Featured Application
With its term mapping capability, MARIE can be used to improve data interoperability between different biomedical institutions. It can also be applied to text data pre-processing or normalization in non-biomedical domains.
Abstract
With growing interest in machine learning, text standardization is becoming an increasingly important aspect of data pre-processing within biomedical communities. As performances of machine learning algorithms are affected by both the amount and the quality of their training data, effective data standardization is needed to guarantee consistent data integrity. Furthermore, biomedical organizations, depending on their geographical locations or affiliations, rely on different sets of text standardization in practice. To facilitate easier machine learning-related collaborations between these organizations, an effective yet practical text data standardization method is needed. In this paper, we introduce MARIE (a context-aware term mapping method with string matching and embedding vectors), an unsupervised learning-based tool, to find standardized clinical terminologies for queries, such as a hospital’s own codes. By incorporating both string matching methods and term embedding vectors generated by BioBERT (bidirectional encoder representations from transformers for biomedical text mining), it utilizes both structural and contextual information to calculate similarity measures between source and target terms. Compared to previous term mapping methods, MARIE shows improved mapping accuracy. Furthermore, it can be easily expanded to incorporate any string matching or term embedding methods. Without requiring any additional model training, it is not only effective, but also a practical term mapping method for text data standardization and pre-processing.
1. Introduction
Due to the growing interest in text mining and natural language processing (NLP) in the biomedical field [1,2,3], data pre-processing is becoming an increasingly crucial issue for many biomedical practitioners. As datasets used in research and practice are often different, it is challenging to directly apply recent advancements in biomedical NLP to existing IT systems without effective data pre-processing. One of the key issues addressed during pre-processing is concept normalization, which refers to the task of aligning different text datasets or corpora into a common standard.
Previous works on concept normalization have relied on either extracting meaningful features from standardized ontologies [4,5,6] or training machine learning models [7,8]. Recently, deep learning models [9,10,11] have further improved normalization accuracy. However, these previous works are limited to a supervised learning setting, in which correctly aligned and standardized training datasets are required. If these manually aligned training datasets are not readily available, the normalization accuracy of these supervised approaches cannot be guaranteed. Without any additional annotated data, previously trained and supervised models cannot be fine-tuned for new datasets that exhibit completely different properties from their annotated counterparts. When the models fail to capture changes in the data distribution, their modeling capabilities cannot be transferred to the new datasets [12,13,14]. Therefore, these previous supervised learning models that were trained from standardized datasets cannot be easily applied in practice, in which mapping between non-standardized datasets is prevalent.
However, creating new annotated or standardized datasets is extremely challenging. Due to geographical, organizational or operational factors, biomedical organizations have their own unique sets of standards and conventions for processing their text data. For example, medical institutions in the United Kingdom and the United States use a systematized nomenclature of medicine, using clinical terms (SNOMED CT) [15] to standardize their clinical data. In South Korea, on the other hand, medical institutions rely on completely different sets of clinical terminologies issued by the Korean Health Insurance Review & Assessment Service, due to its fee-for-service model. Therefore, creating a highly standardized annotated dataset for every mapping need is simply too costly.
Despite its practical importance, unsupervised biomedical term mapping has not been actively explored. Previously, unsupervised learning-based mapping methods have relied on string matching-based approaches [16,17]. Without any model training, they utilize the structural information of the terms to define a similarity score, thereby providing greater applicability in mapping non-standardized or small datasets. However, a similarity measure between terms is often defined beyond their structural similarity. Using word embedding methods [18,19], we can also define a similarity measure between the terms based on their contextual similarity. As shown in the tasks of text classification [20,21], sentiment analysis [22,23] and machine translation [24], the contextual information captured by the embedding methods provides finer granularity to defining term similarities.
In this paper, we introduce MARIE (Python implementation of MARIE is available online at: https://github.com/hank110/MARIE), a context-aware mapping method with string matching and embedding vectors. It is a new, unsupervised concept normalization method that maps biomedical terms from different datasets, based on both their structural and contextual similarities. Its approach for capturing structural similarity is identical to previous string matching methods. However, the term similarity calculated from these sting matching methods is fine-tuned by corresponding term similarity measures, calculated from the word embedding vectors of BERT [25]. By incorporating both structural and contextual similarities, our proposed method improves upon the existing unsupervised term mapping methods, thereby providing an effective tool for aligning and standardizing biomedical text data in practice.
2. Methods
Since MARIE utilizes both string matching methods and BERT, we will briefly discuss these methods. Subsequently, we will provide details of our proposed method.
2.1. String Matching Methods
String matching methods define a similarity score between terms, based on the number of commonly shared letters or substrings. Among various string matching methods, edit distance [26,27], the Jaccard index [28,29] and Ratcliff/Obershelp similarity [30] are some of the most commonly used string matching methods. Given two terms, and , edit distance calculates their distance based on the number of insertions, deletions and substitutions of characters required in transforming into . While edit distance captures the structural information of terms on a character level, the Jaccard index utilizes the token-level, structural information. It computes the similarity between and as the length of the longest common substring in proportion to the overall length of and . Ratcliff/Obershelp (R/O) similarity is regarded as an extension of the Jaccard index. Initially, it finds the longest common substring between and . Subsequently, it finds the next longest common substring from the non-matching regions of and . This process of finding the longest common substring from the non-matching regions is repeated until there is no common substring between and . Finally, it uses the length of all common substrings in proportion to the overall length of and to calculate their similarity.
2.2. BERT
As a word embedding model, BERT learns to generate continuous numerical vector representations of words from text data. To generate these embedding vectors, it relies on a bi-directional transformer architecture [31]. Although Embeddings from Language Model (ELMo) [32] similarly utilizes bi-directional contextual information to train its word vectors, BERT is one of the first embedding methods that simultaneously captures bi-directional contextual information during its training. Furthermore, its multi-head attention mechanisms serve as a computationally efficient solution for retaining long-range contextual information, thereby improving the quality of the embedding vectors, compared with those generated from the recurrent neural network-based models [33]. Additionally, it uses WordPiece embeddings [34] to tokenize the input words during training. Subsequently, a word vector is generated by aggregating these tokenized embedding vectors, thereby addressing the issue of out-of-vocabulary words. Recently, BioBERT [35] has been introduced in the field of biomedical NLP. Despite its near-identical architecture to BERT, it has been trained from biomedical corpora such as PubMed abstracts and PubMed Central full-text articles. Due to its domain-specific training data, it has improved the performance of named entity recognition, relation extraction and question answering within the biomedical domain. Based on its trained word embedding vectors, the similarity between terms and is measured by calculating the cosine distance between their respective embedding vectors.
2.3. MARIE
As described in Equation (1), MARIE calculates a term similarity between terms and as a weighted average of string matching methods and the cosine similarity of their embedding vectors from BioBERT. As long as the range of the resulting similarity score is between 0 and 1, any type of string matching methods can be incorporated into MARIE. To generate word embedding vectors, MARIE extracts the activations from the last few layers of BioBERT. When a term is composed of multiple words or tokens, MARIE uses the average of the constituting word or token embedding vectors to generate a final term vector. As both similarity measures are ranged between 0 and 1, the final similarity score computed from MARIE also lies between 0 and 1. Furthermore, a hyperparameter α enables users to adjust the relative importance of the similarity scores calculated from each of the string matching methods and the BioBERT embedding vectors. Although α is a user-defined hyperparameter, it enables MARIE to flexibly adjust its mapping capability to different datasets. To summarize the entire mapping process of MARIE, Figure 1 provides its overall flow diagram. The MARIE calculation method is as follows:
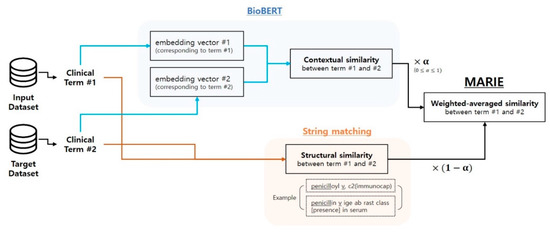
Figure 1.
The overall mapping process of MARIE. To find a mapping between the input and the target datasets, MARIE initially compares a clinical term from the input dataset with a clinical term from the target dataset. To capture the contextual similarity between and , it generates BioBERT embedding vectors for both and and calculates their vector similarity, based on a cosine similarity measure. Along with their structural similarity, computed from the string matching method, MARIE calculates the weighted average similarity score between and . By ranking these final similarity scores computed from MARIE, is mapped to with the highest similarity score.
The string matching methods and BioBERT use distinctively different criteria when calculating a similarity score between terms and . The string matching methods purely rely on the structural similarities between the terms. On the other hand, BioBERT uses the distance between the embedding vectors and . As these embedding vectors are trained to capture contextual dependencies between the terms, the distance between and is determined by the contextual similarities between and . By combining these two approaches, MARIE utilizes both structural and contextual dependencies, thereby employing a richer set of criteria to define a similarity between and .
Previously, word embedding-based edit distance [36] had similarly attempted to incorporate contextual information in edit distance. However, MARIE is a generalized improvement upon this previous work. Besides edit distance, MARIE is capable of incorporating a wider selection of string matching methods, thereby further expanding its applicability in practice. Furthermore, the biggest improvement of MARIE is its choice of term embedding methods. Among various term embedding methods, the previous work relies on word2vec [37] to generate term embedding vectors. However, word2vec does not capture richer bidirectional contextual information between words. Furthermore, it does not employ WordPiece embedding, thereby suffering from an out-of-vocabulary issue. When the embedding vector of a term has not been trained, word2vec is not capable of inferring its embedding vector. For this type of out-of-vocabulary term, word embedding-based edit distance simply ignores the contextual information and reverts back to a string matching method. However, BioBERT relies on WordPiece embedding to overcome this weakness of word2vec. During training, BioBERT learns the embedding vectors of both input words and their subwords. Thus, when it needs to infer the embedding vector of , it breaks into known subwords and estimates its embedding vector by adding up its subword embedding vectors. By using BioBERT instead of other term embedding methods such as word2vec or GloVe [38], MARIE utilizes richer contextual dependencies between terms and is robust to new words. Therefore, MARIE is capable of leveraging contextual information, even when the resources for training term embedding vectors are limited.
3. Results
3.1. Dataset and Experiment Setups
To validate the performance of MARIE, we tested it by mapping 3489 medical terms currently used in Seoul National University Hospital (SNUH corpus) with the Observational Medical Outcomes Partnership Common Data Model (OMOP CDM). Among various code standards available in the CDM, we limited our mapping to SNOMED CT, Logical Observation Identifiers Names and Codes (LOINC), RxNorm and RxNorm Extension [39,40]. With Usagi software and manual reviews from six medical professionals, we created a true mapping between SNUH corpus and the CDM. Given the terms from SNUH corpus as inputs, the performance of MARIE was evaluated on how accurately it could reconstruct this true mapping between SNUH corpus and the CDM. For example, given a term from SNUH corpus, we calculated its similarity scores with all of the terms in a target dataset and selected the term with the highest similarity score as its predicted mapped term. The mapping between and was considered correct if it corresponded to the true mapping between SNUH corpus and the CDM.
For robust evaluation, we tested MARIE on three different artificial target datasets. Each of these target datasets contained 3489 correctly mapped terms, as expressed in the CDM. In addition to these correctly mapped terms, each of these target datasets also contained 5000, 10,000 and 50,000 randomly sampled terms from the CDM, respectively. These random samples included only the terms that were excluded or ignored during the true mapping between SNUH corpus and the CDM. The sampled target datasets increased the number of possible mapping candidates that MARIE needed to explore, thereby introducing additional complexity to accurately mapping the terms in SNUH corpus.
As for BioBERT, we used the BioBERT-Large v1.1 model from its official Github repository. Without additional model training, we extracted the term embedding vectors from its pre-trained weights.
3.2. Experiment Results
Table 1 shows the top 1, 3, 5 and 10 mapping accuracies of MARIE in each of three target datasets. To provide objective comparisons, we also reported the mapping accuracy of the string matching methods and an embedding vector-based mapping method. For string matching methods, we used the Jaccard index, edit distance and R/O similarity. For the embedding vector-based mapping method, we calculated the similarity score between terms and , based on the cosine distance of their respective embedding vectors and . This embedding vector-based mapping is a variant of the concept normalization method investigated by Karadeniz et al. [41]. The only difference is that the embedding vectors were generated from BioBERT instead of word2vec.

Table 1.
Experiment results in biomedical mapping terminologies between Seoul National University Hospital (SNUH corpus) and three artificial target datasets. Compared with other existing mapping methods, MARIE showed superior mapping accuracy, which was further improved by changing the value of α.
The interplay of structural and contextual information within terms enabled MARIE to outperform other mapping methods across all target datasets. Considering the process involved in mapping terms, this outcome is not surprising. Let us assume that a correct mapping between datasets A and B resulted in a term to be mapped to a term . Unless and are identical strings, two types of discrepancies occur between these two terms. Given , either the order of words in will be different, or will be missing some of the words or subwords in . The term embedding vectors and string matching methods are effective in addressing each of these discrepancies, respectively. As an embedding vector of a term is defined by the average of its word vectors, the similarity score computed from the embedding vectors of BioBERT is invariant to the changes in the word orders within a term. Therefore, the contextual information of an entire term is preserved, regardless of the changes in its word order. On the other hand, similarity scores from string matching methods depend on the length of common characters or substrings. Consequently, as long as the lengths of missing words are short, compared with the overall lengths of the terms, string matching methods are effective in overlooking these missing words. As MARIE simultaneously resolves these two discrepancies, it achieves effective mapping performance. This characteristic is also evident in Table 2, which compares the actual mapping outcomes between SNUH local concepts and Observational Health Data Sciences and Informatics (OHDSI) Athena (https://athena.ohdsi.org/) from MARIE, R/O similarity and the embedding vector-based mapping.

Table 2.
Comparison of the top three mapped outcomes from MARIE, the R/O string matching method and embedding vector-based mapping. A bold and underlined term represents a correctly mapped term. (a) MARIE and the embedding vector-based method found the correct mapping for the input word to a target term in RxNorm, even though there was a word order discrepancy. (b) MARIE and the R/O string matching method found the correct mapping for the input word to a target term in LOINC, even when there were missing words between the mapping. (c) MARIE was even capable of finding a mapping missed by the other two methods, thereby correctly mapping to a term in LOINC.
4. Discussion
4.1. Impact of α
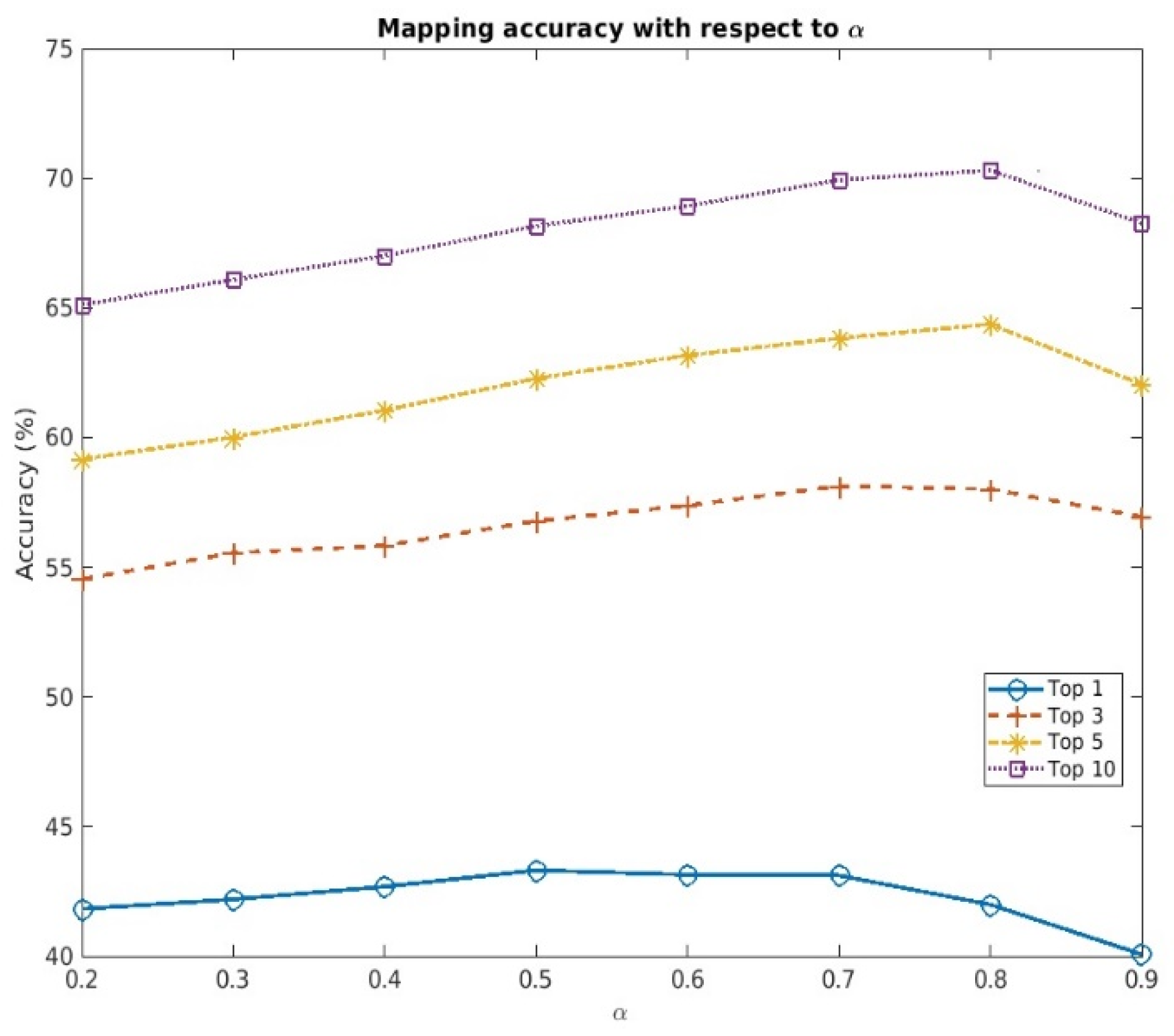
Hyperparameter balances the effect of structural and contextual information on calculating the overall similarity score with MARIE, thereby altering the mapping accuracy, as shown in Table 1. Therefore, we additionally analyzed the changes in the accuracy, with respect to various , in the target dataset with 50,000 random samples. Within the range between 0.2 and 0.9, we changed the value of in increments of 0.1.
As shown in Figure 2, there is an optimal cutoff value of that maximizes the mapping accuracy, thereby suggesting that solely relying on either structural or contextual information is not sufficient for accurate mapping. However, the optimal value of will vary, depending on the input and target datasets, and should be determined based on the type of discrepancy that is prevalent in them. If the word order discrepancy is a major difference between the input and target datasets, an value greater than 0.5 should be used for better mapping accuracy. On the other hand, if the words missing from the corresponding terms are prevalent, an value less than 0.5 is recommended.
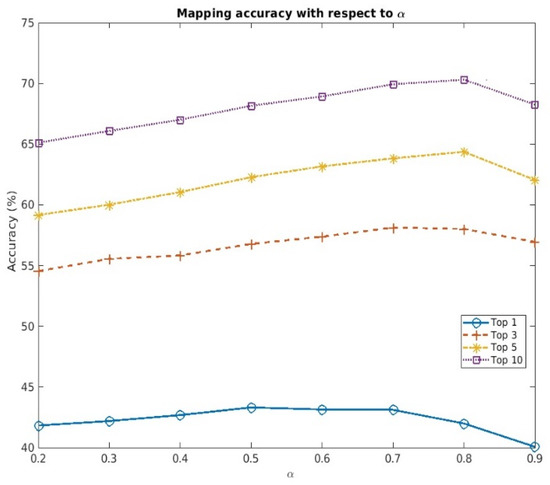
Figure 2.
Changes in the mapping accuracy of MARIE (BioBERT + R/O) with respect to α (target dataset with 50,000 random samples).
4.2. Impact of BioBERT Layers
Depending on the number of BioBERT layers used to represent a word embedding vector, the amount of contextual information captured by the vector can vary. When we utilized multiple layers, the final embedding vector was generated by taking an average of the embedding vectors from each layer. Table 3 summarizes the changes in the mapping accuracy of MARIE when we used the last 2~4 layers of BioBERT to generate a word embedding vector. Although the improvement was not significant, using more layers increased the overall mapping accuracy.

Table 3.
Changes in the mapping accuracy of MARIE (BioBERT + R/O with α = 0.8) with respect to the number of BioBERT layers used to represent an embedding vector.
4.3. Limitations of MARIE
Despite its superior mapping capability, MARIE does suffer from a few limitations. One of its limitations is its inability to handle biomedical terms that contain both English and foreign languages, such as Korean. In order to map these terms possessing multiple languages, we will need a BioBERT model that has been simultaneously trained for these different languages. However, such a heterogeneous model or training corpus for biomedical applications does not yet exist, to the best of our knowledge.
Furthermore, the computation speed of MARIE is bounded by the computation speed of its string matching methods. As MARIE extracts pre-trained BioBERT vectors, the majority of its computation time is spent on applying string matching methods. However, this computation time varies, based on the choice of string matching methods and their implementations.
This computation time issue becomes especially critical when mapping between large datasets. Therefore, we highly recommend using string matching methods with low computational complexities if fast computation time is critical.
5. Conclusions
In this paper, we introduced a new biomedical term mapping method that incorporates both structural and contextual information to calculate a term similarity score. As pre-processing and standardizing datasets are crucial in applying the latest text mining and machine learning models in practice, MARIE will serve as an essential tool for many biomedical professionals. As a generalized mapping method, it does not require any additional model training and can incorporate other string matching and embedding methods not discussed in this paper.
Furthermore, MARIE will make significant contributions to the process of mapping local clinical concept codes to standardized codes. Specifically, this mapping generated from MARIE will be useful for deploying the OMOP CDM outside of the United States or the United Kingdom. Based on the data standardization achieved by MARIE, applications related to health information exchange and personal health records will also benefit from improved interoperability.
For the direction of future research, it will be interesting to incorporate recent advances in machine translation or transfer learning to MARIE, thereby enabling it to map biomedical terms across datasets recorded in different languages. Furthermore, we will also explore ways to apply MARIE to create new standardized biomedical text datasets. By aligning and aggregating biomedical text datasets from numerous organizations, we will be able to create a large, yet highly standardized, training dataset for machine learning applications.
Author Contributions
Conceptualization, H.K.K. and T.K.; methodology, H.K.K. and T.K.; software, H.K.K.; validation, S.W.C., Y.S.B., J.C., H.K. and C.P.L.; formal analysis, J.C., H.K., C.P.L.; investigation, H.K.K., S.W.C. and T.K.; data curation, S.W.C. and Y.S.B.; writing—original draft preparation, H.K.K.; writing—review and editing, H.K.K. and T.K.; visualization, H.K.K. and T.K.; supervision, H.-Y.L.; project administration, H.-Y.L.; funding acquisition, H.-Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, South Korea (grant number: HI19C0572).
Acknowledgments
We would like to gratefully acknowledge the Division of Clinical Bioinformatics in Seoul National University Hospital, led by Hyung-Jin Yoon and Kwangsoo Kim. They have maintained the OMOP Common Data Model and provided the research environment.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Huang, C.C.; Lu, Z. Community challenges in biomedical text mining over 10 years: Success, failure and the future. Brief. Bioinform. 2016, 17, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.H.; Leaman, R.; Lu, Z. Beyond accuracy: Creating interoperable and scalable text-mining web services. Bioinformatics 2016, 32, 1907–1910. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Junge, A.; Jensen, L.J. CoCoScore: Context-aware co-occurrence scoring for text mining applications using distant supervision. Bioinformatics 2020, 36, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Aronson, A.R. Effective mapping of biomedical text to the UMLS Metathesaurus: The MetaMap program. In Proceedings of the AMIA Symposium; American Medical Informatics Association: Bethesda, MD, USA, 2001; p. 17. [Google Scholar]
- Bodenreider, O. The unified medical language system (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 2004, 32, D267–D270. [Google Scholar] [CrossRef] [PubMed]
- Rindflesch, T.C.; Fiszman, M. The interaction of domain knowledge and linguistic structure in natural language processing: Interpreting hypernymic propositions in biomedical text. J. Biomed. Inform. 2003, 36, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Leaman, R.; Islamaj Doğan, R.; Lu, Z. DNorm: Disease name normalization with pairwise learning to rank. Bioinformatics 2013, 29, 2909–2917. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, Z.; Bethard, S. A Generate-and-Rank Framework with Semantic Type Regularization for Biomedical Concept Normalization. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online, 5–10 July 2020; pp. 8452–8464. [Google Scholar]
- Li, H.; Chen, Q.; Tang, B.; Wang, X.; Xu, H.; Wang, B.; Huang, D. CNN-based ranking for biomedical entity normalization. BMC Bioinform. 2017, 18, 79–86. [Google Scholar] [CrossRef]
- Ji, Z.; Wei, Q.; Xu, H. Bert-based ranking for biomedical entity normalization. AMIA Summits Transl. Sci. Proc. 2020, 2020, 269. [Google Scholar]
- Schumacher, E.; Mulyar, A.; Dredze, M. Clinical Concept Linking with Contextualized Neural Representations. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online, 5–10 July 2020; pp. 8585–8592. [Google Scholar]
- Dai, W.; Yang, Q.; Xue, G.R.; Yu, Y. Boosting for transfer learning. In Proceedings of the 24th International Conference on MACHINE Learning; Association for Computing Machinery: New York, NY, USA, 2007; pp. 193–200. [Google Scholar]
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009, 22, 1345–1359. [Google Scholar] [CrossRef]
- Ben-David, S.; Blitzer, J.; Crammer, K.; Kulesza, A.; Pereira, F.; Vaughan, J.W. A theory of learning from different domains. Mach. Learn. 2010, 79, 151–175. [Google Scholar] [CrossRef]
- Donnelly, K. SNOMED-CT: The advanced terminology and coding system for eHealth. Stud. Health Technol. Inform. 2006, 121, 279. [Google Scholar]
- Dogan, R.I.; Lu, Z. An inference method for disease name normalization. In Proceedings of the 2012 AAAI Fall Symposium Series, Arlington, VA, USA, 2–4 November 2012. [Google Scholar]
- Kate, R.J. Normalizing clinical terms using learned edit distance patterns. J. Am. Med. Inform. Assoc. 2016, 23, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Turian, J.; Ratinov, L.; Bengio, Y. Word representations: A simple and general method for semi-supervised learning. In Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics, Uppsala, Sweden, 11–16 July 2010; pp. 384–394. [Google Scholar]
- Collobert, R.; Weston, J.; Bottou, L.; Karlen, M.; Kavukcuoglu, K.; Kuksa, P. Natural language processing (almost) from scratch. J. Mach. Learn. Res. 2011, 12, 2493–2537. [Google Scholar]
- Wang, P.; Xu, B.; Xu, J.; Tian, G.; Liu, C.L.; Hao, H. Semantic expansion using word embedding clustering and convolutional neural network for improving short text classification. Neurocomputing 2016, 174, 806–814. [Google Scholar] [CrossRef]
- Kim, H.K.; Kim, H.; Cho, S. Bag-of-concepts: Comprehending document representation through clustering words in distributed representation. Neurocomputing 2017, 266, 336–352. [Google Scholar] [CrossRef]
- Tang, D.; Wei, F.; Yang, N.; Zhou, M.; Liu, T.; Qin, B. Learning sentiment-specific word embedding for twitter sentiment classification. In Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics, Baltimore, MD, USA, 23–25 June 2014; pp. 1555–1565. [Google Scholar]
- Nikfarjam, A.; Sarker, A.; O’connor, K.; Ginn, R.; Gonzalez, G. Pharmacovigilance from social media: Mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J. Am. Med. Inform. Assoc. 2015, 22, 671–681. [Google Scholar] [CrossRef]
- Xing, C.; Wang, D.; Liu, C.; Lin, Y. Normalized word embedding and orthogonal transform for bilingual word translation. In Proceedings of the 2015 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Denver, CO, USA, 31 May–5 June 2015; pp. 1006–1011. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Wagner, R.A.; Fischer, M.J. The string-to-string correction problem. J. ACM 1974, 21, 168–173. [Google Scholar] [CrossRef]
- Hyyrö, H. Explaining and Extending the Bit-Parallel Approximate String Matching Algorithm of Myers; Technical Report A-2001-10; Department of Computer and Information Sciences, University of Tampere: Tampere, Finland, 2001. [Google Scholar]
- Jaccard, P. Distribution de la flore alpine dans le bassin des dranses et dans quelques régions voisines. Bull. Soc. Vaud. Sci. Nat. 1901, 37, 241–272. [Google Scholar]
- Gower, J.C.; Warrens, M.J. Similarity, dissimilarity, and distance, measures of. In Wiley StatsRef: Statistics Reference Online; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014; pp. 1–11. [Google Scholar]
- Black, P.E. Ratcliff/obershelp pattern recognition. In Dictionary of Algorithms and Data Structures; National Institute of Standards and Technology: Gaithersburg, MD, USA, 2004. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 5998–6008. [Google Scholar]
- Peters, M.E.; Neumann, M.; Iyyer, M.; Gardner, M.; Clark, C.; Lee, K.; Zettlemoyer, L. Deep contextualized word representations. arXiv 2018, arXiv:1802.05365. [Google Scholar]
- Sutskever, I.; Vinyals, O.; Le, Q.V. Sequence to sequence learning with neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Montréal, QC, Canada, 8–13 December 2014; pp. 3104–3112. [Google Scholar]
- Wu, Y.; Schuster, M.; Chen, Z.; Le, Q.V.; Norouzi, M.; Macherey, W.; Dean, J. Google’s neural machine translation system: Bridging the gap between human and machine translation. arXiv 2016, arXiv:1609.08144. [Google Scholar]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef]
- Niu, Y.; Qiao, C.; Li, H.; Huang, M. Word embedding based edit distance. arXiv 2018, arXiv:1810.10752. [Google Scholar]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.S.; Dean, J. Distributed representations of words and phrases and their compositionality. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 5–10 December 2013; pp. 3111–3119. [Google Scholar]
- Pennington, J.; Socher, R.; Manning, C.D. Glove: Global vectors for word representation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1532–1543. [Google Scholar]
- Liu, S.; Ma, W.; Moore, R.; Ganesan, V.; Nelson, S. RxNorm: Prescription for electronic drug information exchange. IT Prof. 2005, 7, 17–23. [Google Scholar] [CrossRef]
- Nelson, S.J.; Zeng, K.; Kilbourne, J.; Powell, T.; Moore, R. Normalized names for clinical drugs: RxNorm at 6 years. J. Am. Med. Inform. Assoc. 2011, 18, 441–448. [Google Scholar] [CrossRef]
- Karadeniz, I.; Özgür, A. Linking entities through an ontology using word embeddings and syntactic re-ranking. BMC Bioinform. 2019, 20, 156. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).