Identification of Risk Factors and Machine Learning-Based Prediction Models for Knee Osteoarthritis Patients
Abstract
1. Introduction
2. Data Description
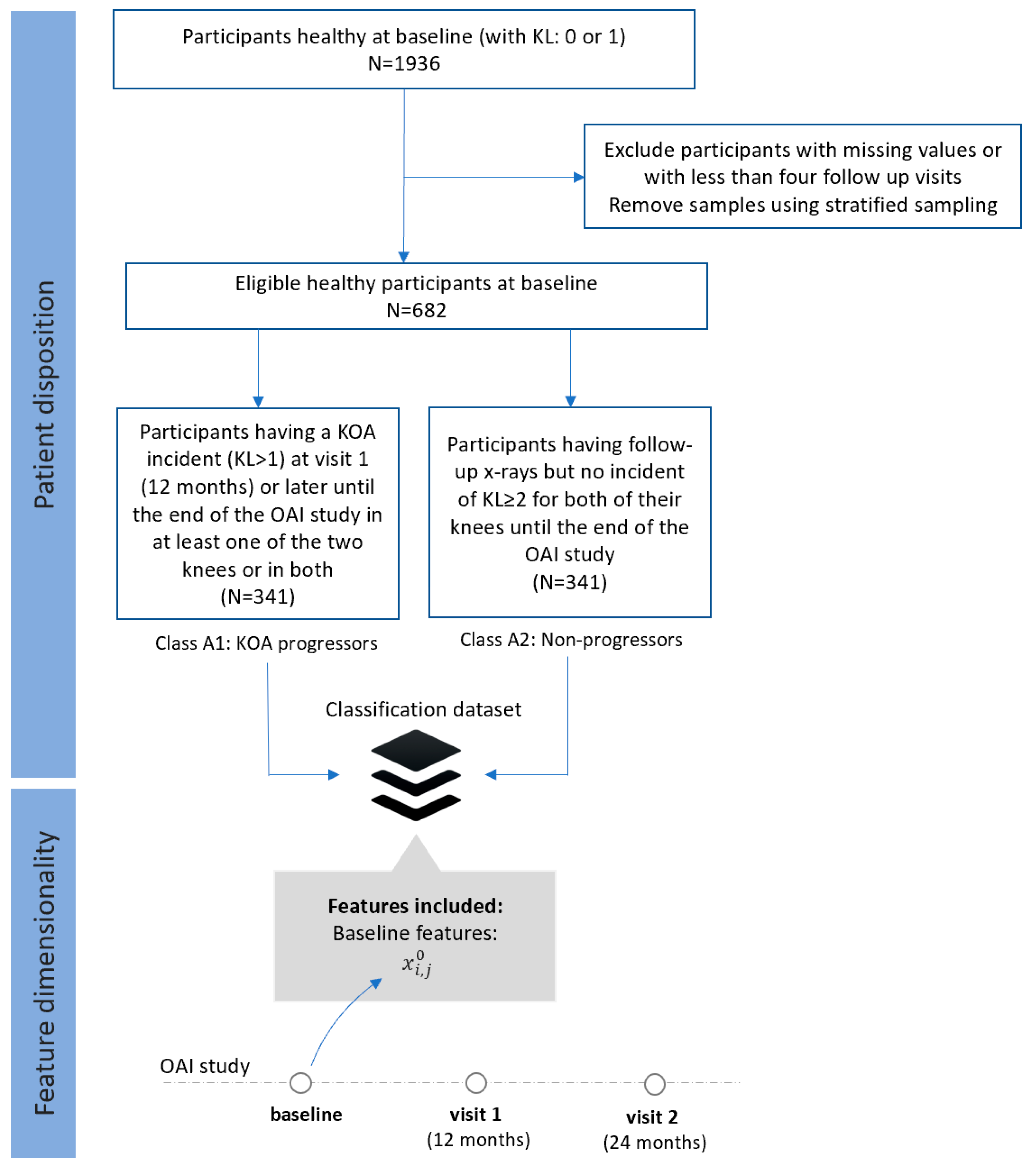
- Dataset A (FS1): Progressors vs. non-progressors using data from the baseline visit
- -
- Class A1 (KOA progressors): This class comprises 341 participants who had KL 0 or 1 at baseline, but they had also some incident of KL ≥ 2 at visit 1 (12 months) or later until the end of the OAI study in at least one of the two knees or in both.
- -
- Class A2 (non-progressors): This class involves 341 participants with KL 0 or 1 at baseline, with follow-up x-rays but no incident of KL ≥ 2 for both of their knees until the end of the OAI study.
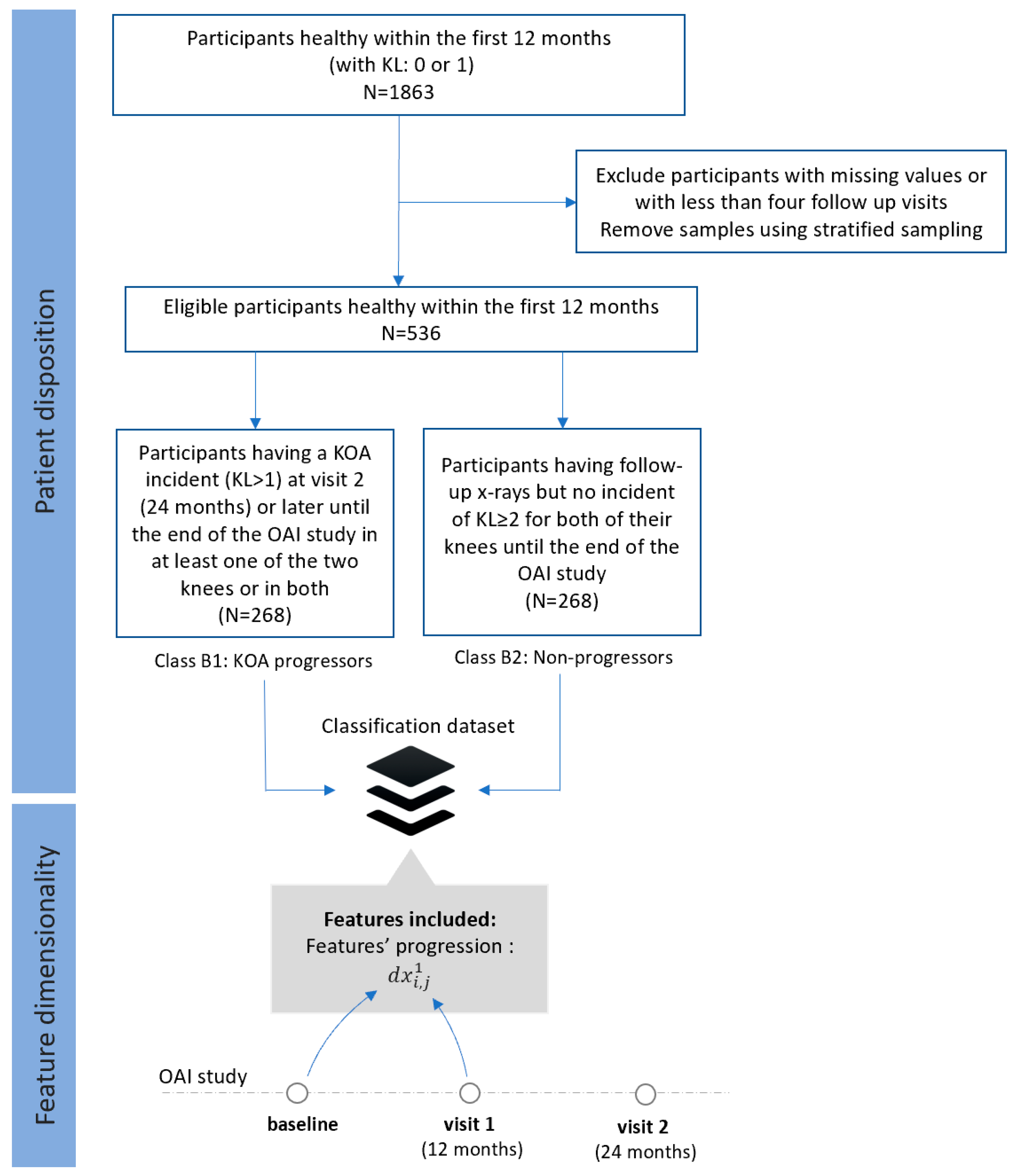
- Dataset B (FS2): Progressors vs. non-progressors using progression data within the first 12 months
- -
- Class B1 (KOA progressors): This class comprises progression data of 268 participants who were healthy (KL 0 or 1) within the first 12 months (both at the baseline and the visit 1), but they had an incident of KL ≥ 2 at the second visit (24 months) or later (until the end of the OAI study).
- -
- Class B2 (non-progressors): This class involves progression data from 268 participants with KL 0 or 1 at the baseline, who had follow-up x-rays with no other incident of KL ≥ 2 in any of their knees until the end of the OAI study.
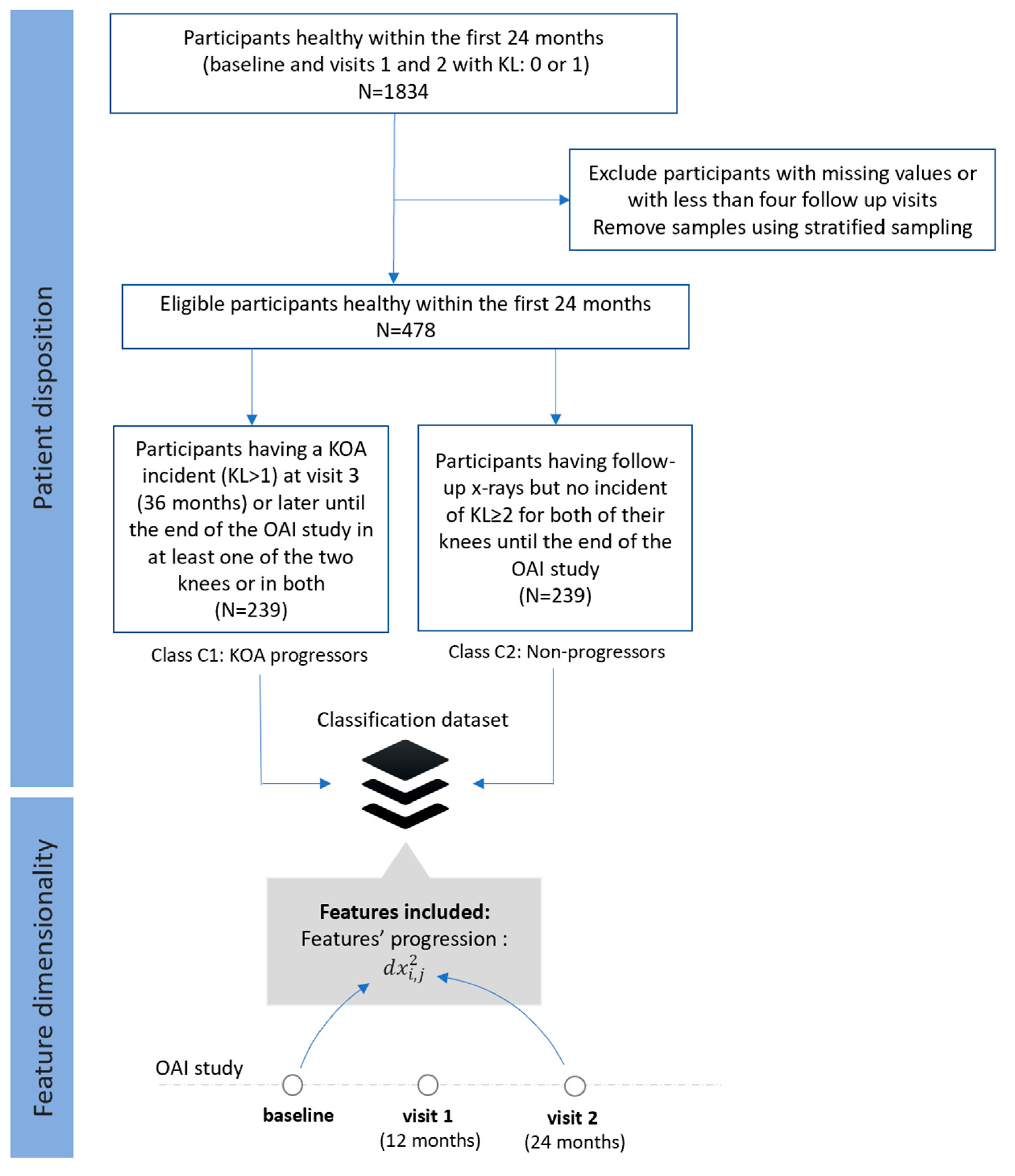
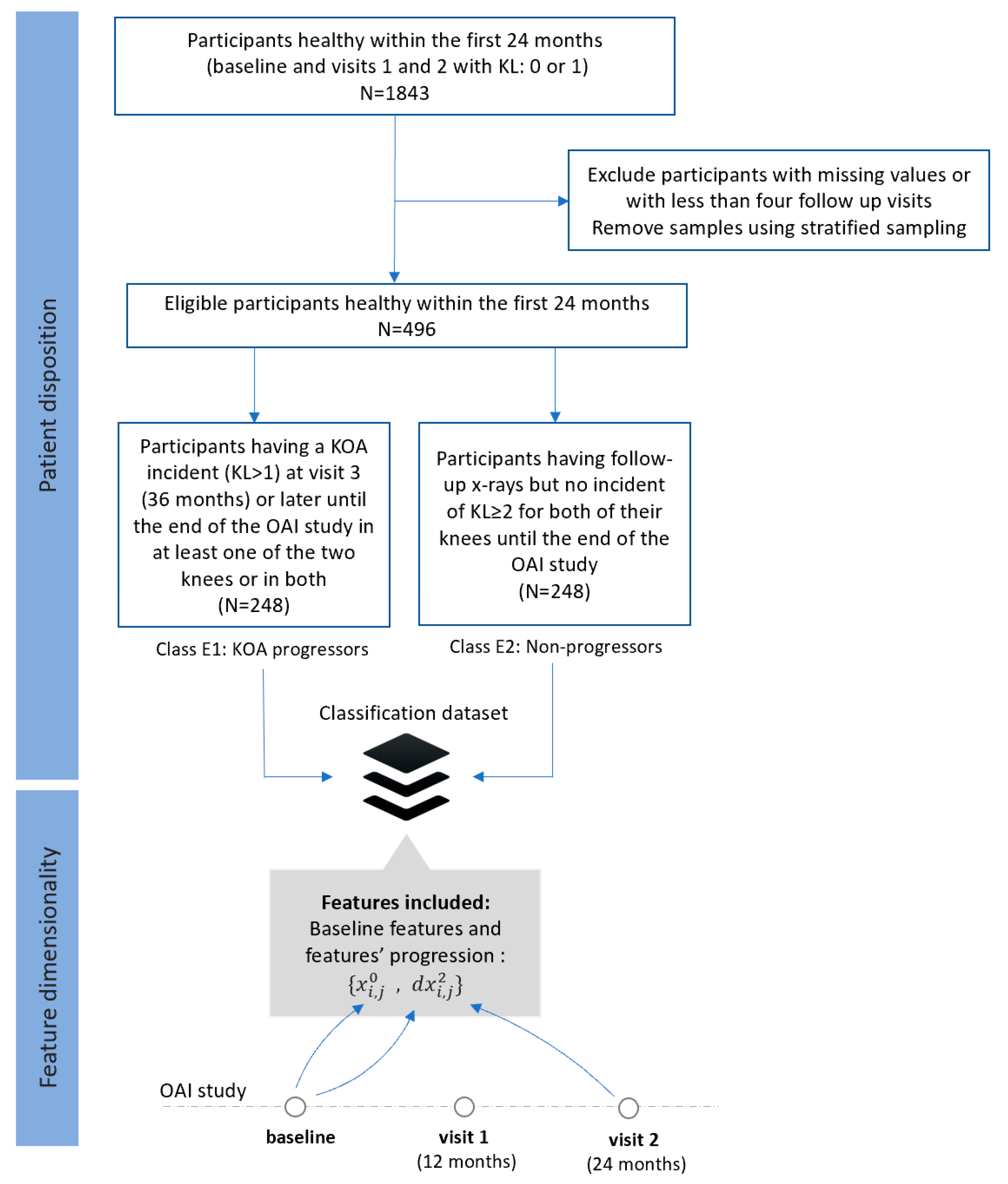
- Dataset C (FS3): Progressors vs. non-progressors using progression data within the first 24 months
- -
- Class C1 (KOA progressors): This class comprises of 239 participants who had KL 0 or 1 during the first 24 months, whereas a KOA incident (KL ≥ 2) observed at visit 3 (36 months) or later during the OAI course in at least one of the two knees or in both.
- -
- Class C2 (non-progressors): This class involves 239 participants with KL grade 0 or 1 at baseline, with follow-up X-rays and no further incidents (KL ≥ 2) for both of their knees.
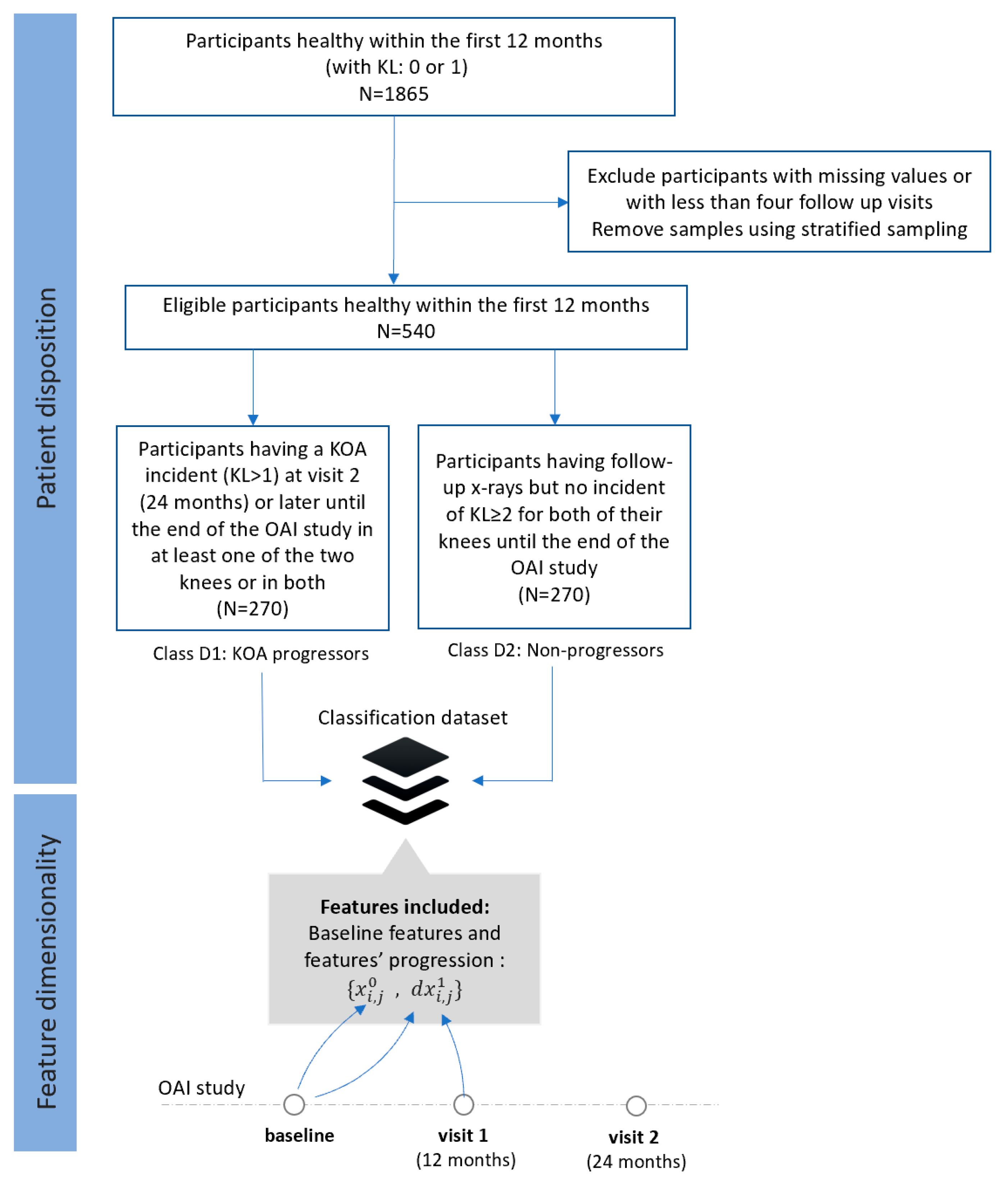
- Dataset D (FS4): Progressors vs. non-progressors using data from the baseline visit along with progression data within the first 12 months
- -
- Class D1 (KOA progression): This class comprises 270 participants (KL 0 or 1) who were heathy during the first 12 months (with no incident at the baseline and the first visit) and then they had an incident (KL ≥ 2) recorded at their second visit (24 months) or later until the end of the OAI study.
- -
- Class D2 (non-KOA): This class involves 270 healthy participants with KL0 or 1 at baseline with no further incidents in both of their knees until the end of the OAI data collection.
- Dataset E (FS5): Progressors vs. non-progressors using data from the baseline visit along with progression data within the first 24 months
- -
- Class E1 (KOA progression): This class comprises 248 participants who were healthy (KL 0 or 1) in the first 24 months, but they had a KOA incident (KL ≥ 2) at the third visit (36 months) or later until the end of the OAI study in at least one of the two knees or in both.
- -
- Class E2 (non-KOA): This class involves 248 healthy participants (KL0 or 1) with no further progression of KOA in both of their knees until the end of the OAI study.
3. Methodology
3.1. Pre-Processing
3.2. Feature Selection (FS)
3.3. Learning Process
3.4. Validation
4. Results
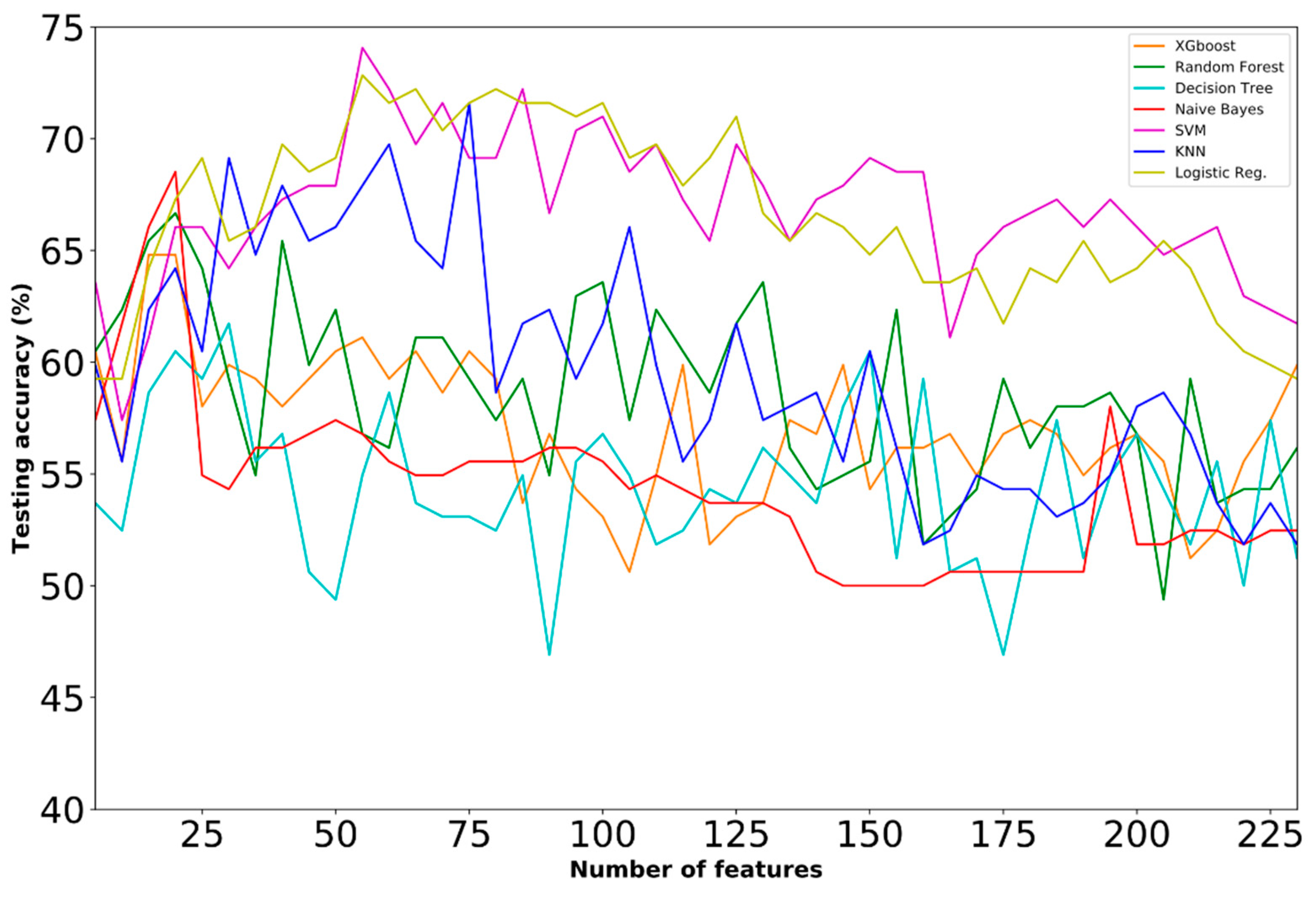
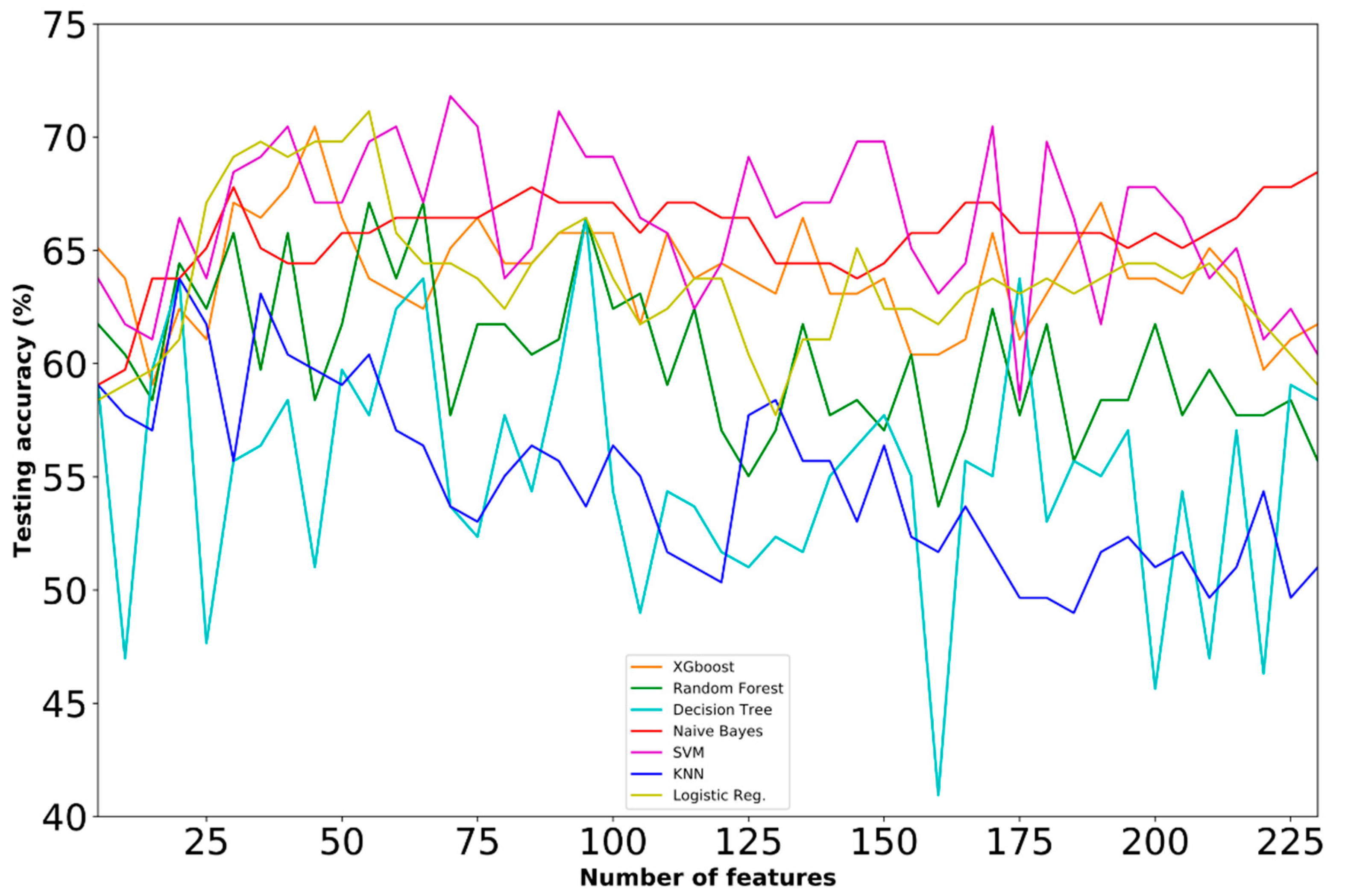
4.1. Prediction Performance
- Dataset A
- Dataset B
- Dataset C
- Dataset D
- Dataset E
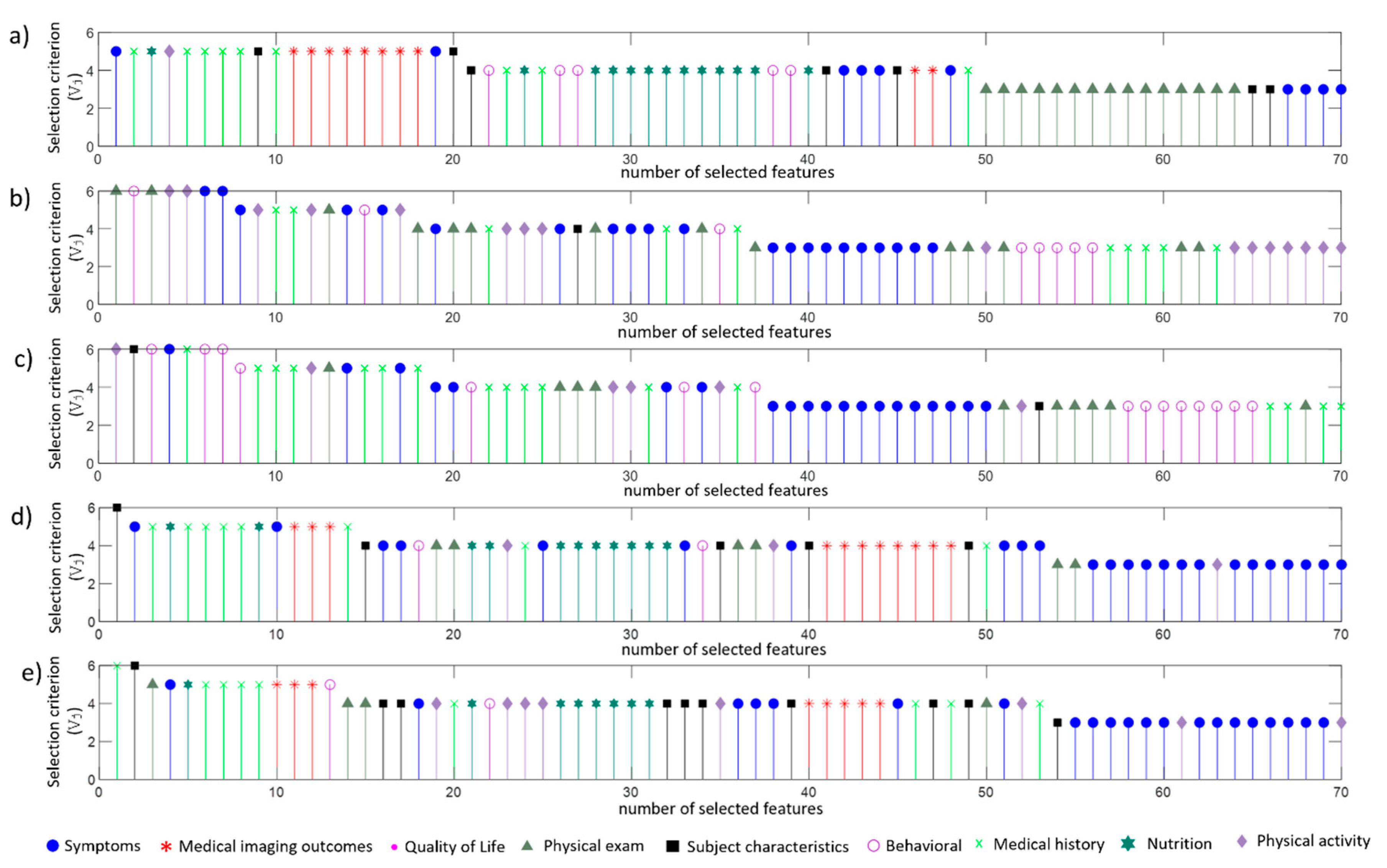
4.2. Selected Features
4.3. Comparative Analysis
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Feature | Description | Category | |
|---|---|---|---|
| 1 | P02WTGA | Above weight cut-off for age/gender group (calc, used for study eligibility) | Subject characteristics |
| 2 | V00WPRKN2 | Right knee pain: stairs, last 7 days | Symptoms |
| 3 | V00RXANALG | Rx Analgesic use indicator (calc) | Medical history |
| 4 | V00PCTSMAL | Block Brief 2000: error flag, percent of foods marked as small portion (calc) | Nutrition |
| 5 | V00GLUC | Used glucosamine for joint pain or arthritis, past 6 months | Medical history |
| 6 | V00GLCFQCV | Glucosamine frequency of use, past 6 months (calc) | Medical history |
| 7 | V00CHON | Used chondroitin sulfate for joint pain or arthritis, past 6 months | Medical history |
| 8 | V00CHNFQCV | Chondroitin sulfate frequency of use, past 6 months (calc) | Medical history |
| 9 | V00BAPCARB | Block Brief 2000: daily % of calories from carbohydrate, alcoholic beverages excluded from denominator (kcal) (calc) | Nutrition |
| 10 | P02KPNRCV | Right knee pain, aching or stiffness: more than half the days of a month, past 12 months (calc, used for study eligibility) | Symptoms |
| 11 | P01XRKOA | Baseline radiographic knee OA status by person (calc) | Medical imaging outcome |
| 12 | P01SVLKOST | Left knee baseline x-ray: evidence of knee osteophytes (calc) | Medical imaging outcome |
| 13 | P01OAGRDL | Left knee baseline x-ray: composite OA grade (calc) | Medical imaging outcome |
| 14 | P01GOUTCV | Doctor said you had gout (calc) | Medical history |
| 15 | V00WTMAXKG | Maximum adult weight, self-reported (kg) (calc) | Subject characteristics |
| 16 | V00WSRKN1 | Right knee stiffness: in morning, last 7 days | Symptoms |
| 17 | V00WOMSTFR | Right knee: WOMAC Stiffness Score (calc) | Symptoms |
| 18 | V00SF1 | In general, how is health | Behavioural |
| 19 | V00RKMTTPN | Right knee exam: medial tibiofemoral pain/tenderness present on exam | Physical exam |
| 20 | V00RFXCOMP | Isometric strength: right knee flexion, able to complete (3) measurements | Physical exam |
| 21 | V00PCTFAT | Block Brief 2000: daily percent of calories from fat (kcal) (calc) | Nutrition |
| 22 | V00PCTCARB | Block Brief 2000: daily percent of calories from carbohydrate (kcal) (calc) | Nutrition |
| 23 | V00PASE | Physical Activity Scale for the Elderly (PASE) score (calc) | Physical activity |
| 24 | V00LUNG | Charlson Comorbidity: have emphysema, chronic bronchitis or chronicobstructive lung disease (also called COPD) | Medical history |
| 25 | V00KSXLKN1 | Left knee symptoms: swelling, last 7 days | Symptoms |
| 26 | V00FFQSZ16 | Block Brief 2000: rice/dishes made with rice, how much each time | Nutrition |
| 27 | V00FFQSZ14 | Block Brief 2000: white potatoes not fried, how much each time | Nutrition |
| 28 | V00FFQSZ13 | Block Brief 2000: french fries/fried potatoes/hash browns, how much each time | Nutrition |
| 29 | V00FFQ69 | Block Brief 2000: regular soft drinks/bottled drinks like Snapple (not diet drinks), drink how often, past 12 months | Nutrition |
| 30 | V00FFQ59 | Block Brief 2000: ice cream/frozen yogurt/ice cream bars, eat how often, past 12 months | Nutrition |
| 31 | V00FFQ37 | Block Brief 2000: fried chicken, at home or in a restaurant, eat how often, past 12 months | Nutrition |
| 32 | V00DTCAFFN | Block Brief 2000: daily nutrients from food, caffeine (mg) (calc) | Nutrition |
| 33 | V00DILKN11 | Left knee difficulty: socks off, last 7 days | Symptoms |
| 34 | V00CESD13 | CES-D: how often talked less than usual, past week | Behavioural |
| 35 | V00ABCIRC | Abdominal circumference (cm) (calc) | Subject characteristics |
| 36 | TIMET1 | 20-m walk: trial 1 time to complete (sec.hundredths/sec) | Physical exam |
| 37 | STEPST1 | 20-m walk: trial 1 number of steps | Physical exam |
| 38 | PASE6 | Leisure activities: muscle strength/endurance, past 7 days | Physical activity |
| 39 | P02KPNLCV | Left knee pain, aching or stiffness: more than half the days of a month, past 12 months (calc, used for study eligibility) | Symptoms |
| 40 | P01WEIGHT | Average current scale weight (kg) (calc) | Subject characteristics |
| 41 | P01SVRKOST | Right knee baseline x-ray: evidence of knee osteophytes (calc) | Medical imaging outcome |
| 42 | P01SVRKJSL | Right knee baseline x-ray: evidence of knee lateral joint space narrowing (calc) | Medical imaging outcome |
| 43 | P01RXRKOA2 | Right knee baseline x-ray: osteophytes and JSN (calc) | Medical imaging outcome |
| 44 | P01RXRKOA | Right knee baseline radiographic OA (definite osteophytes, calc, used in OAI definition of symptomatic knee OA) | Medical imaging outcome |
| 45 | P01RSXKOA | Right knee baseline symptomatic OA status (calc) | Medical imaging outcome |
| 46 | P01OAGRDR | Right knee baseline x-ray: composite OA grade (calc) | Medical imaging outcome |
| 47 | P01LXRKOA2 | Left knee baseline x-ray: osteophytes and JSN (calc) | Medical imaging outcome |
| 48 | P01LXRKOA | Left knee baseline radiographic OA (definite osteophytes, calc, used in OAI definition of symptomatic knee OA) | Medical imaging outcome |
| 49 | P01BMI | Body mass index (calc) | Subject characteristics |
| 50 | P01ARTDRCV | Seeing doctor/other professional for knee arthritis (calc) | Medical history |
| 51 | KSXRKN2 | Right knee symptoms: feel grinding, hear clicking or any other type of noise when knee moves, last 7 days | Symptoms |
| 52 | KPRKN1 | Right knee pain: twisting/pivoting on knee, last 7 days | Symptoms |
| 53 | DIRKN7 | Right knee difficulty: in car/out of car, last 7 days | Symptoms |
| 54 | rkdefcv | Right knee exam: alignment varus or valgus (calc) | Physical exam |
| 55 | lkdefcv | Left knee exam: alignment varus or valgus (calc) | Physical exam |
References
- Silverwood, V.; Blagojevic-Bucknall, M.; Jinks, C.; Jordan, J.; Protheroe, J.; Jordan, K. Current evidence on risk factors for knee osteoarthritis in older adults: A systematic review and meta-analysis. Osteoarthr. Cartil. 2015, 23, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, I.N.; Kemp, J.L.; Crossley, K.M.; Culvenor, A.G.; Hinman, R.S. Hip and Knee Osteoarthritis Affects Younger People, Too. J. Orthop. Sports Phys. Ther. 2017, 47, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Lespasio, M.J.; Piuzzi, N.S.; Husni, M.E.; Muschler, G.F.; Guarino, A.; Mont, M.A. Knee osteoarthritis: A primer. Perm. J. 2017, 21. [Google Scholar] [CrossRef] [PubMed]
- Kokkotis, C.; Moustakidis, S.; Papageorgiou, E.; Giakas, G.; Tsaopoulos, D. Machine Learning in Knee Osteoarthritis: A Review. Osteoarthr. Cartil. Open 2020, 100069. [Google Scholar] [CrossRef]
- Lazzarini, N.; Runhaar, J.; Bay-Jensen, A.C.; Thudium, C.S.; Bierma-Zeinstra, S.M.A.; Henrotin, Y.; Bacardit, J. A machine learning approach for the identification of new biomarkers for knee osteoarthritis development in overweight and obese women. Osteoarthr. Cartil. 2017, 25, 2014–2021. [Google Scholar] [CrossRef]
- Halilaj, E.; Le, Y.; Hicks, J.L.; Hastie, T.J.; Delp, S.L. Modeling and predicting osteoarthritis progression: Data from the osteoarthritis initiative. Osteoarthr. Cartil. 2018, 26, 1643–1650. [Google Scholar] [CrossRef]
- Pedoia, V.; Haefeli, J.; Morioka, K.; Teng, H.L.; Nardo, L.; Souza, R.B.; Ferguson, A.R.; Majumdar, S. MRI and biomechanics multidimensional data analysis reveals R2 -R1rho as an early predictor of cartilage lesion progression in knee osteoarthritis. J. Magn. Reson. Imaging JMRI 2018, 47, 78–90. [Google Scholar] [CrossRef]
- Abedin, J.; Antony, J.; McGuinness, K.; Moran, K.; O’Connor, N.E.; Rebholz-Schuhmann, D.; Newell, J. Predicting knee osteoarthritis severity: Comparative modeling based on patient’s data and plain X-ray images. Sci. Rep. 2019, 9, 5761. [Google Scholar] [CrossRef]
- Nelson, A.; Fang, F.; Arbeeva, L.; Cleveland, R.; Schwartz, T.; Callahan, L.; Marron, J.; Loeser, R. A machine learning approach to knee osteoarthritis phenotyping: Data from the FNIH Biomarkers Consortium. Osteoarthr. Cartil. 2019, 27, 994–1001. [Google Scholar] [CrossRef]
- Tiulpin, A.; Klein, S.; Bierma-Zeinstra, S.M.; Thevenot, J.; Rahtu, E.; van Meurs, J.; Oei, E.H.; Saarakkala, S. Multimodal machine learning-based knee osteoarthritis progression prediction from plain radiographs and clinical data. Sci. Rep. 2019, 9, 20038. [Google Scholar] [CrossRef]
- Widera, P.; Welsing, P.M.; Ladel, C.; Loughlin, J.; Lafeber, F.P.; Dop, F.P.; Larkin, J.; Weinans, H.; Mobasheri, A.; Bacardit, J. Multi-classifier prediction of knee osteoarthritis progression from incomplete imbalanced longitudinal data. arXiv 2019, arXiv:1909.13408. [Google Scholar] [CrossRef] [PubMed]
- Alexos, A.; Moustakidis, S.; Kokkotis, C.; Tsaopoulos, D. Physical Activity as a Risk Factor in the Progression of Osteoarthritis: A Machine Learning Perspective. In International Conference on Learning and Intelligent Optimization; Springer: Cham, Switzerland, 2020; pp. 16–26. [Google Scholar]
- Ashinsky, B.G.; Bouhrara, M.; Coletta, C.E.; Lehallier, B.; Urish, K.L.; Lin, P.C.; Goldberg, I.G.; Spencer, R.G. Predicting early symptomatic osteoarthritis in the human knee using machine learning classification of magnetic resonance images from the osteoarthritis initiative. J. Orthop. Res. Official Publ. Orthop. Res. Soc. 2017, 35, 2243–2250. [Google Scholar] [CrossRef] [PubMed]
- Donoghue, C.; Rao, A.; Bull, A.M.J.; Rueckert, D. Manifold learning for automatically predicting articular cartilage morphology in the knee with data from the osteoarthritis initiative (OAI). Proc. Prog. Biomed. Opt. Imaging Proc. SPIE 2011, 7962, 79620E. [Google Scholar]
- Marques, J.; Genant, H.K.; Lillholm, M.; Dam, E.B. Diagnosis of osteoarthritis and prognosis of tibial cartilage loss by quantification of tibia trabecular bone from MRI. Magn. Reson. Med. 2013, 70, 568–575. [Google Scholar] [CrossRef] [PubMed]
- Yoo, T.K.; Kim, S.K.; Choi, S.B.; Kim, D.Y.; Kim, D.W. Interpretation of movement during stair ascent for predicting severity and prognosis of knee osteoarthritis in elderly women using support vector machine. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 192–196. [Google Scholar] [CrossRef]
- Moustakidis, S.; Christodoulou, E.; Papageorgiou, E.; Kokkotis, C.; Papandrianos, N.; Tsaopoulos, D. Application of machine intelligence for osteoarthritis classification: A classical implementation and a quantum perspective. Quantum Mach. Intell. 2019. [Google Scholar] [CrossRef]
- Juszczak, P.; Tax, D.; Duin, R.P. Feature scaling in support vector data description. Proc. Asci 2002, 95–102. [Google Scholar]
- Dodge, Y.; Commenges, D. The Oxford Dictionary of Statistical Terms; Oxford University Press: Oxford, UK, 2006. [Google Scholar]
- Biesiada, J.; Duch, W. Feature selection for high-dimensional data—A Pearson redundancy based filter. In Computer Recognition Systems 2; Springer: Berlin/Heidelberg, Germany, 2007; pp. 242–249. [Google Scholar]
- Thaseen, I.S.; Kumar, C.A. Intrusion detection model using fusion of chi-square feature selection and multi class SVM. J. King Saud Univ. Comput. Inf. Sci. 2017, 29, 462–472. [Google Scholar]
- Xiong, M.; Fang, X.; Zhao, J. Biomarker identification by feature wrappers. Genome Res. 2001, 11, 1878–1887. [Google Scholar] [CrossRef]
- Nie, F.; Huang, H.; Cai, X.; Ding, C.H. Efficient and robust feature selection via joint ℓ2, 1-norms minimization. In Proceedings of the Advances in neural information processing systems, Vancouver, BC, Canada, 6–9 December 2010; pp. 1813–1821. [Google Scholar]
- Zhou, Q.; Zhou, H.; Li, T. Cost-sensitive feature selection using random forest: Selecting low-cost subsets of informative features. Knowl. Based Syst. 2016, 95, 1–11. [Google Scholar] [CrossRef]
- Al Daoud, E. Comparison between XGBoost, LightGBM and CatBoost Using a Home Credit Dataset. Int. J. Comput. Inf. Eng. 2019, 13, 6–10. [Google Scholar]
- Rockel, J.S.; Zhang, W.; Shestopaloff, K.; Likhodii, S.; Sun, G.; Furey, A.; Randell, E.; Sundararajan, K.; Gandhi, R.; Zhai, G. A classification modeling approach for determining metabolite signatures in osteoarthritis. PLoS ONE 2018, 13, e0199618. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Kannari, T.; Horiuchi, H.; Matsui, N.; Ito, T.; Nojin, K.; Kakuse, K.; Okawa, M.; Yamanaka, M. Predictors affecting balance performances in patients with knee osteoarthritis using decision tree analysis. Osteoarthr. Cartil. 2019, 27, S243. [Google Scholar] [CrossRef]
- Peterson, L.E. K-nearest neighbor. Scholarpedia 2009, 4, 1883. [Google Scholar] [CrossRef]
- Gornale, S.S.; Patravali, P.U.; Marathe, K.S.; Hiremath, P.S. Determination of Osteoarthritis Using Histogram of Oriented Gradients and Multiclass SVM. Int. J. Image Graph. Signal Process. 2017, 9. [Google Scholar] [CrossRef]
- Kotti, M.; Duffell, L.D.; Faisal, A.A.; McGregor, A.H. Detecting knee osteoarthritis and its discriminating parameters using random forests. Med. Eng. Phys. 2017, 43, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Torlay, L.; Perrone-Bertolotti, M.; Thomas, E.; Baciu, M. Machine learning–XGBoost analysis of language networks to classify patients with epilepsy. Brain Inform. 2017, 4, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Shan, J.; Zhang, M. Knee osteoarthritis prediction on MR images using cartilage damage index and machine learning methods. In Proceedings of the Proceedings—2017 IEEE International Conference on Bioinformatics and Biomedicine, BIBM, Kansas City, MO, USA, 13–16 November 2017; pp. 671–677. [Google Scholar]
- Du, Y.; Almajalid, R.; Shan, J.; Zhang, M. A Novel Method to Predict Knee Osteoarthritis Progression on MRI Using Machine Learning Methods. IEEE Trans. Nanobiosci. 2018. [Google Scholar] [CrossRef]

| Timeline of Visit | ||||
|---|---|---|---|---|
| Category | Description | Baseline | 12 Months | 24 Months |
| Subject characteristics | Anthropometric parameters including height, weight, BMI, abdominal circumference, etc. | ● | ● | ● |
| Behavioural | Participants’ social behaviour and quality level of daily routine | ● | ● | ● |
| Medical history | Questionnaire data regarding a Participant’s arthritis-related and general health histories and medications | ● | - | - |
| Medical imaging outcome | Medical imaging outcomes (e.g., osteophytes and joint space narrowing) | ● | - | - |
| Nutrition | Block Food Frequency questionnaire | ● | - | - |
| Physical activity | Questionnaire results regarding leisure activities, etc. | ● | ● | ● |
| Physical exam | Physical measurements of participants, including isometric strength, knee and hand exams, walking tests and other performance measures | ● | ● | ● |
| Symptoms | Arthritis symptoms and general arthritis or health-related function and disability | ● | ● | ● |
| ML Models | Hyperparameters | Description |
|---|---|---|
| XGboost | Gamma | Minimum loss reduction required to make a further partition on a leaf node of the tree. |
| Maximal depth | Maximum depth of a tree. Increasing this value will make the model more complex and more likely to overfit. | |
| Minimum child and Weight | Minimum sum of instance weight (hessian) needed in a child. If the tree partition step results in a leaf node with the sum of instance weight less than min_child_weight, then the building process will give up further partitioning. | |
| Random Forest | Criterion | The function to measure the quality of a split. |
| Minimum samples leaf | The minimum number of samples required to be at a leaf node. | |
| Number of estimators | The number of trees in the forest. | |
| Decision Trees | Maximal features | The number of features to consider when looking for the best split. |
| Minimum samples split | The minimum number of samples required to split an internal node | |
| Minimum number of leafs | The minimum number of samples required to be at a leaf node. | |
| SVMs | C | Regularization parameter. The strength of the regularization is inversely proportional to C. |
| Kernel | Specifies the kernel type to be used in the algorithm. | |
| KNN | k-parameter | Number of neighbors to use by default for k neighbors queries. |
| Logistic Regression | Penalty | Used to specify the norm used in the penalization. |
| C | Inverse of regularization strength; must be a positive float. |
| Actual Classes | |||
|---|---|---|---|
| Positive | Negative | ||
| Predicted classes | Positive | True Positive | False Positive |
| Negative | False Negative | True Negative | |
| Models | Accuracy (%) | Confusion Matrix | Features | Parameters | ||
|---|---|---|---|---|---|---|
| Logistic Regression | 71.71 | A1 | A2 | 50 | Penalty: l1, C: 1.0 | |
| A1 | 73 | 28 | ||||
| A2 | 30 | 74 | ||||
| Naive Bayes | 70.73 | A1 | A2 | 55 | GaussianNB | |
| A1 | 72 | 29 | ||||
| A2 | 31 | 73 | ||||
| SVM | 70.73 | A1 | A2 | 45 | C = 2, kernel = sigmoid | |
| A1 | 75 | 26 | ||||
| A2 | 34 | 70 | ||||
| KNN | 66.83 | A1 | A2 | 145 | leaf_size: 1, n_neighbors: 12, weights: distance | |
| A1 | 78 | 23 | ||||
| A2 | 45 | 59 | ||||
| Decision Tree | 65.85 | A1 | A2 | 85 | max_features: log2, min_samples_leaf: 4, min_samples_split: 11 | |
| A1 | 68 | 33 | ||||
| A2 | 37 | 67 | ||||
| Random Forest | 68.78 | A1 | A2 | 30 | criterion: gini, min_samples_leaf: 3, min_samples_split: 7, n_estimators: 15 | |
| A1 | 71 | 30 | ||||
| A2 | 34 | 70 | ||||
| XGboost | 67.8 | A1 | A2 | 45 | gamma: 0, max_depth: 1, min_child_weight: 4 | |
| A1 | 69 | 32 | ||||
| A2 | 34 | 70 | ||||
| Models | Accuracy (%) | Confusion Matrix | Features | Parameters | ||
|---|---|---|---|---|---|---|
| Logistic Regression | 63.98 | B1 | B2 | 25 | Penalty: l1, C: 1.0 | |
| B1 | 48 | 22 | ||||
| B2 | 36 | 55 | ||||
| Naive Bayes | 63.98 | B1 | B2 | 35 | GaussianNB | |
| B1 | 50 | 20 | ||||
| B2 | 38 | 53 | ||||
| SVM | 61.49 | B1 | B2 | 35 | C: 6, kernel: linear | |
| B1 | 46 | 24 | ||||
| B2 | 38 | 53 | ||||
| KNN | 57.76 | B1 | B2 | 15 | leaf_size: 1, n_neighbors: 16, weights: uniform | |
| B1 | 63 | 7 | ||||
| B2 | 61 | 30 | ||||
| Decision Tree | 58.39 | B1 | B2 | 15 | max_features: auto, min_samples_leaf: 1, min_samples_split: 6 | |
| B1 | 41 | 29 | ||||
| B2 | 38 | 53 | ||||
| Random Forest | 62.11 | B1 | B2 | 15 | criterion: gini, min_samples_leaf: 2, min_samples_split: 7, n_estimators: 30 | |
| B1 | 48 | 22 | ||||
| B2 | 39 | 52 | ||||
| XGboost | 60.25 | B1 | B2 | 40 | gamma: 0.4, max_depth: 7, min_child_weight: 5 | |
| B1 | 44 | 26 | ||||
| B2 | 38 | 53 | ||||
| Models | Accuracy (%) | Confusion Matrix | Features | Parameters | ||
|---|---|---|---|---|---|---|
| Logistic Regression | 61.11 | C1 | C2 | 35 | Penalty: l1, C: 1.0 | |
| C1 | 49 | 15 | ||||
| C2 | 41 | 39 | ||||
| Naive Bayes | 59.03 | C1 | C2 | 160 | GaussianNB | |
| C1 | 23 | 41 | ||||
| C2 | 18 | 62 | ||||
| SVM | 65.28 | C1 | C2 | 65 | C: 5, kernel: rbf | |
| C1 | 48 | 16 | ||||
| C2 | 34 | 46 | ||||
| KNN | 61.11 | C1 | C2 | 120 | leaf_size: 1, n_neighbors: 5, weights: uniform | |
| C1 | 55 | 9 | ||||
| C2 | 47 | 33 | ||||
| Decision Tree | 66.67 | C1 | C2 | 225 | max_features: auto, min_samples_leaf: 2, min_samples_split: 8 | |
| C1 | 44 | 20 | ||||
| C2 | 28 | 52 | ||||
| Random Forest | 59.72 | C1 | C2 | 140 | criterion: gini, min_samples_leaf’: 1, min_samples_split: 5, n_estimators: 25 | |
| C1 | 37 | 27 | ||||
| C2 | 31 | 49 | ||||
| XGboost | 62.5 | C1 | C2 | 150 | n_estimators = 100, max_depth = 8, learning_rate = 0.1, subsample = 0.5 | |
| C1 | 44 | 20 | ||||
| C2 | 34 | 46 | ||||
| Models | Accuracy (%) | Confusion Matrix | Features | Parameters | ||
|---|---|---|---|---|---|---|
| Logistic Regression | 72.84 | D1 | D2 | 55 | Penalty: l1, C: 1.0 | |
| D1 | 54 | 27 | ||||
| D2 | 17 | 64 | ||||
| Naive Bayes | 68.52 | D1 | D2 | 20 | GaussianNB | |
| D1 | 44 | 37 | ||||
| D2 | 14 | 67 | ||||
| SVM | 74.07 | D1 | D2 | 55 | C: 0.1, kernel: linear | |
| D1 | 56 | 25 | ||||
| D2 | 17 | 64 | ||||
| KNN | 71.6 | D1 | D2 | 75 | algorithm: auto, leaf_size: 1, n_neighbors: 17, weights: uniform | |
| D1 | 55 | 26 | ||||
| D2 | 20 | 61 | ||||
| Decision Tree | 61.73 | D1 | D2 | 30 | max_features: auto, min_samples_leaf: 3, min_samples_split: 10 | |
| D1 | 56 | 25 | ||||
| C2 | 37 | 44 | ||||
| Random Forest | 66.67 | D1 | D2 | 20 | criterion: gini, min_samples_leaf: 3, min_samples_split: 3, n_estimators: 25 | |
| D1 | 47 | 34 | ||||
| D2 | 20 | 61 | ||||
| XGboost | 64.81 | D1 | D2 | 15 | gamma: 0.6, max_depth: 1, min_child_weight: 8 | |
| D1 | 51 | 30 | ||||
| D2 | 27 | 54 | ||||
| Models | Accuracy (%) | Confusion Matrix | Features | Parameters | ||
|---|---|---|---|---|---|---|
| Logistic Regression | 71.14 | E1 | E2 | 55 | Penalty: l1, C: 1.0 | |
| E1 | 50 | 17 | ||||
| E2 | 26 | 56 | ||||
| Naive Bayes | 68.46 | E1 | E2 | 230 | GaussianNB | |
| E1 | 48 | 19 | ||||
| E2 | 28 | 54 | ||||
| SVM | 71.81 | E1 | E2 | 70 | C: 1, kernel: sigmoid | |
| E1 | 50 | 17 | ||||
| E2 | 25 | 57 | ||||
| KNN | 63.76 | E1 | E2 | 20 | algorithm: auto, leaf_size: 1, n_neighbors: 16, weights: uniform | |
| E1 | 48 | 19 | ||||
| E2 | 35 | 47 | ||||
| Decision Tree | 66.44 | E1 | E2 | 95 | max_features: auto, min_samples_leaf: 2, min_samples_split: 12 | |
| E1 | 45 | 22 | ||||
| E2 | 28 | 54 | ||||
| Random Forest | 67.11 | E1 | E2 | 55 | criterion: gini, min_samples_leaf: 1, min_samples_split: 3, n_estimators: 30 | |
| E1 | 42 | 25 | ||||
| E2 | 24 | 58 | ||||
| Xgboost | 70.47 | E1 | E2 | 45 | gamma: 0.6, max_depth: 2, min_child_weight: 1 | |
| E1 | 43 | 24 | ||||
| E2 | 20 | 62 | ||||
| Dataset | Data Used in the Training | Best Testing Performance (%) Achieved | Best Model | Num. of Selected Features | ||
|---|---|---|---|---|---|---|
| Baseline | M12 Progress Wrt Beseline | M24 Progress Wrt Baseline | ||||
| A | ● | 71.71 | Logistic Regression | 50 | ||
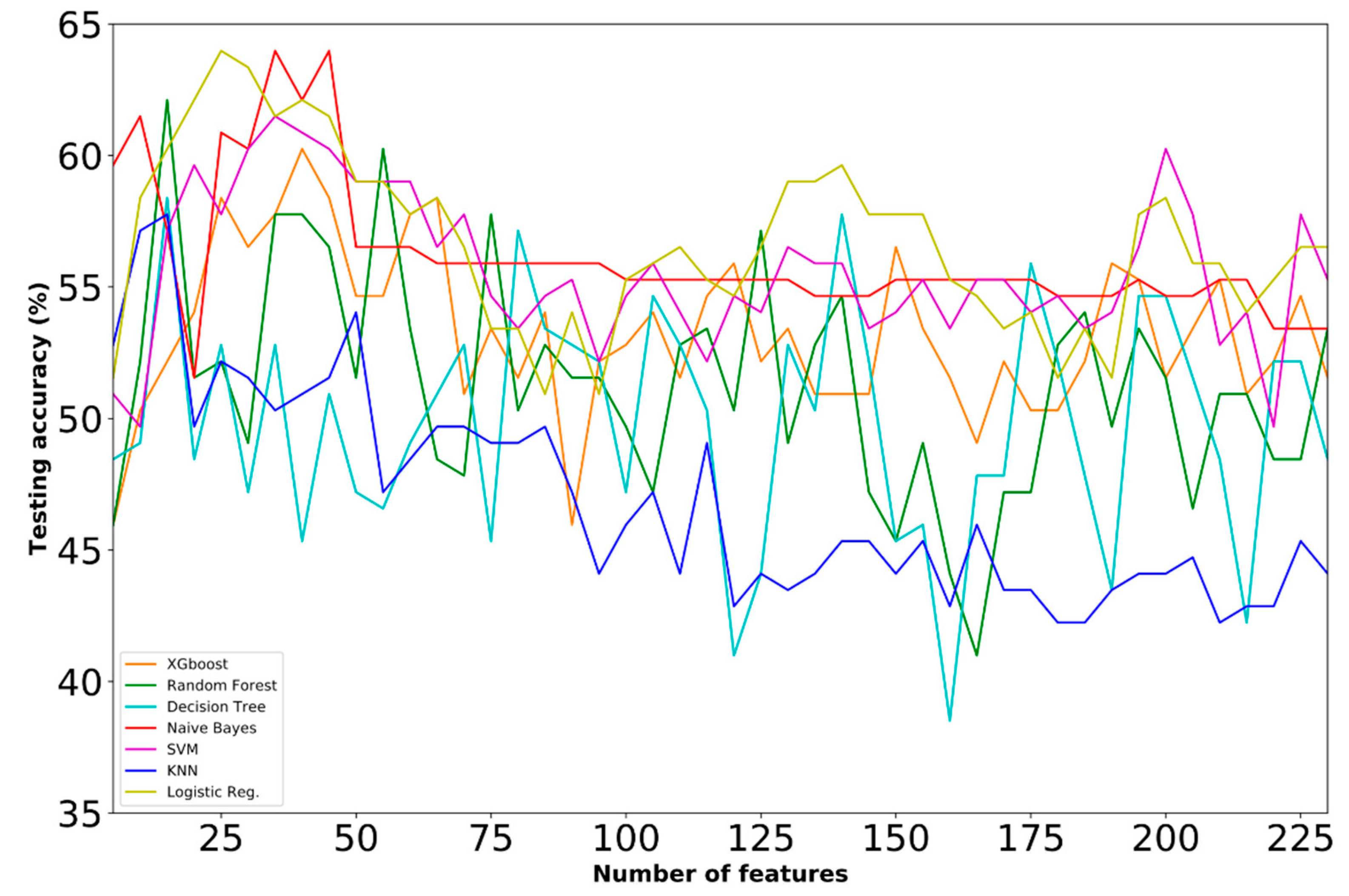
| B | ● | 63.98 | Logistic Regression | 25 | ||
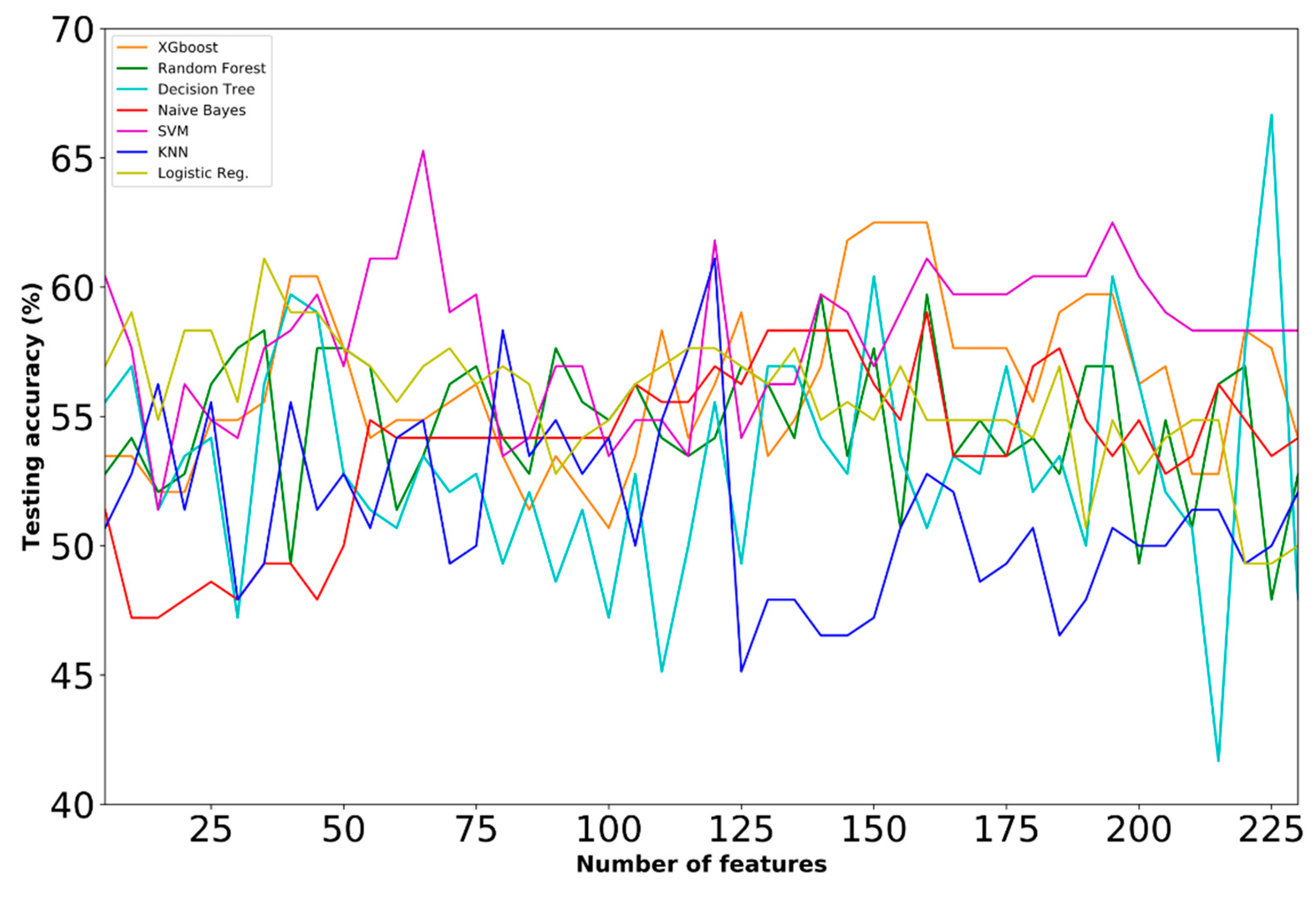
| C | ● | 66.67 | Decision Tree | 225 | ||
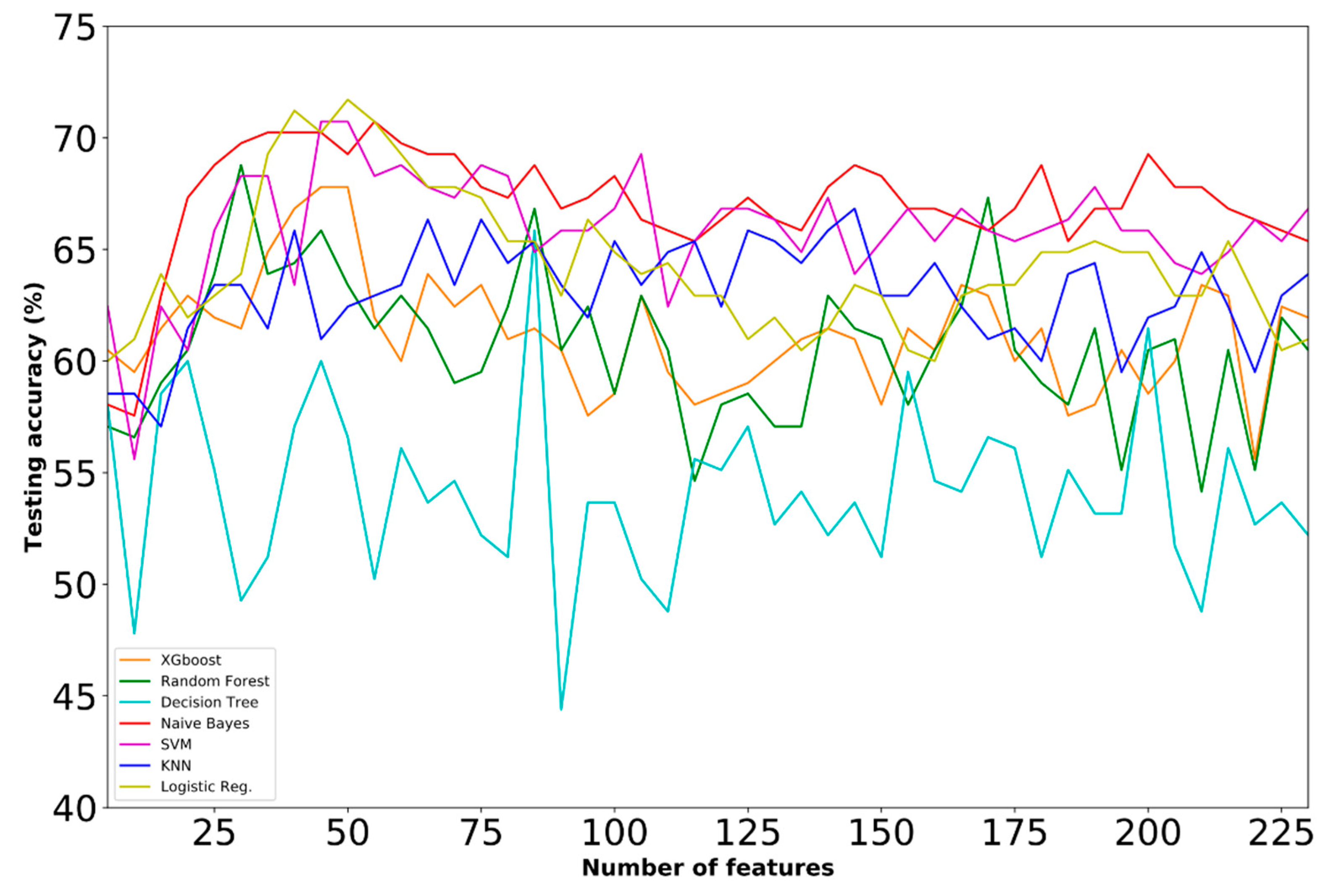
| D | ● | ● | 74.07 | SVM | 55 | |
| E | ● | ● | 71.81 | SVM | 70 | |
| FS Criteria | |||||||
|---|---|---|---|---|---|---|---|
| Filter Algorithms | Wrapper Algorithms | Embedded | Proposed FS Criterion | ||||
| Features | Chi-2 | Pearson | Logistic Regression | Logistic Regression (L2) | Random Forest | LightGBM | |
| 5 | 58.02 | 62.35 | 62.96 | 54.32 | 45.68 | 56.17 | 63.58 |
| 10 | 63.58 | 63.58 | 59.88 | 51.23 | 48.77 | 50.00 | 57.41 |
| 15 | 61.11 | 58.02 | 51.85 | 50.62 | 50.62 | 53.70 | 61.11 |
| 20 | 53.09 | 61.11 | 57.41 | 48.77 | 50.62 | 50.00 | 66.05 |
| 25 | 60.49 | 65.43 | 60.49 | 51.85 | 56.79 | 53.70 | 66.05 |
| 30 | 64.81 | 70.37 | 70.37 | 60.49 | 58.02 | 51.23 | 64.2 |
| 35 | 66.67 | 65.43 | 62.96 | 56.79 | 58.02 | 53.70 | 66.05 |
| 40 | 59.26 | 66.67 | 65.43 | 60.49 | 60.49 | 54.32 | 67.28 |
| 45 | 64.81 | 67.90 | 69.75 | 54.32 | 58.02 | 46.30 | 67.9 |
| 50 | 63.58 | 67.28 | 68.52 | 55.56 | 60.49 | 48.77 | 67.9 |
| 55 | 64.81 | 69.75 | 64.81 | 53.09 | 59.88 | 53.09 | 74.07 |
| 60 | 69.75 | 67.28 | 65.43 | 55.56 | 59.88 | 55.56 | 72.22 |
| 65 | 61.73 | 64.81 | 70.99 | 60.49 | 58.64 | 54.94 | 69.75 |
| 70 | 68.52 | 66.67 | 74.07 | 56.17 | 56.17 | 54.32 | 71.6 |
| 75 | 68.52 | 64.81 | 72.22 | 54.32 | 51.85 | 59.26 | 69.14 |
| 80 | 66.05 | 66.67 | 69.14 | 58.02 | 58.02 | 59.88 | 69.14 |
| 85 | 66.05 | 66.67 | 72.84 | 53.70 | 59.26 | 57.41 | 72.22 |
| 90 | 67.90 | 56.79 | 73.46 | 58.64 | 62.96 | 53.09 | 66.67 |
| 95 | 66.67 | 56.79 | 69.14 | 59.88 | 61.11 | 55.56 | 70.37 |
| 100 | 62.96 | 59.88 | 72.22 | 61.73 | 56.79 | 55.56 | 70.99 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokkotis, C.; Moustakidis, S.; Giakas, G.; Tsaopoulos, D. Identification of Risk Factors and Machine Learning-Based Prediction Models for Knee Osteoarthritis Patients. Appl. Sci. 2020, 10, 6797. https://doi.org/10.3390/app10196797
Kokkotis C, Moustakidis S, Giakas G, Tsaopoulos D. Identification of Risk Factors and Machine Learning-Based Prediction Models for Knee Osteoarthritis Patients. Applied Sciences. 2020; 10(19):6797. https://doi.org/10.3390/app10196797
Chicago/Turabian StyleKokkotis, Christos, Serafeim Moustakidis, Giannis Giakas, and Dimitrios Tsaopoulos. 2020. "Identification of Risk Factors and Machine Learning-Based Prediction Models for Knee Osteoarthritis Patients" Applied Sciences 10, no. 19: 6797. https://doi.org/10.3390/app10196797
APA StyleKokkotis, C., Moustakidis, S., Giakas, G., & Tsaopoulos, D. (2020). Identification of Risk Factors and Machine Learning-Based Prediction Models for Knee Osteoarthritis Patients. Applied Sciences, 10(19), 6797. https://doi.org/10.3390/app10196797







