclei segmentation methods trained using MIC17 and BNS datasets over the images acquired during the HistoDSSP project. For brevity in Figure 4, we reported the visual results obtained over a single patch, where the segmentation result of each model (in green colour) was superimposed on the original image.
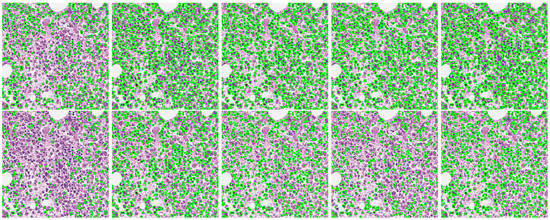
Figure 4.
Visual segmentation results of the CNN-based methods trained using MIC17 and BNS datasets over an image used in the HistoDSSP project.
As can be seen, both the models trained with the MIC17 and the BNS datasets underestimated the number of cells, but in different ways. Indeed, the models trained with the MIC17 dataset undersegmented the images by producing vast regions incorporating more cells and some background parts. On the contrary, the models trained with the BNS dataset produced smaller regions by segmenting only portions of cells and completely neglecting other cells. The leading cause of these results lies in the characteristics of the public datasets used for training, whose images, even if coming from different tissues, were all acquired at a magnification of 40×. Thus, the obtained models were not able to generalise to images acquired with different scale (magnification of 20×), not even the HED and DIMAN methods, which are multi-scale by nature.
All the attained results demonstrate that much effort is still needed to be able to create more generalising methods and, therefore, methods that are suitable for real applications. Indeed, even if some nuclei segmentation methods can achieve relatively good performances in cross-dataset scenarios when the target images are similar to the ones used for training, much effort must be devoted to guaranteeing higher invariance in colour and scale.
6. Conclusions
In this work, we evaluated the performances of several state-of-the-art nuclei segmentation methods based on CNNs, focusing on a challenging, real-world application scenario in which a LIS database is updated daily with images of different domains and no manually annotated images are available. To this aim, we simulated a real application scenario, performing several cross-dataset experiments by training each model on one source dataset and then testing it on a different target dataset. Our results show that some of the existing CNN models can also achieve a relatively good performance in cross-dataset scenarios, but this happens only when the target images are similar to the ones used for training, while their performance is considerably worse when target and source significantly differ in colours and scale.
The large gap between same- and cross-dataset performances demonstrated that more efforts must be devoted to creating more generalising methods and, therefore, methods that are suitable for real applications. The attained results also suggest to avoid focusing future work on improving the cell nuclei segmentation accuracy on benchmark datasets under the same-domain scenario and to address the efforts towards achieving a higher invariance in colour and scale.
As a possible solution to improve cross-domain effectiveness when no manually annotated data from the target scene are available, or when few images (even unlabelled) of the same domain are available to perform a DA, we envisage the use of a pre-processing procedure on the target images, in order to modify them to be more similar to the source images used for training the models. We are currently investigating this approach, and preliminary results will soon be submitted for consideration in a journal.
Author Contributions
Investigation, L.P.; software, L.P.; supervision, G.F.; writing—original draft, L.P.; writing—review and editing, G.F. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the project Histopathology Decision Support Systems (HistoDSSP) based on whole slide imaging analysis, funded by Regione Autonoma della Sardegna (POR FESR Sardegna 2014–2020 Asse 1 Azione 1.1.3).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; nor in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| CAD | computer-aided diagnosis |
| DSSP | decision support system for pathologists |
| LIS | laboratory information system |
| WSI | Whole Slide Image |
| H&E | hematoxylin & eosin |
| GBM | glioblastoma |
| LGG | lower grade glioma |
| HNSC | head and neck squamous cell carcinoma |
| LUSC | lung squamous cell carcinoma |
| DA | domain adaptation |
| UDA | unsupervised domain adaptation |
| CNN | convolutional neural network |
| RCNN | regional CNN |
| FCN | fully connected network |
| HED | holistically-nested edge detector |
| DIMAN | Deep Interval-Marker-Aware Network |
| TP | true positive |
| FN | false negative |
| FP | false positive |
| F1 | F1-score |
| P | precision |
| R | recall |
| ODI | object-level Dice index |
| OHD | object-level Hausdorff distance |
References
- Qi, X.; Xing, F.; Foran, D.J.; Yang, L. Robust segmentation of overlapping cells in histopathology specimens using parallel seed detection and repulsive level set. IEEE Trans. Biomed. Eng. 2012, 59, 754–765. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Yang, L. Robust Nucleus/Cell Detection and Segmentation in Digital Pathology and Microscopy Images: A Comprehensive Review. IEEE Rev. Biomed. Eng. 2016, 9, 234–263. [Google Scholar] [CrossRef] [PubMed]
- Dey, P. Cancer Nucleus: Morphology and Beyond. Diagn. Cytopathol. 2010, 38, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Gurcan, M.N.; Boucheron, L.; Can, A.; Madabhushi, A.; Rajpoot, N.; Yener, B. Histopathological image analysis: A review. IEEE Rev. Biomed. Eng. 2009, 2, 147–171. [Google Scholar] [CrossRef]
- Nawaz, S.; Yuan, Y. Computational pathology: Exploring the spatial dimension of tumor ecology. Cancer Lett. 2015, 380, 296–303. [Google Scholar] [CrossRef]
- Chen, J.-M.; Qu, A.-P.; Wang, L.-W.; Yuan, J.-P.; Yang, F.; Xiang, Q.-M.; Maskey, N.; Yang, G.-F.; Liu, J.; Li, Y. New breast cancer prognostic factors identified by computer-aided image analysis of he stained histopathology images. Sci. Rep. 2015, 5, 10690. [Google Scholar] [CrossRef][Green Version]
- Gandomkar, Z.; Brennan, P.C.; Mello-Thoms, C. Computer-based image analysis in breast pathology. J. Pathol. Inform. 2016, 7, 43. [Google Scholar] [CrossRef]
- Veta, M.; Pluim, J.P.W.; van Diest, P.J.; Viergever, M.A. Breast cancer histopathology image analysis: A review. IEEE Trans. Biomed. Eng. 2014, 61, 1400–1411. [Google Scholar] [CrossRef]
- Veta, M.; Diest, P.J.V.; Willems, S.M.; Wang, H.; Madabhushi, A.; Cruz-Roa, A.; Gonzalez, F.; Larsen, A.B.; Vestergaard, J.S.; Dahl, A.B.; et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med. Image Anal. 2015, 20, 237–248. [Google Scholar] [CrossRef]
- Alturkistani, H.A.; Tashkandi, F.M.; Mohammedsaleh, Z.M. Histological stains: A literature review and case study. Glob. J. Health Sci. 2016, 8, 72–79. [Google Scholar] [CrossRef]
- Kapuscinski, J. DAPI: A DNA-Specific Fluorescent Probe. Biotech. Histochem. 1995, 70, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Titford, M. The long history of hematoxylin. Biotech. Histochem. 2005, 80, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Bándi, P.; Geessink, O.; Manson, Q.; van Dijk, M.; Balkenhol, M.; Hermsen, M.; Bejnordi, B.E.; Lee, B.; Paeng, K.; Zhong, A.; et al. From detection of individual metastases to classification of lymph node status at the patient level: The camelyon17 challenge. IEEE Trans. Med. Imaging 2018, 38, 550–560. [Google Scholar] [CrossRef]
- Titford, M.; Bowman, B. What May the Future Hold for Histotechnologists? Who Will Be Affected? What Will Drive the Change? Lab. Med. 2012, 43, 5–10. [Google Scholar] [CrossRef]
- Pantanowitz, L.; Szymas, J.; Yagi, Y.; Wilbur, D. Whole slide imaging for educational purposes. J. Pathol. Inform. 2012, 3, 46. [Google Scholar] [CrossRef]
- Stoler, M.H.; Ronnett, B.M.; Joste, N.E.; Hunt, W.C.; Cuzick, J.; Wheeler, C.M. The Interpretive Variability of Cervical Biopsies and Its Relationship to HPV Status. Am. J. Surg. Pathol. 2015, 39, 729–736. [Google Scholar] [CrossRef]
- Kowal, M.; Filipczuk, P.; Obuchowicz, A.; Korbicz, J.; Monczak, R. Computer-aided diagnosis of breast cancer based on fine needle biopsy microscopic images. Comput. Biol. Med. 2013, 43, 1563–1572. [Google Scholar] [CrossRef]
- Sirinukunwattana, K.; Raza, S.E.A.; Tsang, Y.-w.; Snead, D.R.J.; Cree, I.A.; Rajpoot, N.M. Locality Sensitive Deep Learning for Detection and Classification of Nuclei in Routine Colon Cancer Histology Images. IEEE Trans. Med. Imaging 2016, 35, 1196–1206. [Google Scholar] [CrossRef]
- Xing, F.; Xie, Y.; Yang, L. An automatic learning-based framework for robust nucleus segmentation. IEEE Trans. Med. Imaging 2016, 35, 550–566. [Google Scholar] [CrossRef]
- Gurcan, M.N.; Pan, T.; Shimada, H.; Saltz, J. Image analysis for neuroblastoma classification: Segmentation of cell nuclei. In Proceedings of the International Conference of the IEEE Engineering in Medicine and Biology Society, New York, NY, USA, 30 August–3 September 2006. [Google Scholar] [CrossRef]
- Zhang, C.; Sun, C.; Pham, T.D. Segmentation of clustered nuclei based on concave curve expansions. J. Microsc. 2013, 251, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Verma, R.; Sharma, S.; Bhargava, S.; Vahadane, A.; Sethi, A. A dataset and a technique for generalised nuclear segmentation for computational pathology. IEEE Trans. Med. Imaging 2017, 36, 1550–1560. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Qi, J.; Pan, L.; Wali, S. Integrating deep convolutional neural networks with marker-controlled watershed for overlapping nuclei segmentation in histopathology images. Neurocomputing 2020, 376, 166–179. [Google Scholar] [CrossRef]
- Al-Kofahi, Y.; Lassoued, W.; Lee, W.; Roysam, B. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans. Biomed. Eng. 2010, 57, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef]
- Xu, J.; Xiang, L.; Liu, Q.; Gilmore, H.; Wu, J.; Tang, J.; Madabhushi, A. Stacked sparse autoencoder (ssae) for nuclei detection on breast cancer histopathology images. IEEE Trans. Med. Imaging 2016, 35, 119–130. [Google Scholar] [CrossRef]
- Irshad, H.; Veillard, A.; Roux, L.; Racoceanu, D. Methods for Nuclei Detection, Segmentation, and Classification in Digital Histopathology: A Review—Current Status and Future Potential. IEEE Rev. Biomed. Eng. 2014, 7, 97–114. [Google Scholar] [CrossRef]
- Naylor, P.; Laé, M.; Reyal, F.; Walter, T. Nuclei segmentation in histopathology images using deep neural networks. In Proceedings of the IEEE 14th International Symposium on Biomedical Imaging, Melbourne, Australia, 18–21 April 2017; pp. 933–936. [Google Scholar]
- Hou, L.; Agarwal, A.; Samaras, D.; Kurc, T.M.; Gupta, R.R.; Saltz, J.H. Robust histopathology image analysis: To label or to synthesise? In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 8533–8542. [Google Scholar]
- Liu, D.; Zhang, D.; Song, Y.; Zhang, F.; ODonnell, L.; Huang, H.; Chen, M.; Cai, W. Unsupervised Instance Segmentation in Microscopy Images via Panoptic Domain Adaptation and Task Re-Weighting. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 16–18 June 2020; pp. 4242–4251. [Google Scholar]
- Yi, F.; Huang, J.; Yang, L.; Xie, Y.; Xiao, G. Automatic extraction of cell nuclei from H&E-stained histopathological images. J. Med. Imaging 2017, 4, 027502. [Google Scholar]
- Boyle, D.P.; McArt, D.G.; Irwin, G.; Wilhelm-Benartzi, C.S.; Lioe, T.F.; Sebastian, E.; McQuaid, S.; Hamilton, P.W.; James, J.A.; Mullan, P.B.; et al. The prognostic significance of the aberrant extremes of p53 immunophenotypes in breast cancer. Histopathology 2014, 65, 340–352. [Google Scholar] [CrossRef]
- Li, J.; Yang, S.; Huang, X.; Da, Q.; Yang, X.; Hu, Z.; Duan, Q.; Wang, C.; Li, H. Signet ring cell detection with a semi-supervised learning framework. In Proceedings of the International Conference on Information Processing in Medical Imaging, Hong Kong, China, 2–7 June 2019; pp. 842–854. [Google Scholar]
- Zhou, Y.; Dou, Q.; Chen, H.; Qin, J.; Heng, P.-A. Sfcn-opi: Detection and fine-grained classification of nuclei using sibling fcn with objectness prior interaction. In Proceedings of the Thirty-Second AAAI Conference on Artificial Intelligence, New Orleans, LA, USA, 2–7 February 2018. [Google Scholar]
- Qu, H.; Riedlinger, G.; Wu, P.; Huang, Q.; Yi, J.; De, S.; Metaxas, D. Joint segmentation and fine-grained classification of nuclei in histopathology images. In Proceedings of the International Symposium on Biomedical Imaging, Venice, Italy, 8–11 April 2019; pp. 900–904. [Google Scholar]
- Naylor, P.; Laé, M.; Reyal, F.; Walter, T. Segmentation of nuclei in histopathology images by deep regression of the distance map. IEEE Trans. Med. Imaging 2018, 38, 448–459. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. Automatica 1975, 11, 23–27. [Google Scholar] [CrossRef]
- Huang, D.-C.; Hung, K.-D.; Chan, Y.-K. A computer assisted method for leukocyte nucleus segmentation and recognition in blood smear images. J. Syst. Softw. 2012, 85, 2104–2118. [Google Scholar] [CrossRef]
- Nawandhar, A.A.; Yamujala, L.; Kumar, N. Image segmentation using thresholding for cell nuclei detection of colon tissue. In Proceedings of the International Conference on Advances in Computing, Communications and Informatics (ICACCI), Kochi, India, 10–13 August 2015. [Google Scholar] [CrossRef]
- Meyer, F.; Beucher, S. Morphological segmentation. J. Vis. Commun. Image Rep. 1990, 1, 21–46. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, X.; Wong, S. Automated segmentation, classification, and tracking of cancer cell nuclei in time-lapse microscopy. IEEE Trans. Biomed. Eng. 2006, 53, 762–766. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, H.; Zhou, X. Nuclei Segmentation Using Marker-Controlled Watershed, Tracking Using Mean-Shift, and Kalman Filter in Time-Lapse Microscopy. IEEE Trans. Circuits Syst. 2006, 53, 2405–2414. [Google Scholar] [CrossRef]
- Fatakdawala, H.; Basavanhally, A.; Xu, J.; Bhanot, G.; Ganesan, S.; Feldman, M.; Tomaszewski, J.; Madabhushi, A. Expectation maximisation driven geodesic active contour with overlap resolution (EMaGACOR): Application to lymphocyte segmentationl on breast cancer histopathology. IEEE Trans. Biomed. Eng. 2010, 57, 1676–1689. [Google Scholar] [CrossRef]
- Yan, P.; Zhou, X.; Shah, M.; Wong, S. Automatic segmentation of high-throughput RNAi fluorescent cellular images. IEEE Trans. Inf. Technol. Biomed. 2008, 12, 109–117. [Google Scholar] [CrossRef]
- Kong, H.; Gurcan, M.; Belkacem-Boussaid, K. Partitioning histopathological images: An integrated framework for supervised color-texture segmentation and cell splitting. IEEE Trans. Med. Imaging 2011, 30, 1661–1677. [Google Scholar] [CrossRef]
- Plissiti, M.L.E.; Nikou, C. Overlapping cell nuclei segmentation using a spatially adaptive active physical model. IEEE Trans. Image Process. 2012, 21, 4568–4580. [Google Scholar] [CrossRef]
- Chen, H.; Qi, X.; Yu, L.; Dou, Q.; Qin, J.; Heng, P.-A. Dcan: Deep contour-aware networks for object instance segmentation from histology images. Med. Image Anal. 2017, 36, 135–146. [Google Scholar] [CrossRef]
- Zhu, J.-Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017. [Google Scholar]
- Mahmood, F.; Borders, D.; Chen, R.; McKay, G.N.; Salimian, K.J.; Baras, A.; Durr, N.J. Deep adversarial training for multi-organ nuclei segmentation in histopathology images. IEEE Trans. Med. Imaging 2019, 39, 3257–3267. [Google Scholar] [CrossRef]
- Ganin, Y.; Lempitsky, V. Unsupervised domain adaptation by backpropagation. In Proceedings of the International Conference on Machine Learning (ICML), Lille, France, 6–11 July 2015. [Google Scholar]
- Kim, T.; Jeong, M.; Kim, S.; Choi, S.; Kim, C. Diversify and match: A domain adaptive representation learning paradigm for object detection. In Proceedings of the Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 16–20 June 2019; pp. 12456–12465. [Google Scholar]
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
- Tzeng, E.; Hoffman, J.; Saenko, K.; Darrell, T. Adversarial discriminative domain adaptation. In Proceedings of the Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 7167–7176. [Google Scholar]
- Chen, C.; Dou, Q.; Chen, H.; Qin, J.; Heng, P.-A. Synergistic image and feature adaptation: Towards cross-modality domain adaptation for medical image segmentation. In Proceedings of the Association for the Advancement of Artificial Intelligence (AAAI), Honolulu, HI, USA, 29–31 January 2019; pp. 865–872. [Google Scholar]
- Dou, Q.; Ouyang, C.; Chen, C.; Chen, H.; Heng, P.-A. Unsupervised cross-modality domain adaptation of convnets for biomedical image segmentations with adversarial loss. arXiv 2018, arXiv:1804.10916. [Google Scholar]
- Zhang, Y.; Miao, S.; Mansi, T.; Liao, R. Task driven generative modeling for unsupervised domain adaptation: Application to x-ray image segmentation. arXiv 2018, arXiv:1806.07201. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. [Google Scholar] [CrossRef]
- Xie, S.; Tu, Z. Holistically-nested edge detection. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Xie, X.; Li, Y.; Zhang, M.; Shen, L. Robust segmentation of nucleus in histopathology images via Mask R-CNN. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11383. [Google Scholar]
- Wilcoxon, F. Individual comparisons by ranking methods. Biometrics 1945, 1, 80–83. [Google Scholar] [CrossRef]
- Demšar, J. Statistical Comparisons of Classifiers over Multiple Data Sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).