1. Introduction
Convolution neural network (CNN) has achieved great success in processing data on Euclidean domains (grid structure) [
1,
2], such as image, speech, and video [
3,
4,
5,
6]. Therefore, many recent studies have devoted to the extension of convolution operations to the data on non-Euclidean domains and proposed the graph convolution network (GCN) [
7,
8,
9].Then GCN has been successfully used to achieve improvements in several research fields, such as protein interface [
10], action recognition [
11], and traffic data processing [
12]. The GCN mainly includes the spectral-based and spatial-based methods, which can leverage the topology information of the graph data to aggregate the local node features, and then automatically learn the embeddings for graph nodes. In many of the graph-related tasks, such as recommender systems [
13,
14], chemical research [
15,
16] and natural language processing [
17], the GCN exhibits outstanding performance.
The research on graph pooling methods is much less than that of the graph convolution models. In fact, the graph pooling technique is very important for obtaining the scaled-down graphs and graph-level embeddings. The most primitive graph pooling methods use graph topology information for node partitioning and graph coarsening [
8,
18,
19]. Recently, a differentiable graph pooling has been proposed to obtain hierarchical graphs by learning end-to-end [
20]. Moreover, there are more advanced graph pooling methods that can automatically learn the scores of nodes or edges and then pool nodes with high scores or merge nodes connected by edges with high scores to achieve the coarsening graph [
21,
22,
23,
24].
However, how to pool nodes using node scores is not well understood and discussed, which may affect effective graph coarsening. If pooling only the highest-scoring nodes as the existing papers might throw away whole neighbourhoods of nodes and therefore information, because it can be intuitively seen from the GCN formulation that similar nodes in feature-space or topological space are assigned similar scores (we have proven it in the following section). The coarsening graph with pooling top nodes is difficult to uniformly inherit the representative features of the original graph, which in turn affects the representation capacity of the GCN.
Here, we proposed a new graph pooling method UGPool with a new point-of-view on selecting pooled nodes, which assigns a score to each node, and uniformly pool the neighboring nodes in score-space instead of pooling top nodes as in existing papers. Pooling only the highest-scoring nodes might throw away whole neighbourhoods of nodes and therefore information, because of that the neighboring nodes in feature-space are also adjacent in score-space, which can be intuitively seen from the GCN formulation and has been proven in the following section. Specifically, we introduced a fully connected layer, taking node features or topology information as input, and learning the score of each graph node. Next, we sort the nodes according to their scores and perform normal pooling for one-dimensional data. Finally, update the adjacency matrix of the nodes according to the coarsening graphs. Our method can learn more comprehensive graph representations in an end-to-end fashion, which may improve the classification performance of the GCN. In short, we transformed the graph pooling problem into a popular one-dimensional data pooling problem by introducing a node automatic sorting mechanism. The implementation principle of our method is simple, and the performance or efficiency of our method is significantly better than the previous graph pooling on several standard benchmark datasets.Specifically, in multiple graph classification tasks, the proposed UGPool method achieves an average accuracy gain of 1.08% compared with other node-based pooling methods and achieves performance comparable to edge-based EdgePool with an average reduction in runtime of 64%. Moreover, UGPool method can be integrated with multiple GCN architectures and effectively improve the classification accuracy by 1.63% on average on multiple benchmark datasets.
To sum up, the main contributions of this work are:
We proposed a novel pooling method UGPool with a new point-of-view in pooling graph nodes that uniformly pooling the neighboring nodes in score-space instead of pooling top nodes as usual, which could uniformly coarsen the graph nodes.
The proposed UGPool could learn hierarchical graph embeddings, which outperforms other node-based graph pooling methods while maintaining high efficiency in multiple graph classification tasks.
The proposed UGPool method could be integrated with multiple GCN models to effectively improve performance compared to no pooling.
The rest of this paper is organized as follows:
Section 2 includes an overview of related work of graph convolution and graph pooling.
Section 3 describes the proposed UGPool methods.
Section 4 evaluates the UGPool method and compares it to other graph pooling methods with extensive experiments. Finally,
Section 5 concludes our work.
2. Related Work
To facilitate background understanding, we make brief reviews on the related work of graph convolution and graph pooling, respectively.
2.1. Graph Convolution
Graph convolution operation can aggregate local features of the data on a non-Euclidean domain and has powerful representation ability on graph data [
7,
25]. In general, graph convolution includes spectral-based and spatial-based models. For spectral-based models, the graph is transformed into a Fourier spectral-domain through graph Laplacian, and then spectral filers are used to achieve graph convolution [
8,
26,
27]. For example, [
8] proposed a spectral graph convolution by applying Chebyshev filters. For spatial-based models, features of locally adjacent nodes are directly aggregated and then propagated to the next layer [
7,
25,
28]. Different aggregation and propagation rules have spawned different graph convolution models.
2.2. Graph Pooling
The pooling layer enables CNN to reduce trainable parameters to overcome the overfitting problem while facilitating the network to learn multi-scale graph embeddings [
1,
29]. Finding the proper pooling layer for GCN is also important but more challenging because the graph nodes are not distributed in the regular grid domains. At present, graph pooling could be group into the following three categories [
23]: topology-based, global, and hierarchical pooling. Topology-based graph pooling method directly clusters nodes to coarse graph using graph theory such as spectral clustering algorithms [
8,
18,
30]. However, these methods often introduce a lot of computation, especially when the node size is large. The global pooling method applies summation and neural network to integrate the node features of each layer into graph-level representations, which can be directly used for graph classification tasks [
31,
32,
33]. Hierarchical pooling can obtain assignments of coarse nodes and their new topology information through neural networks, so that graph convolution can be performed on hierarchical graphs, similar to CNN being executed on multi-scale images. Most neural network-based pooling methods can be used for both global pooling and hierarchical pooling architectures, both of which can improve graph classification.
Ying et al. [
20] proposed a differentiable graph pooling method called DiffPool, which can learn the node assignments in end-to-end fashion using specialized graph convolutional layers. The node assignment matrix
of
layer is generated by input node features
and adjacency matrix
:
Then the node assignments matrix is used to update the node features and adjacency matrix:
where
denotes the output embeddings of
graph convolution layer. Please note that DiffPool learns a dense assignment matrix requiring a large storage complexity of
, where
and
k denote vertices and pooling ratio, respectively. DiffPool introduces more training parameters, making it easier to overfitting.
Gao et al. [
21] proposed a simple gPool method (also known as TopKPool) that uses a linear layer to generate a node score with node feature as input, and then pools the top K nodes with the highest score. Subsequently, gPool was applied for graph classification, and achieved performance comparable to that of DiffPool but required lower storage complexity of
, where
and
denote vertices and edges, respectively [
22]. The procedure in gPool can be described by the following equation:
where
y denotes the node score,
p denotes the parameter of the linear layer,
means selecting the top
nodes and
is the index of the top nodes.
Lee et al. [
23] introduced self-attention graph pooling (SAGPool), which uses a graph neural network (GNN) to provide self-attention scores. SAGPool is a variant of TopKPool and it combines node features and topology information when calculating node scores.
Recently, Diehl et al. [
24] proposed an EdgePool method based on the notion of edge contraction. Compared to the previous method based on node scores, EdgePool selects the combination of two connected node features as input, and calculates the edge score using the neural network. Then the model chooses a highest-scoring edge to contract by merging its nodes. The procedure in EdgePool can be described by the following equation:
where
denotes the node feature,
W and
b are learned parameters,
is the raw edge score,
is the final edge score, and
is the new node feature.
2.3. Summary
To sum up, the above node-based graph pooling methods both pool top K nodes with the highest score to achieve the coarsening graph, which has a limitation that the pooled graph cannot uniformly inherit the node features of the previous layer, because pooling only the highest-scoring nodes might throw away whole neighbourhoods of nodes and therefore information. Discarding all the low-score nodes may miss the useful features for graph embeddings and graph classification. Although EdgePool tries to uniformly extract graph features by contracting edges and merging the connected nodes, its runtime scales linearly in the number of edges, which is generally more complex than that of node-based pooling methods scaling linearly in the number of nodes. Therefore, it is necessary to develop a pooling method that can uniformly coarsen graph data to improve graph classification while maintaining high efficiency.
3. The Proposed Method
Compared with the study of the GCN model, the graph pooling method is still relatively few. To further improve the graph pooling, we propose an UGPool method to uniformly extract node features, which assigns a score to each node and uniformly pools the adjacent nodes in the score-space. Our method outperforms other node-based pooling methods and achieves performance comparable to edge-based EdgePool in a more efficient runtime.
3.1. Uniform Graph Pooling
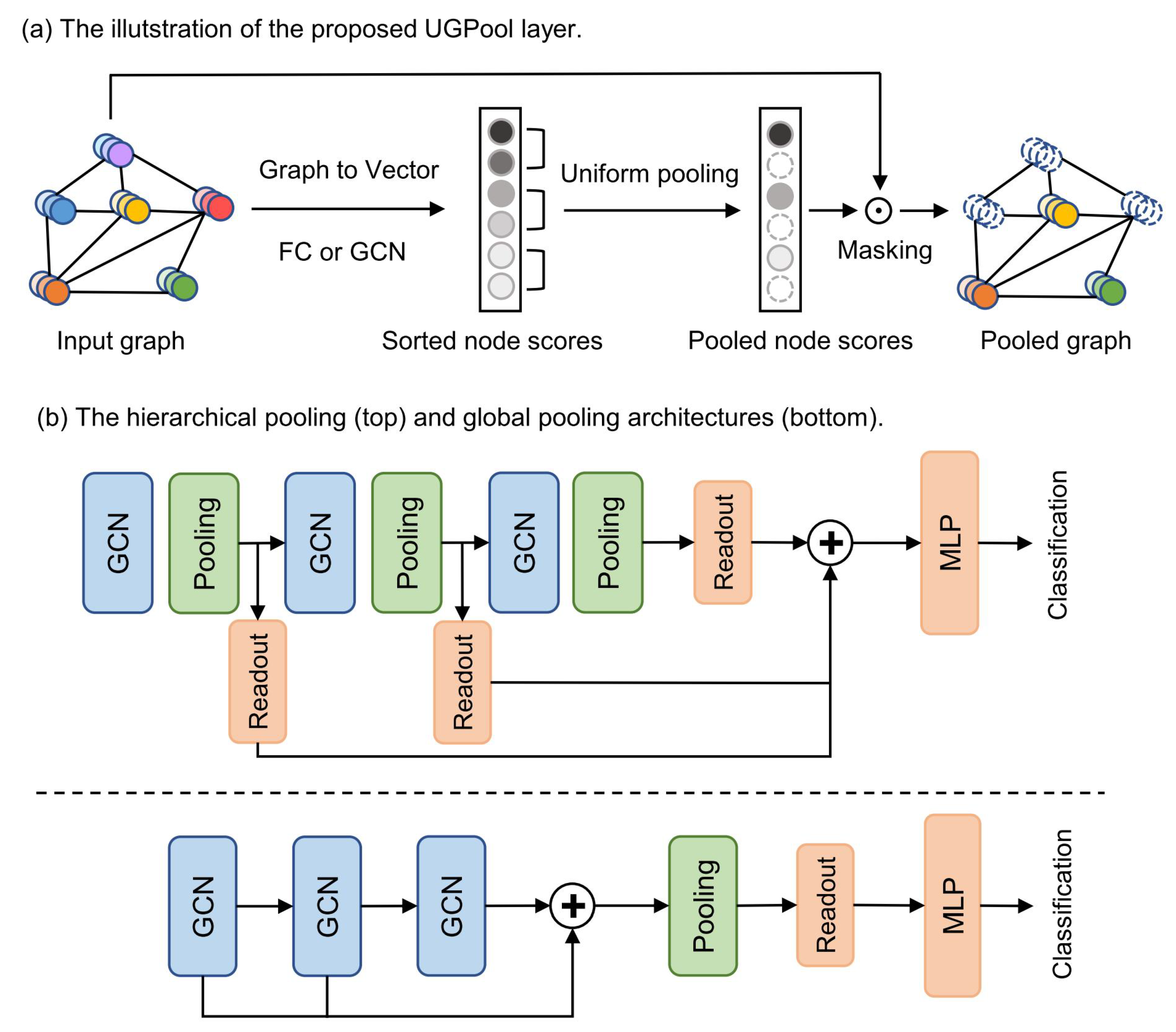
Our UGPool method is to uniformly retain node features, just like pooling on grid-like domain data. UGPool uses neural networks to learn node scores with node features or topological information as input, and ranks nodes based on their scores. Subsequently, a normal one-dimensional pooling is performed on the sorted nodes to achieve a coarse graph, and then the adjacency matrix is updated based on the pooled node.
Graph to Vector. We introduce a layer of neural network that uses node features as input to learning the node scores. If a simple linear layer is applied, the learned node score is equivalent to a one-dimensional mapping of the node features. If the graph convolutional layer is applied, the learned node score reflects both the feature and topology information of the node [
23]. The node score is generated by the following formula:
where
denotes the normalized adjacency matrix,
X is the node features, and
p is the learnable weights of the neural network. Please note that if
is set to the identity matrix, the linear layer is used, otherwise, the graph convolutional layer is used. Next, ranking the node based on the node score for further pooling.
Uniform Node Pooling. We can prove that the nodes with similar features or topology information will be assigned similar scores. This property allows us to perform a normal one-dimensional pooling on the sorted node vector, such as max pooling or average pooling. That is, we can use the overall statistical characteristics of the neighboring node features of a certain node as new features of the node, while the adjacent nodes are discarded. Finally, we applied a gate operation to control the information flow to avoid the gradients from the loss could not be backpropagated. The following equation roughly describes the pooling procedure in UGPool:
where
is the operation of node ranking,
denotes the sorted index for node scores
y,
X denotes the node features.
and
denote the features and scores of the ranked nodes, respectively.
is the normal pooling on one-dimensional data,
is the index for pooled nodes. UGPool retains multiple types of nodes, rather than just high-scoring nodes, which allows the pooled graph to uniformly inherit the representative features of the original graph.
Adjacency Matrix Updating. After the pooled graph is calculated, the adjacency matrix between the pooled nodes also needs to be updated. Using the original adjacency information of the pooled nodes is a solution, but this may cause the problem of sparse edges and isolated nodes. To overcome this problem, we consider the second-order connection between nodes, as done in [
23]. The update of the adjacency matrix is as follows:
where
denotes the index for pooled nodes. It is easy to know that
generates the second-order connection between nodes.
Theorem 1. Let be any graph node features and be output of Equation (9), respectively, then as long as (i.e., as long as the node features are similar, the learned node scores are similar). Proof of Theorem 1. , if , then . □
3.2. Model Architecture
To make fair comparisons and reduce the variables, we applied the same network architecture as the previous studies [
22,
23,
33], and compared our UGPool method to the baseline on the same network architecture.
Graph convolution layer. Graph convolution is the basis of learning graph embedding. While there are many types of graph convolution models, we applied the most widely used model proposed by Kipf et al. [
7], and its network layer function is as follows:
where
is the node features of the
layer,
is the trainable weight matrix and
is the adjacency matrix with added self-connections.
is a diagonal matrix to normalize the adjacency matrix with
. We adopt the widely Rectified Linear Unit (ReLU) as an activation function
.
Readout layer. The readout layer can embed the node features into the embedding of the whole graph, and the readout layer should be able to handle graphs with different scales and have permutation invariance. We used a readout method that calculates the sum and maximum of all node features as the representation of the graph level [
22].
where
N is the nodes number,
is the feature of
node, and ∥ denotes concatenation operation.
Global and hierarchical pooling. In the present study, we compared the global pooling and hierarchical pooling architectures as done in [
22,
23]. The hierarchical pooling architecture includes three graph convolution layers, each followed by a pooling layer (
Figure 1). In contrast, the global pooling architecture consists of three graph convolution layers, followed by a pooling layer after the last graph convolution layer. The output of each pooling layer passes through a readout layer, and the outputs of all readout layers are summed as the final output of the whole GCN. Finally, there are three fully connected layers as prediction layers.
4. Experiments
In this section, we applied the widely used GCNConv model with global pooling or hierarchical pooling architecture for multiple graph classification tasks. Please note that we directly used non-Euclidean structure data to evaluate our UGPool method, because GCN specializes in processing non-Euclidean structure data and has advantages in processing such data. We compared UGPool with other advanced graph pooling methods based on the same GCNConv architecture and under the same experimental conditions to demonstrate the effectiveness and efficiency of our model. The graph pooling methods selected for comparison include DiffPool, gPool, SAGPool, and EdgePool. Moreover, we integrated our UGPool with different GCNs, including GATConv [
9], GCNConv [
7], SAGEConv [
28], and SGCConv [
34], to improve classification performance. Specifically, the core code of UGPool was released at
https://github.com/Qin-J/Uniform-graph-pooling.
4.1. Experimental Settings
For a fair comparison, we performed the same training strategy and hyperparameter optimization strategy for each model. Specifically, we evaluated the generalization of the model by using 10-fold cross-validation, and the final accuracy was the average of the 10 testing results. In the training session, 10 percent of the training data was randomly selected as validation data. The remaining 90% of the graph data and graph labels were used for the supervised training of network parameters. We use the same early stopping criterion as done in [
35] to reduce over-fitting, i.e., when the loss of the validation data is not improved within 50 epochs with a maximum of 100k epochs, the training is stopped. We applied Adam as an optimization method and used a grid search as a hyperparameter search strategy (see
Table 1). To ensure the simplicity of the network and to minimize the modification, the layers of batch normalization and dropout architecture were not used in our model.
4.2. Graph Classification on Benchmark Datasets
From benchmark datasets [
36], we selected three protein structure datasets including D&D [
37], PROTEINS [
38] and ENZYMES [
39], and selected two biological datasets including NCI1 and NCI109 [
40]. D&D and PROTEINS contain large numbers of graph samples (>1 k) associated with two categories, and ENZYMES contains 500 graphs divided into six categories. Both NCI1 and NCI109 maintain more than 4 k graph samples with two categories. See
Table 2 for details of all the datasets.
In this experiment, we trained and tested the GCNConv with global pooling and hierarchical pooling architectures on the selected five datasets. we compared our UGPool method to other pooling methods based on the same GCNConv architecture. In addition, we counted the runtime for each graph pooling method to run an epoch to evaluate its execution efficiency. Please note that the runtime was measured on a V100 GPU. Moreover, because global pooling architecture achieved better classification accuracy in the benchmark datasets than hierarchical architecture, we integrated global UGPool architecture with different GCN models to evaluate whether our pooling model can effectively improve the existing GCN models.
Table 3 lists the classification accuracy of each dataset using GCNConv with different pooling methods. Please note that based on the same GCNConv architecture, our UGPool method dramatically outperforms other node-based graph pooling methods. While the performance of our method is worse than the edge-based pooling method (EdgePool) in ENZYMES, NCI1, and NCI109, the efficiency of our method is significantly superior to EdgePool, as shown in
Figure 2. The runtime of EdgePool is related to the number of edges, and the runtime of our UGPool is only related to the number of nodes. Therefore, the efficiency advantage of our algorithm will be more prominent in the larger graphs with larger edges. These results show that our UGPool that can uniformly pool node features is more reasonable than other pooling methods that only retains high-scoring nodes.
Table 4 lists classification accuracy based on different GCN models with and without UGPool. The results show that our method can be easily integrated into the general GCN models to improve classification performance.
4.3. Graph Classification on Brain Connectivity
fMRI-based functional connectivity is an effective measure of brain function, which reflects the correlation of brain signals between brain regions [
41]. Functional connectivity are often used as features for gender classification or diagnostic classification of brain disorders [
42,
43,
44,
45]. However, in previous studies, the functional connectivity matrix was simply arranged as a vector ignoring the spatial information between brain regions. In fact, the brain regions were selected on the irregular brain cortex and the topological relationship between the brain regions is on non-Euclidean domains, which can be described by a graph structure. Therefore, in this experiment, we will perform pattern analysis on brain connectivity from a new perspective, i.e., GCN model with the global pooling architecture of UGPool. We selected two widely used publicly available fMRI datasets including Human Connectome Project (HCP) [
46] and Autism Brain Imaging Data Exchange (ABIDE) [
47] for gender classification and autism classification, respectively. HCP data consists of resting-state fMRI brain images from 500 males (1944 images) and 596 females (2289 images). ABIDE data consist of resting-state fMRI brain images from 1003 patients (1199 images) and 1166 healthy controls (1415 images). Please note that some subjects have multiple scans images. To increase the samples as much as possible, we used all the images for training and testing the models, and divided the training set and test set based on subjects rather than images to prevent the problem of peeking at samples.
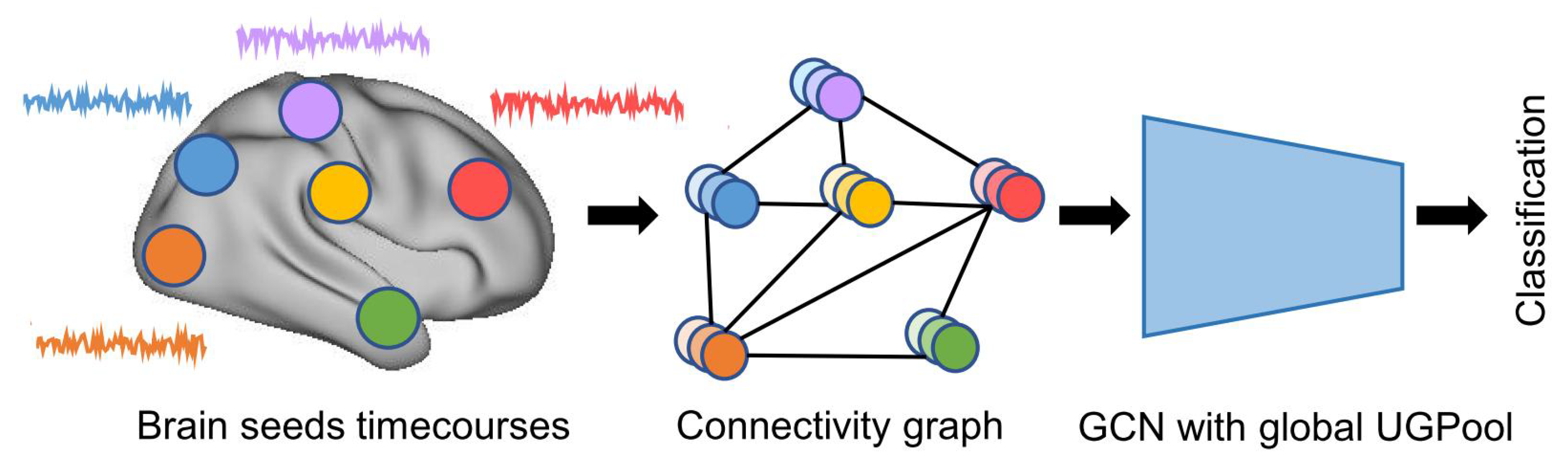
As shown in the illustration of the experiment in
Figure 3, the steps for processing the fMRI data include data preprocessing, selecting the brain region template, extracting the time series of brain regions, calculating the functional connectivity graph, and classifying connectivity graphs using GCN with global UGPool architecture. The data preprocessing pipeline was the same as we did in our previous works [
43,
48]. We used 808 ROIs as the brain template which was generated via spatially constrained spectral clustering [
49]. Pearson’s correlation coefficients between pairs of ROI-based time series were calculated, resulting in an
symmetric connectivity matrix for each subject. We used a k-nn graph
to describe the brain connectivity graph, where its node
represents a brain ROI, and functional connectivity of all the ROIs serve as node signals
and the graph edge
represents the correlation distance between corresponding node signals. We calculated the average functional connectivity of training data to estimate the average adjacency matrix of the connectivity graph, which will be commonly applied to the GCN model for each subject. Please note that using the functional metric to represent the graph edges may capture more accurate brain functional architecture rather than using structural or spatial metrics [
50]. Based on the connectivity graphs, we performed a GCN model with global UGPool architecture for gender classification and autism classification tasks, respectively. To make a comparison, we also evaluated the classification via the GCN model without the pooling method.
Table 5 lists the gender classification accuracy and autism classification accuracy via the GCN models with and without global UGPool on the HCP and ABIDE datasets, respectively. Based on different GCN models, our UGPool method can significantly improve the classification accuracy of the connectivity graphs. The experimental results show that the GCN model is effective in representing the brain connectivity graph. Moreover, our UGPool method can be integrated into general GCN models to improve graph embedding and classification performance.
4.4. Main Results
To sum up, we evaluated UGPool method with multiple benchmark datasets including protein structure datasets, biological datasets, and brain imaging datasets. By applying GCNConv model with different global pooling or hierarchical pooling methods for multiple graph classification tasks, we demonstrated that our UGPool method outperforms other node-based pooling methods with an average accuracy gain of 1.08% and achieves performance comparable to edge-based EdgePool with an average reduction in runtime of 64%. Moreover, we showed that UGPool method can be easily integrated with multiple GCN architectures and effectively improve the classification accuracy by 1.63% on average on multiple benchmark datasets.
4.5. Strengths and Possible Limits
The proposed UGPool could learn hierarchical graph embeddings, which outperforms other node-based graph pooling methods while maintaining high efficiency in multiple graph classification tasks. Moreover, UGPool could be integrated with multiple graph convolution networks to effectively improve performance compared to no pooling. However, the proposed method also has possible limitations, i.e., the feature of graph edges is not used when calculating node scores and pooling nodes, which may not be conducive to processing graphs with rich edge information. Please note that the proposed UGPool method could achieve almost comparable performance to the state-of-the-art method (EdgePool) with a more efficient runtime. The principles of these two pooling methods are quite different. The EdgePool is an edge-based pooling method that coarsens the graph by contracting edges and merging connected nodes. In contrast, the UGPool is a node-based pooling method that coarsens the graph by assigning a score to each node and uniformly pooling the adjacent nodes in the score-space. The runtime of EdgePool is related to the number of edges, and the runtime of our UGPool is only related to the number of nodes. Therefore, the operating efficiency of our UGPool is significantly higher than that of EdgePool, as graph data generally has much more edges than nodes. This advantage will be more prominent in processing large-scale graphs with dense edges.
4.6. Threats to Validity
Please note that there are still several threats to the validity of the experiment. First, although we have evaluated the proposed methods with multiple graph classification tasks, including the protein graphs, the biological graphs, and the brain connectivity graphs, there is still no theory to ensure that the classification performance of the proposed algorithm for other types of graph data, such as social network graphs and citation graphs, will definitely be better. Second, we only evaluated the proposed method on general-scale graph data, and the performance of the method on very large-scale graph data still needs further verification.
5. Conclusions
We proposed a novel pooling method UGPool with a new point-of-view in selecting pooled nodes, which assigns a score to each node, and uniformly pool the neighboring nodes in score-space instead of pooling top nodes as in existing papers. UGPool method has permutation invariance to graph nodes and uses a consistent number of parameters regardless of the input graph size. By uniformly preserving the representative node features, our UGPool method outperforms other node-based pooling methods with an average accuracy gain of 1.08% and achieves performance comparable to edge-based EdgePool with an average reduction in runtime of 64%. The efficiency advantage of UGPool is more prominent in processing large-scale graphs with dense edges. Moreover, UGPool can be integrated with multiple GCN architectures and effectively improve the classification accuracy by 1.63% on average on multiple benchmark datasets.
Our work provides a new perspective for the pooling method, which will promote uniformly coarsening graph-structured data, learning hierarchical graph embedding, and classifying the whole graph. For example, the proposed UGPool can be applied to coarsen and learn the hierarchical structure of graph data such as protein graphs, social network graphs, citation graphs, and brain network graphs. UGPool can also be applied to improve classification tasks of the above graph data by being integrated with different graph convolution models. In the future, it will be interesting to automatically generate the graph edges, instead of manually calculating it as currently done. Moreover, the pooling of dynamic graphs will be also the future direction. In that case, the graph structure and graph nodes are dynamically changing, which requires the pooling method to have dynamic scalability.
Author Contributions
Conceptualization, J.Q., L.L., and H.S.; methodology, J.Q. and L.L.; software, J.Q.; formal analysis, J.Q.; writing—original draft preparation, J.Q.; writing—review and editing, L.L., H.S., and D.H.; funding acquisition, H.S., and D.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (grant numbers 61773391 and 61722313), the National Key Research and Development Program (2018YFB1305101), and the Science and Technology Innovation Program of Hunan Province (2018RS3080).
Conflicts of Interest
The authors declare no conflict of interest.
References
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436. [Google Scholar] [CrossRef]
- Mammone, N.; Ieracitano, C.; Morabito, F.C. A deep CNN approach to decode motor preparation of upper limbs from time–frequency maps of EEG signals at source level. Neural Netw. 2020, 124, 357–372. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Stateline, NV, USA, 3–8 December 2012; pp. 1097–1105. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Hinton, G.; Deng, L.; Yu, D.; Dahl, G.; Mohamed, A.R.; Jaitly, N.; Senior, A.; Vanhoucke, V.; Nguyen, P.; Kingsbury, B.; et al. Deep Neural Networks for Acoustic Modeling in Speech Recognition. IEEE Signal Process. Mag. 2012, 29, 82–97. [Google Scholar] [CrossRef]
- Karpathy, A.; Toderici, G.; Shetty, S.; Leung, T.; Sukthankar, R.; Li, F.-F. Large-scale Video Classification with Convolutional Neural Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Columbus, OH, USA, 24–27 June 2014. [Google Scholar]
- Kipf, T.N.; Welling, M. Semi-supervised classification with graph convolutional networks. In Proceedings of the International Conference on Learning Representations, Toulon, France, 24–26 April 2017; pp. 1–14. [Google Scholar]
- Defferrard, M.; Bresson, X.; Vandergheynst, P. Convolutional neural networks on graphs with fast localized spectral filtering. In Proceedings of the Advances in Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016; pp. 3844–3852. [Google Scholar]
- Veličković, P.; Cucurull, G.; Casanova, A.; Romero, A.; Lio, P.; Bengio, Y. Graph attention networks. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Fout, A.; Byrd, J.; Shariat, B.; Ben-Hur, A. Protein interface prediction using graph convolutional networks. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 6530–6539. [Google Scholar]
- Li, B.; Li, X.; Zhang, Z.; Wu, F. Spatio-temporal graph routing for skeleton-based action recognition. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 8561–8568. [Google Scholar]
- Guo, K.; Hu, Y.; Qian, Z.; Liu, H.; Zhang, K.; Sun, Y.; Gao, J.; Yin, B. Optimized Graph Convolution Recurrent Neural Network for Traffic Prediction. IEEE Trans. Intell. Transp. Syst. 2020, 1–12. [Google Scholar] [CrossRef]
- Berg, R.v.d.; Kipf, T.N.; Welling, M. Graph convolutional matrix completion. arXiv 2017, arXiv:1706.02263. [Google Scholar]
- Yao, L.; Mao, C.; Luo, Y. Graph convolutional networks for text classification. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 7370–7377. [Google Scholar]
- You, J.; Liu, B.; Ying, Z.; Pande, V.; Leskovec, J. Graph convolutional policy network for goal-directed molecular graph generation. In Proceedings of the Advances in Neural Information Processing Systems, Montréal, QC, Canada, 2–8 December 2018; pp. 6410–6421. [Google Scholar]
- Zitnik, M.; Agrawal, M.; Leskovec, J. Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics 2018, 34, i457–i466. [Google Scholar] [CrossRef]
- Peng, H.; Li, J.; He, Y.; Liu, Y.; Bao, M.; Wang, L.; Song, Y.; Yang, Q. Large-scale hierarchical text classification with recursively regularized deep graph-cnn. In Proceedings of the 2018 World Wide Web Conference. International World Wide Web Conferences Steering Committee, Lyon, France, 23–27 April 2018; pp. 1063–1072. [Google Scholar]
- Rhee, S.; Seo, S.; Kim, S. Hybrid approach of relation network and localized graph convolutional filtering for breast cancer subtype classification. In Proceedings of the 27th International Joint Conference on Artificial Intelligence, Stockholm, Sweden, 13–19 July 2018; pp. 3527–3534. [Google Scholar]
- Ma, Y.; Wang, S.; Aggarwal, C.C.; Tang, J. Graph Convolutional Networks with EigenPooling. arXiv 2019, arXiv:1904.13107. [Google Scholar]
- Ying, Z.; You, J.; Morris, C.; Ren, X.; Hamilton, W.; Leskovec, J. Hierarchical graph representation learning with differentiable pooling. In Proceedings of the Advances in Neural Information Processing Systems, Montréal, QC, Canada, 2–8 December 2018; pp. 4800–4810. [Google Scholar]
- Gao, H.; Ji, S. Graph U-Nets. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019; pp. 2083–2092. [Google Scholar]
- Cangea, C.; Veličković, P.; Jovanović, N.; Kipf, T.; Liò, P. Towards sparse hierarchical graph classifiers. In Proceedings of the NeurIPS Workshop on Relational Representation Learning, Montréal, QC, Canada, 8 December 2018. [Google Scholar]
- Lee, J.; Lee, I.; Kang, J. Self-Attention Graph Pooling. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019; pp. 3734–3743. [Google Scholar]
- Diehl, F.; Brunner, T.; Michael, T.L.; Knoll, A. Towards Graph Pooling by Edge Contraction. In Proceedings of the ICML Workshop on Learning and Reasoning with Graph-Structured Data, Los Angeles, CA, USA, 15 June 2019. [Google Scholar]
- Xu, K.; Hu, W.; Leskovec, J.; Jegelka, S. How powerful are graph neural networks? In Proceedings of the International Conference on Learning Representations, New Orleans, LA, USA, 6–9 May 2019. [Google Scholar]
- Bruna, J.; Zaremba, W.; Szlam, A.; LeCun, Y. Spectral networks and locally connected networks on graphs. In Proceedings of the International Conference on Learning Representations, Banff, AB, Canada, 14–16 April 2014. [Google Scholar]
- Henaff, M.; Bruna, J.; LeCun, Y. Deep convolutional networks on graph-structured data. arXiv 2015, arXiv:1506.05163. [Google Scholar]
- Hamilton, W.; Ying, Z.; Leskovec, J. Inductive representation learning on large graphs. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 1024–1034. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT press: Cambridge, MA, USA, 2016. [Google Scholar]
- Dhillon, I.S.; Guan, Y.; Kulis, B. Weighted graph cuts without eigenvectors a multilevel approach. IEEE Trans. Pattern Anal. Mach. Intell. 2007, 29, 1944–1957. [Google Scholar] [CrossRef]
- Gilmer, J.; Schoenholz, S.S.; Riley, P.F.; Vinyals, O.; Dahl, G.E. Neural message passing for quantum chemistry. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–22 August 2017; pp. 1263–1272. [Google Scholar]
- Vinyals, O.; Bengio, S.; Kudlur, M. Order matters: Sequence to sequence for sets. In Proceedings of the International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Zhang, M.; Cui, Z.; Neumann, M.; Chen, Y. An end-to-end deep learning architecture for graph classification. In Proceedings of the Thirty-Second AAAI Conference on Artificial Intelligence, New Orleans, LA, USA, 2–7 February 2018. [Google Scholar]
- Wu, F.; Souza, A.; Zhang, T.; Fifty, C.; Yu, T.; Weinberger, K. Simplifying Graph Convolutional Networks. In Proceedings of the 36th International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 10–15 June 2019; pp. 6861–6871. [Google Scholar]
- Shchur, O.; Mumme, M.; Bojchevski, A.; Günnemann, S. Pitfalls of graph neural network evaluation. arXiv 2018, arXiv:1811.05868. [Google Scholar]
- Kersting, K.; Kriege, N.M.; Morris, C.; Mutzel, P.; Neumann, M. Benchmark Data Sets for Graph Kernels. 2016. Available online: http://graphkernels.cs.tu-dortmund.de (accessed on 8 April 2016).
- Dobson, P.D.; Doig, A.J. Distinguishing enzyme structures from non-enzymes without alignments. J. Mol. Biol. 2003, 330, 771–783. [Google Scholar] [CrossRef]
- Borgwardt, K.M.; Ong, C.S.; Schönauer, S.; Vishwanathan, S.; Smola, A.J.; Kriegel, H.P. Protein function prediction via graph kernels. Bioinformatics 2005, 21, i47–i56. [Google Scholar] [CrossRef]
- Feragen, A.; Kasenburg, N.; Petersen, J.; de Bruijne, M.; Borgwardt, K. Scalable kernels for graphs with continuous attributes. In Proceedings of the Advances in Neural Information Processing Systems, Stateline, NV, USA, 5–10 December 2013; pp. 216–224. [Google Scholar]
- Wale, N.; Watson, I.A.; Karypis, G. Comparison of descriptor spaces for chemical compound retrieval and classification. Knowl. Inf. Syst. 2008, 14, 347–375. [Google Scholar] [CrossRef]
- Sporns, O. The human connectome: A complex network. Ann. N. Y. Acad. Sci. 2011, 1224, 109–125. [Google Scholar] [CrossRef] [PubMed]
- Dosenbach, N.U.F.; Nardos, B.; Cohen, A.L.; Fair, D.A.; Power, J.D.; Church, J.A.; Nelson, S.M.; Wig, G.S.; Vogel, A.C.; Lessovschlaggar, C.N. Prediction of Individual Brain Maturity Using fMRI. Science 2010, 329, 1358–1361. [Google Scholar] [CrossRef]
- Zeng, L.L.; Shen, H.; Liu, L.; Wang, L.; Li, B.; Fang, P.; Zhou, Z.; Li, Y.; Hu, D. Identifying major depression using whole-brain functional connectivity: a multivariate pattern analysis. Brain 2012, 135, 1498–1507. [Google Scholar] [CrossRef]
- Hou, C.; Zeng, L.L.; Hu, D. Safe Classification With Augmented Features. IEEE Trans. pattern Anal. Mach. Intell. 2018, 41, 2176–2192. [Google Scholar] [CrossRef]
- Yuan, L.; Chen, F.; Zeng, L.; Wang, L.; Hu, D. Gender Identification of Human Brain Image with A Novel 3D Descriptor. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 15, 551–561. [Google Scholar] [CrossRef]
- Smith, S.M.; Beckmann, C.F.; Andersson, J.; Auerbach, E.J.; Bijsterbosch, J.; Douaud, G.; Duff, E.; Feinberg, D.A.; Griffanti, L.; Harms, M.P. Resting-state fMRI in the human connectome project. Neuroimage 2013, 80, 144–168. [Google Scholar] [CrossRef]
- Di Martino, A.; Yan, C.G.; Li, Q.; Denio, E.; Castellanos, F.X.; Alaerts, K.; Anderson, J.S.; Assaf, M.; Bookheimer, S.Y.; Dapretto, M. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry 2014, 19, 659. [Google Scholar] [CrossRef]
- Shen, H.; Wang, L.; Liu, Y.; Hu, D. Discriminative analysis of resting-state functional connectivity patterns of schizophrenia using low dimensional embedding of fMRI. Neuroimage 2010, 49, 3110–3121. [Google Scholar] [CrossRef]
- Craddock, R.C.; James, G.A.; Holtzheimer III, P.E.; Hu, X.P.; Mayberg, H.S. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum. Brain Mapp. 2012, 33, 1914–1928. [Google Scholar] [CrossRef] [PubMed]
- Ktena, S.I.; Parisot, S.; Ferrante, E.; Rajchl, M.; Lee, M.; Glocker, B.; Rueckert, D. Metric learning with spectral graph convolutions on brain connectivity networks. NeuroImage 2018, 169, 431–442. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).