1. Introduction
The macular region is located in the center of the retina and is the most critical and sensitive part where more than 90% of the cones are concentrated, being responsible for determining the light, shape, and color vision of the human. Macular edema occurs in the macular region, and it is one of the three major retinal diseases. It is mainly caused by the rupture of vascular epithelial cells and mural cells between the blood vessels and the retina, causing damage to the retinal fluid transport cells, which can cause unclear vision and vision loss, deformation of sight, and even blindness in severe cases.
Spectral domain optical coherence tomography (SD-OCT) is a non-invasive imaging mode that is typically used to achieve a high-resolution (6 µm) cross-section scan of biological tissue with a depth of penetration (0.5–2 mm) [
1].
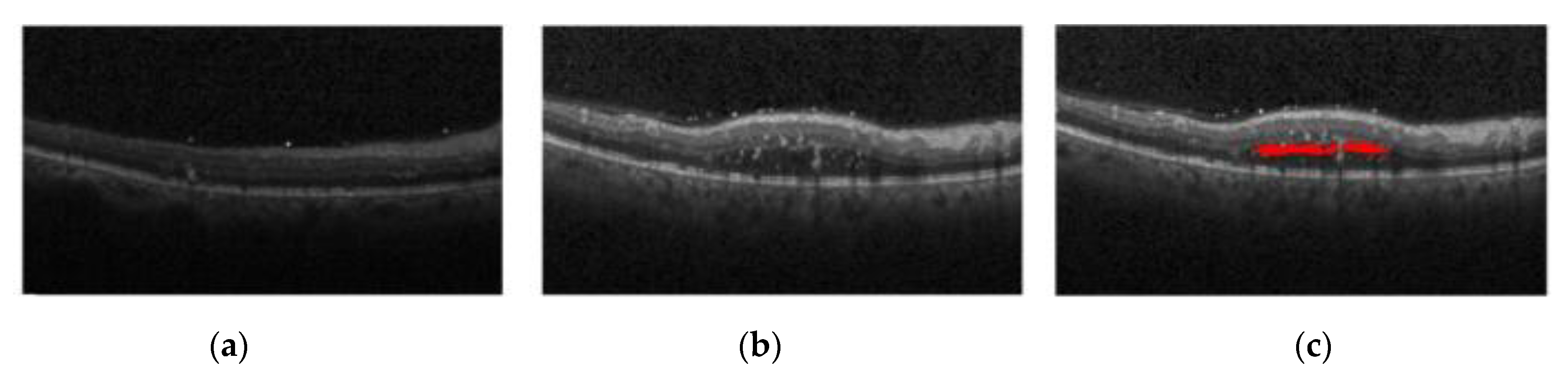
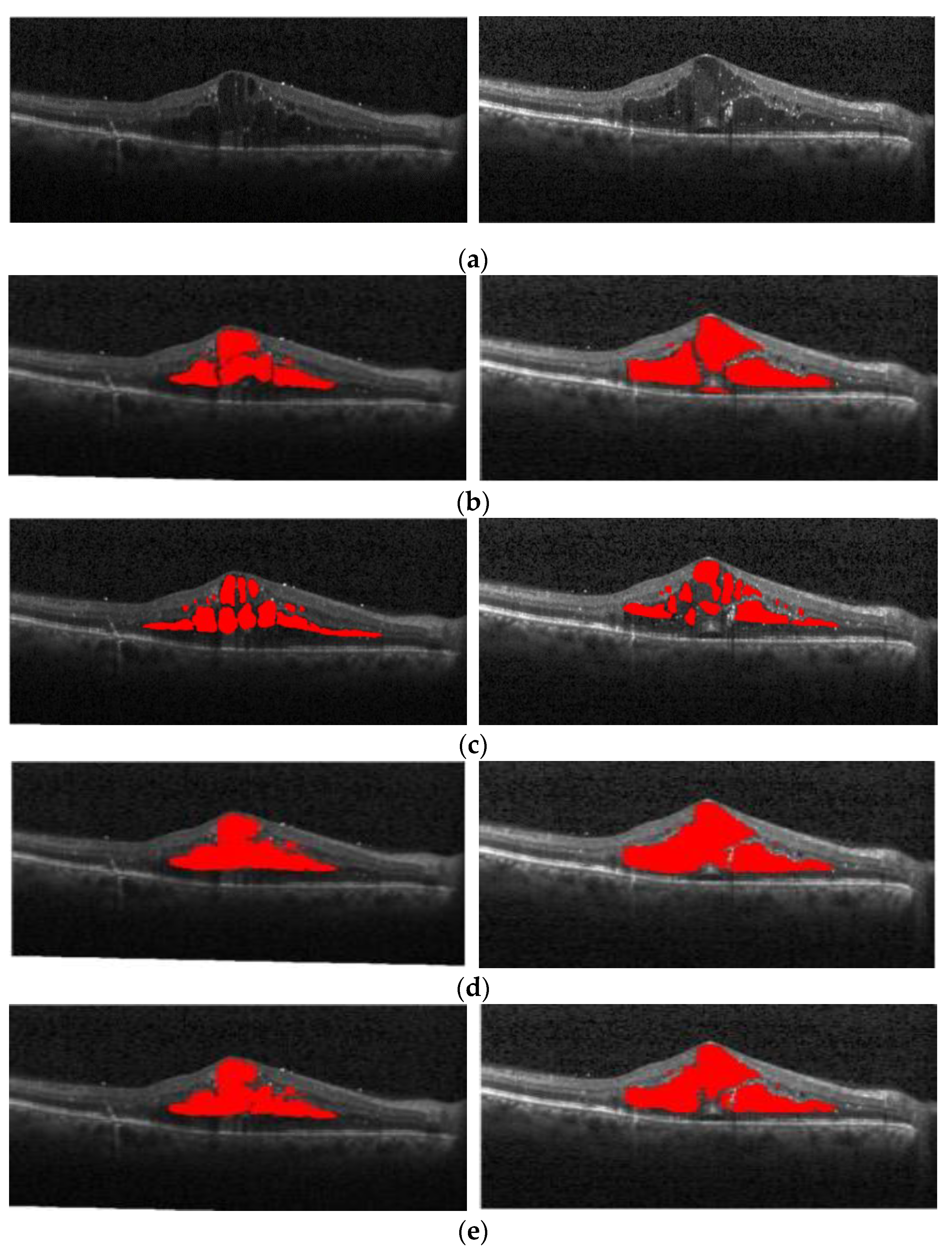
Figure 1a shows the normal retina optical coherence tomography (OCT) image,
Figure 1b shows the OCT image of macular edema, and
Figure 1c shows red regions in the image that are annotations of edema lesions. For the past two decades, optical coherence tomography has been recognized as one of the most important instruments in ophthalmic diagnosis. Ophthalmologists can screen and diagnose ocular patients by the number and size of cysts in the macular region.
However, the segmentation of macular edema in OCT images has always been a challenge for clinicians because of the high acquisition rate of the OCT system, which generates a large amount of 3D data, and each 3D volume contains hundreds of 2D B-scan images [
2]. It requires a long processing time to manually annotate each B-scan image and is unrealistic for clinical applications. In addition to the above difficulties, both the image noise and severe retinopathy can lead to the boundary between healthy tissue and diseased tissue to spread highly [
3]. This diffusion may lead to inconsistent manual annotation results among the different experts, and manual segmentation of edema regions will become very subjective. Therefore, in the past 10 years, many scholars have conducted important research on segmentation and detection of macular edema in retinal OCT images, obtaining many remarkable semi-automatic and automatic segmentation methods. There are two main categories: (1) those based on traditional methods, and (2) those based on deep learning.
For the traditional method, in 2005, Fernandez used a deformable model to delineate filled regions in OCT images of patients with the age related macular degeneration [
4]. In 2010, Quellec and Sonka et al. reported a three-dimensional analysis method of retinal layer texture, which can identify fluid-filled regions in SD-OCT of the macula [
5]. In 2013, Zheng et al. proposed a four-step process of first segmenting all low-reflection regions in an image as candidate regions, and then performed pre-processing, coarse segmentation, fine segmentation, and quantitative analysis on the candidate regions to automatically segment the intraretinal fluid (IRF) and the subretinal fluid (SRF) [
6]. In 2015, Xu et al. well used a layer-dependent stratified sampling strategy to segment the intraretinal and subretinal fluid in OCT images [
7]. Chiu et al. modified the dynamic programming-based approach to classify the layers and segment fluid using the kernel regression-based approach, and then fine-estimated them using the graph-based dynamic programming approach in the second phase [
2]. Following a similar path, in 2016, on the basis of graph segmentation technology, Karri et al. proposed a structured random forest method to learn layer-specific edges and to enhance segmentation [
8]. In the same year, Chiu et al. modified a method on the basis of dynamic programming by considering the spatial dependence of a frame and its two adjacent frames [
9]. This has made some great contributions to the segmentation of macular edema in retinal OCT images.
With the improvement of graphics processing unit performance and the proposal of new deep neural network models, deep neural network has been widely used in medical image segmentation, as well as in other fields. In 2015, full convolutional neural network (FCN) [
10] presented excellent performance in image segmentation tasks. In 2017, on the basis of FCN and fully connected conditional random field (CRF), Bai et al. proposed a new method for segmentation of macular edema in OCT images [
11]. The framework uses a convolution layer to extract and predict the pixel category of the input image, using fine-tuning to exploit the learning convolution kernel of the classification network to complete the segmentation task; then, according to the fully connected CRF model of image and FCN, the feature vector of heat map is outputted, including the pixel positions and values in different channels, and finally each pixel is classified, generating an amended prediction map as the final output.
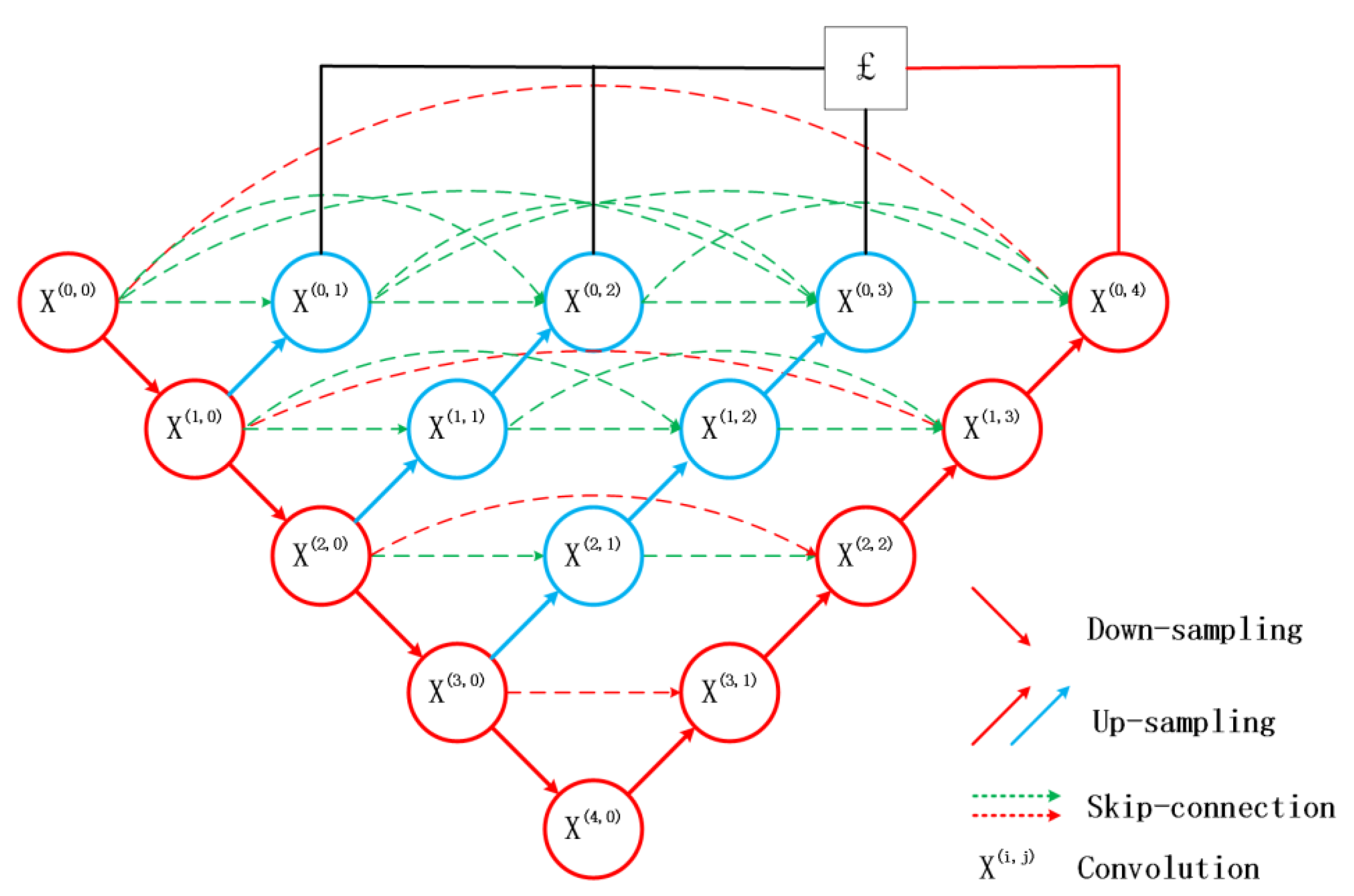
In 2015, U-Net came out, wherein its skip connection structure combines deep and shallow features, improves segmentation accuracy, and is widely used in the field of medical images [
12]. The application effect is better than other frameworks in small datasets of medical image segmentation, such as neural structures, kidneys [
13], and liver tumor [
14]. In 2017, Roy et al. proposed ReLayNet [
15], which is suitable for OCT macular edema segmentation by the U-Net as a framework. The network segmented the edema region with OCT images and achieved good segmentation results. In 2018, Venhuizen et al. proposed a two-stage full convoluted neural network based on U-Net [
16]—the first stage network was used to extract retinal regional features, and the second stage network was used to combine retinal information extracted in the previous stage for edema segmentation. In 2019, Li et al. improved the network parameters and loss function of U-Net in order to solve the imbalance problem if the categories in retinal OCT images, and added batch normalization (BN) between the convolution block and the activation function, which further improved the accuracy [
17]. In 2020, on the basis of U-Net, Chen et al. used multi-scale convolution instead of ordinary 3 × 3 convolution to achieve the adaptive field of the image. The channel attention module is embedded in the model, so that it can ignore irrelevant information and focus on key information in the channels. [
18]. In 2020, Liu et al. took advantage of the multi-scale input, multi-scale side output, and dual attention mechanism, presenting an enhanced nested U-Net architecture (MDAN-UNet) and achieving great performance with multi-layer segmentation and multi-fluid segmentation [
19]. In same year, Xie et al. used image enhancement and the improved 3D U-Net to achieve a fast and automated hyperreflective foci segmentation method, obtaining good segmentation results [
20]. Recently, Mohamed et al. integrated the segmentation of region of interest (RoI), proposing a HyCAD hybrid learning system for classification of diabetic macular edema, choroidal neovascularization, and drusen disorders [
21].
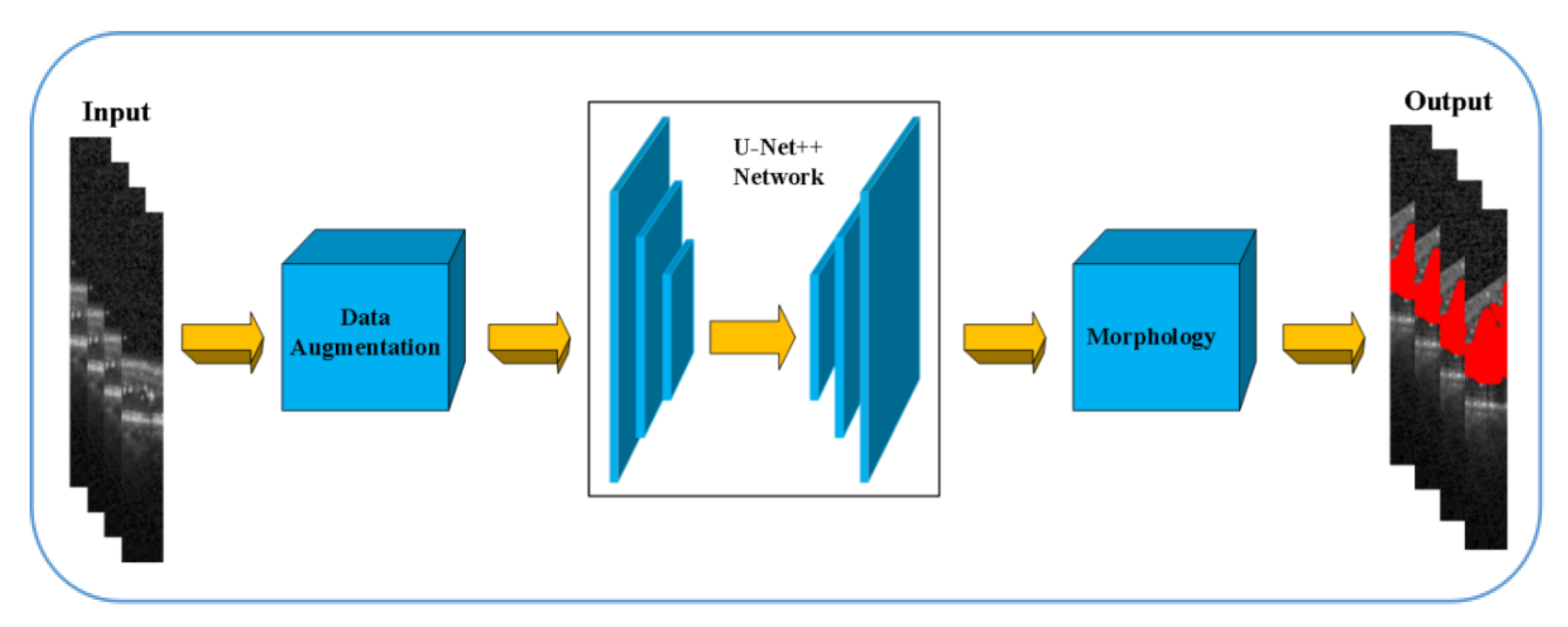
At present, due to the loss of shallow features and the influence of image noise, the existing segmentation methods have not achieved a more ideal segmentation effect on the whole and diverse edema lesions. Therefore, this paper proposes a new method for automatic segmentation of macular edema regions in retinal OCT images on the basis of improved U-Net++. This method makes full use of U-Net++’s redesigned skip connections and dense convolutional blocks, reducing the semantic loss of feature maps in the encoder/decoder subnetwork. By improving the U-Net++ network and using ResNet as the backbone network, the overall segmentation accuracy is not only further improved, but also the accuracy of segmentation for diverse edemas is significantly improved because the ResNeSt is used to reduce the loss of features in the transmission process.
The paper is organized as follows:
Section 2 introduces the fully convolutional neural network U-Net++ and explains the proposed method,
Section 3 describes the experimental settings and shows experimental results and comparative analysis,
Section 4 presents the discussion, and
Section 5 draws the conclusion of the study.
4. Discussion
In terms of diverse edemas in multi-regions,
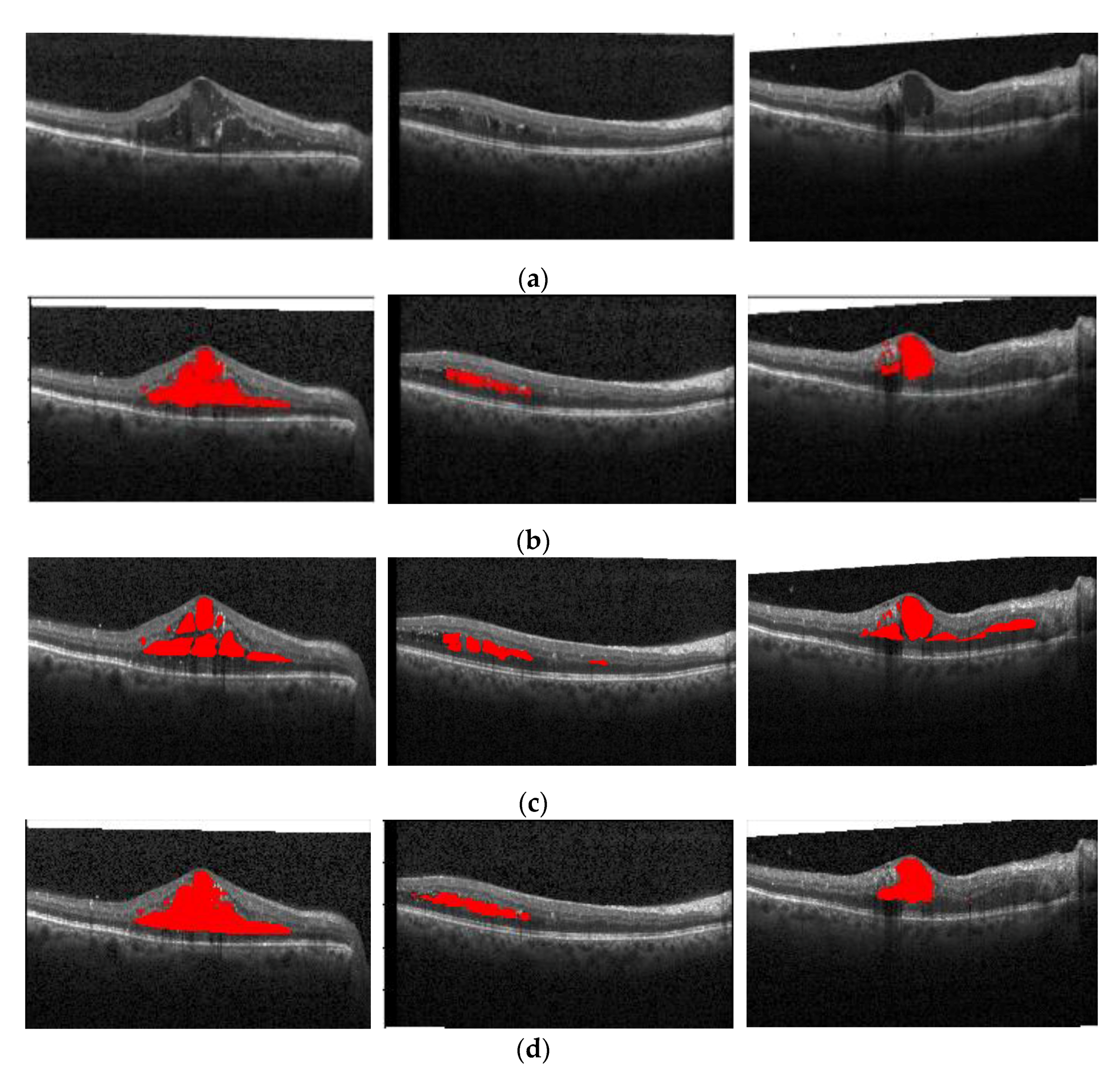
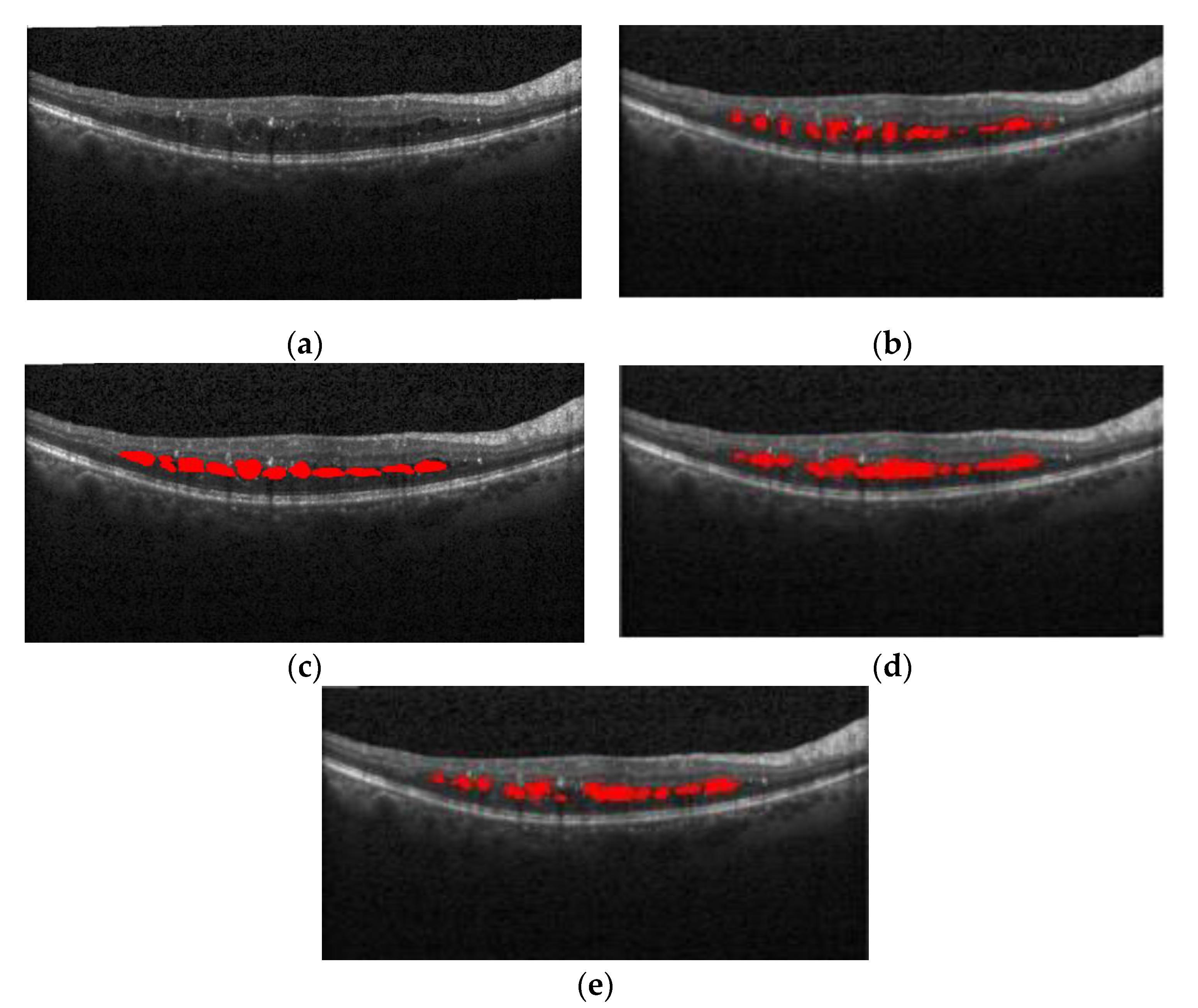
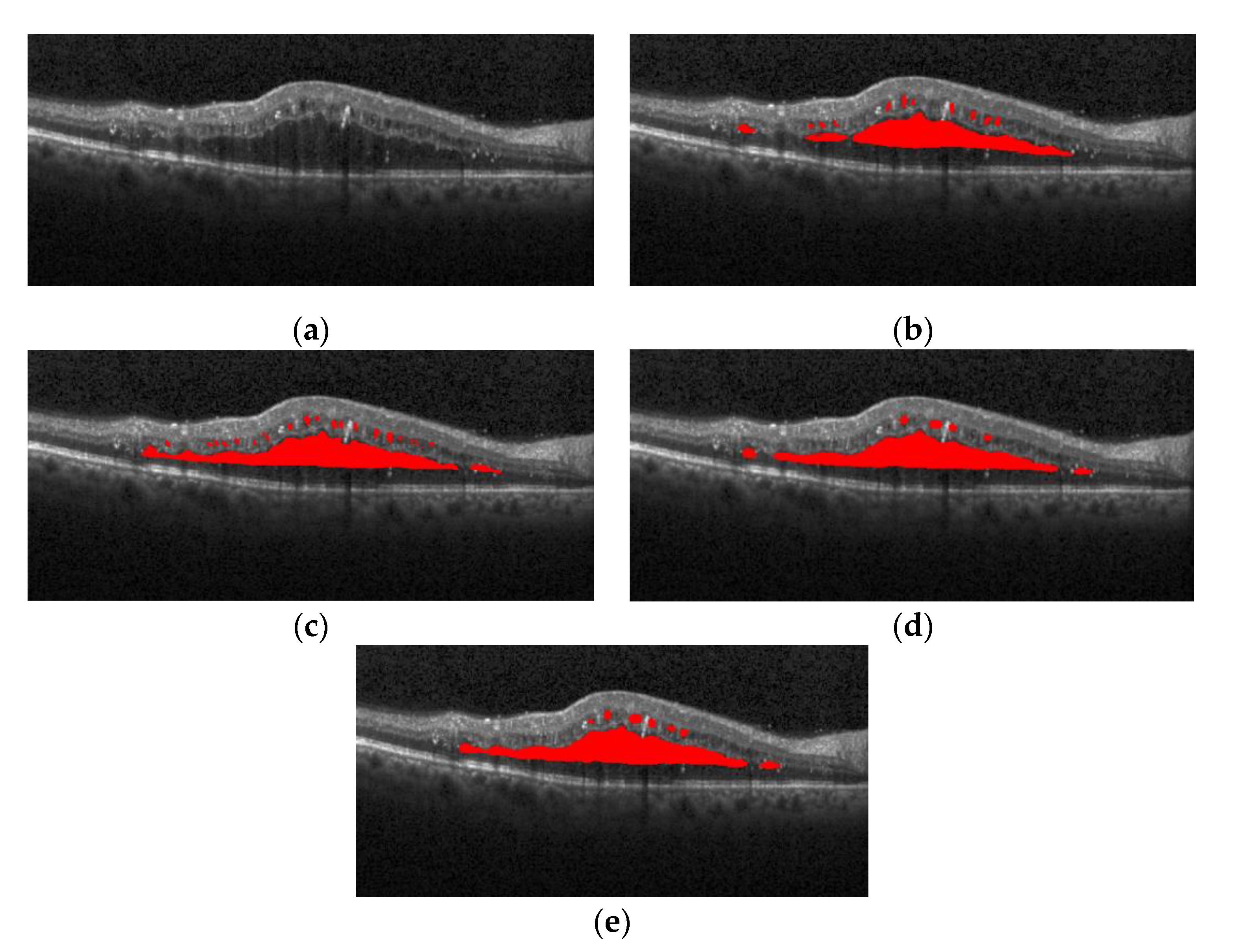
Figure 16 shows the segmentation effect of the proposed method and ReLayNet on the one B-scan image.
Figure 16a is the original OCT image,
Figure 16b corresponds to the 13 annotated edema regions from expert 1,
Figure 16c corresponds to the 11 annotated edema regions from expert 2,
Figure 16d shows the only seven predicted edema regions by the ReLayNet, and
Figure 16e shows the 11 predicted edema regions by the proposed method. Compared with ReLayNet, the proposed method had obvious advantages in the segmentation of multi-regions edema. It could better identify and segment diverse edema regions, and the merged situation of edema regions was reduced.
For diverse edemas in the multi-region, by the above 50 B-scan test images,
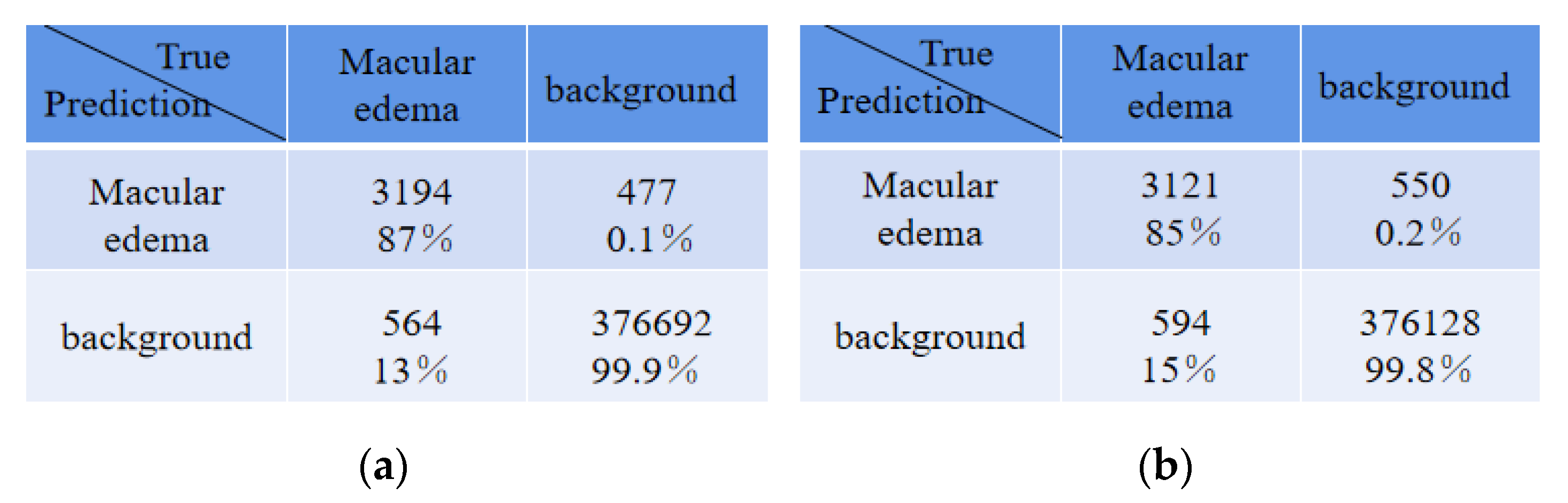
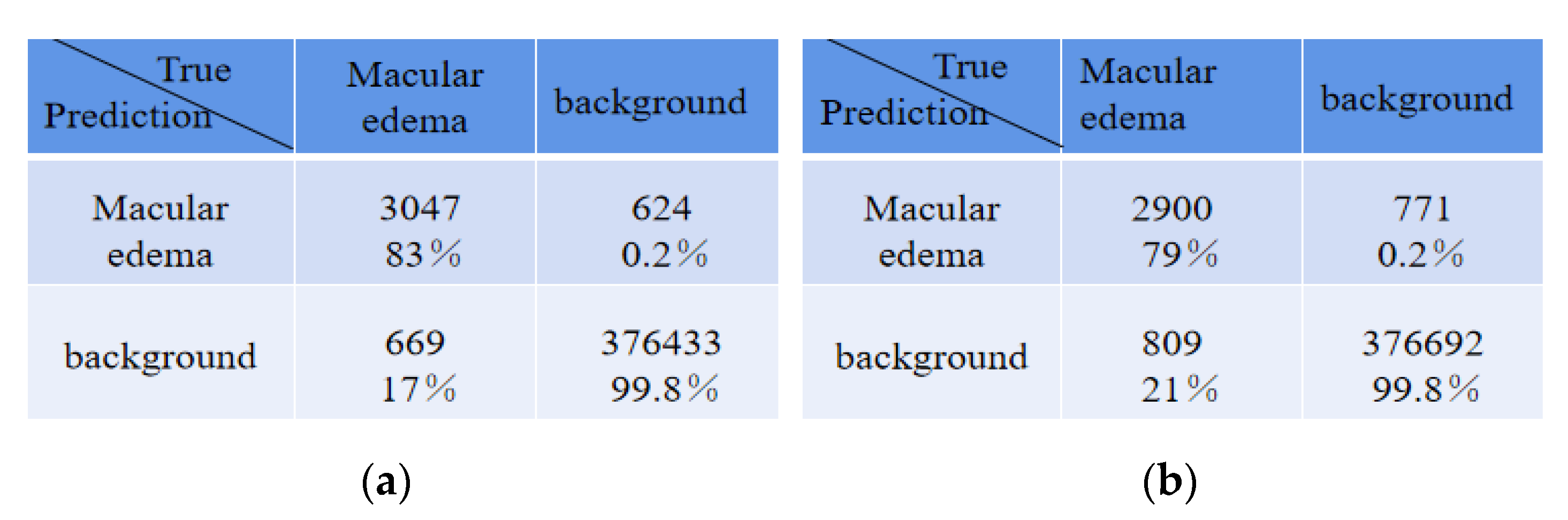
Table 5 shows the average absolute error of the number for edema regions between expert 1 and the expert 2, expert 1 and ReLayNet, and expert 1 and the proposed method. In comparison, the results show that the proposed method achieved the least average absolute error of 2.87 for the number of edemas, and is 1.15 and 2.30 edemas less than the ReLayNet and the expert 2, respectively.
There were some disagreements in the annotations between expert 1 and expert 2 due to their subjectivity, and thus we conducted an experiment that exchanged the two expert roles, namely, the expert 2 annotations were used as the ground truth to train and evaluate the proposed network model. The comparison of quantitative results is shown in
Table 6. Compared with ReLayNet, Dice, Iou, Recall, and precision were improved by 0.02, 0.01, 0.02, and 0.01, respectively, in the proposed method. The experiment demonstrated that the proposed method in this paper has a good generalization ability.
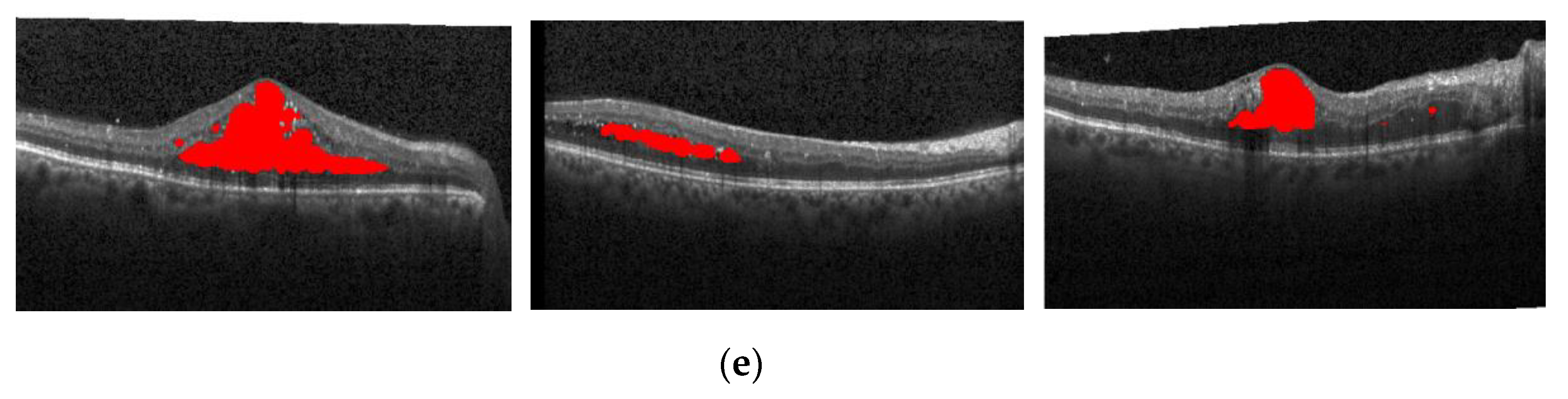
Having the expert 2 annotations as the ground truth,
Figure 17 also shows the segmentation effect of the proposed method and ReLayNet on the one B-scan image.
Figure 17a is the original OCT image,
Figure 17b corresponds to the 14 annotated edema regions from expert 1,
Figure 17c corresponds to the 23 annotated edema regions from expert 2,
Figure 17d shows the only seven predicted edema regions by the ReLayNet, and
Figure 17e shows the eight predicted edema regions by the proposed method. Compared with the ReLayNet, the proposed method can not only effectively predict the pixels of true edema as edema pixels, but also can better identify and segment diverse edema regions, being closer to the annotations from two experts.
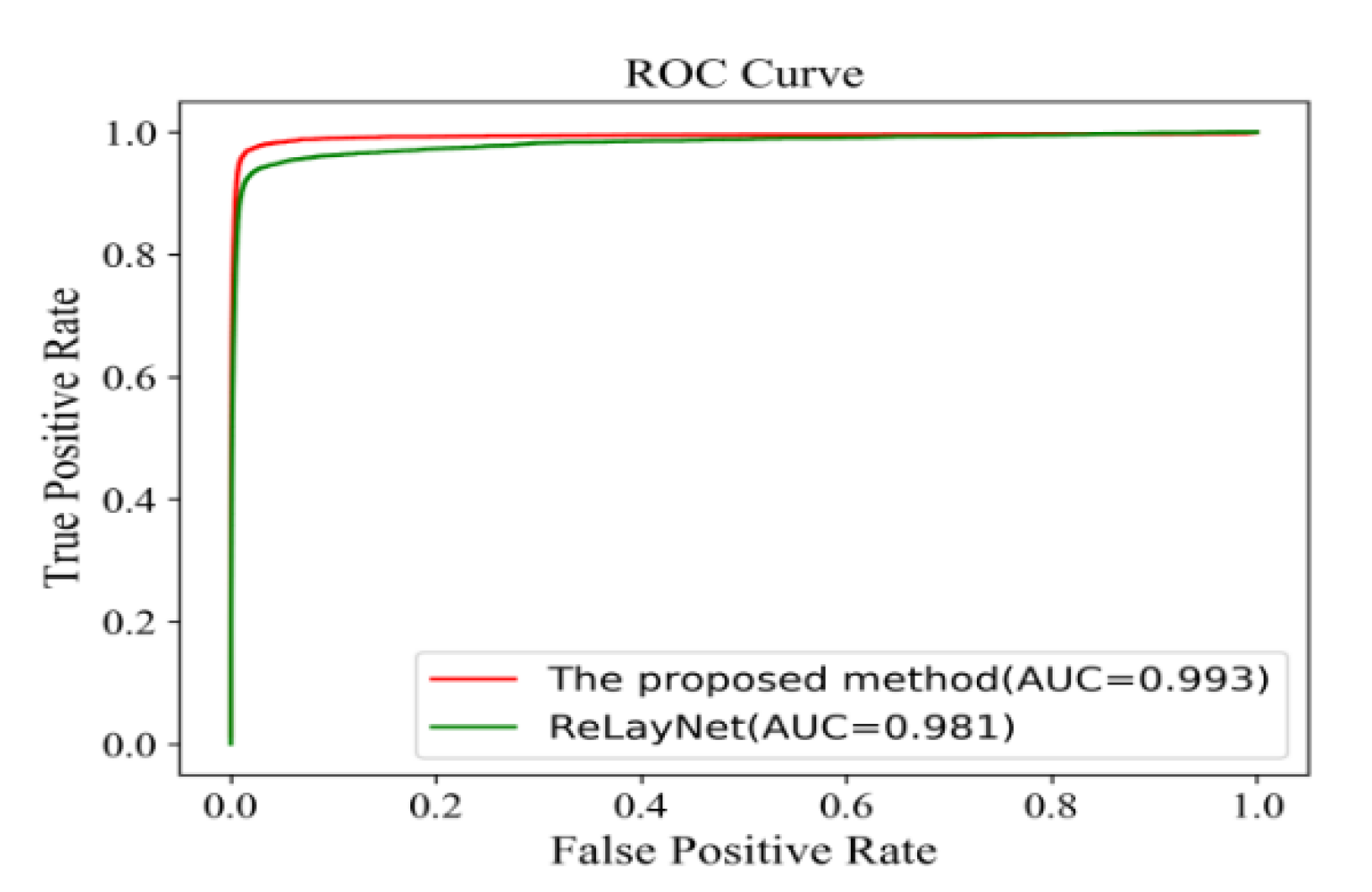
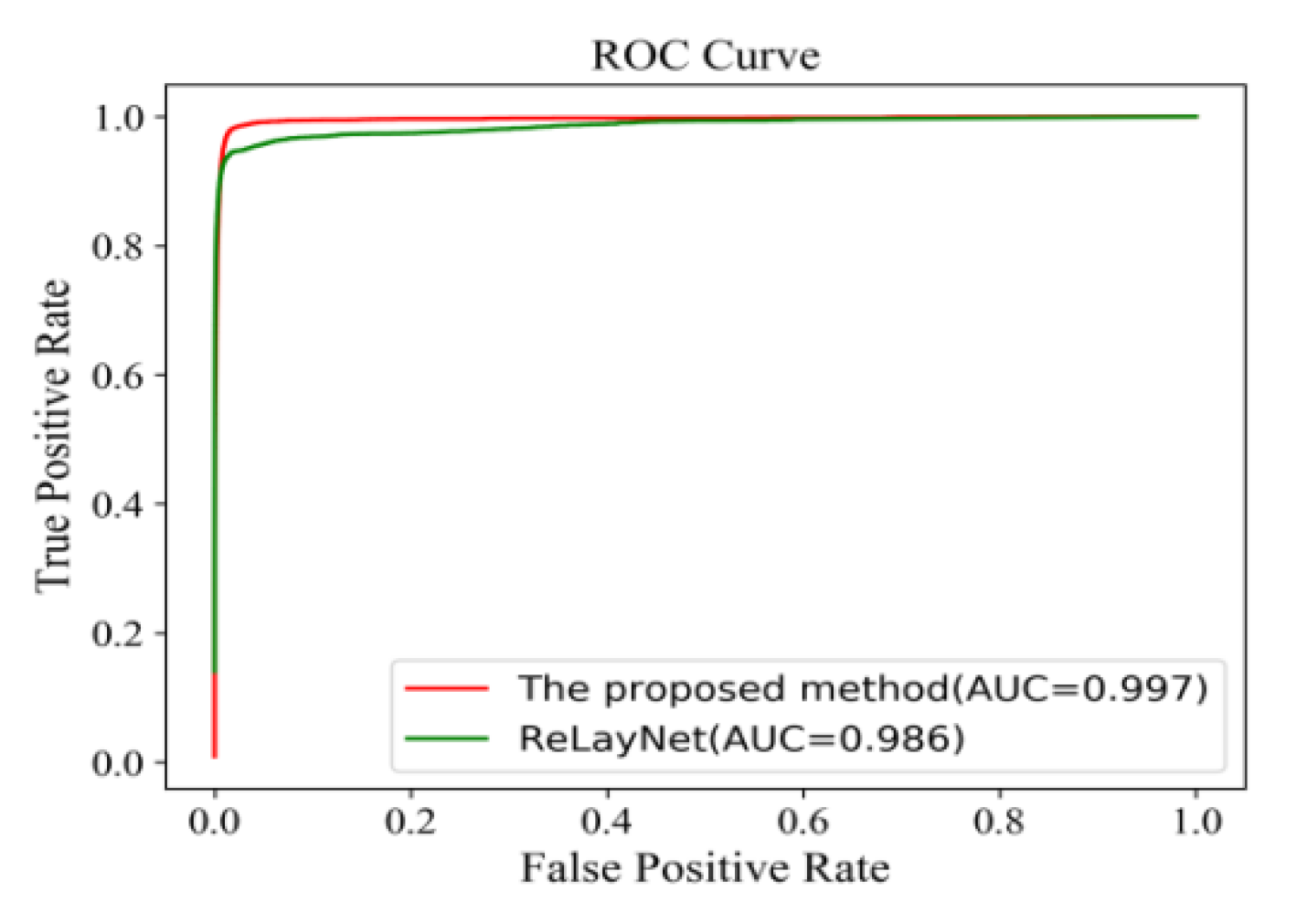
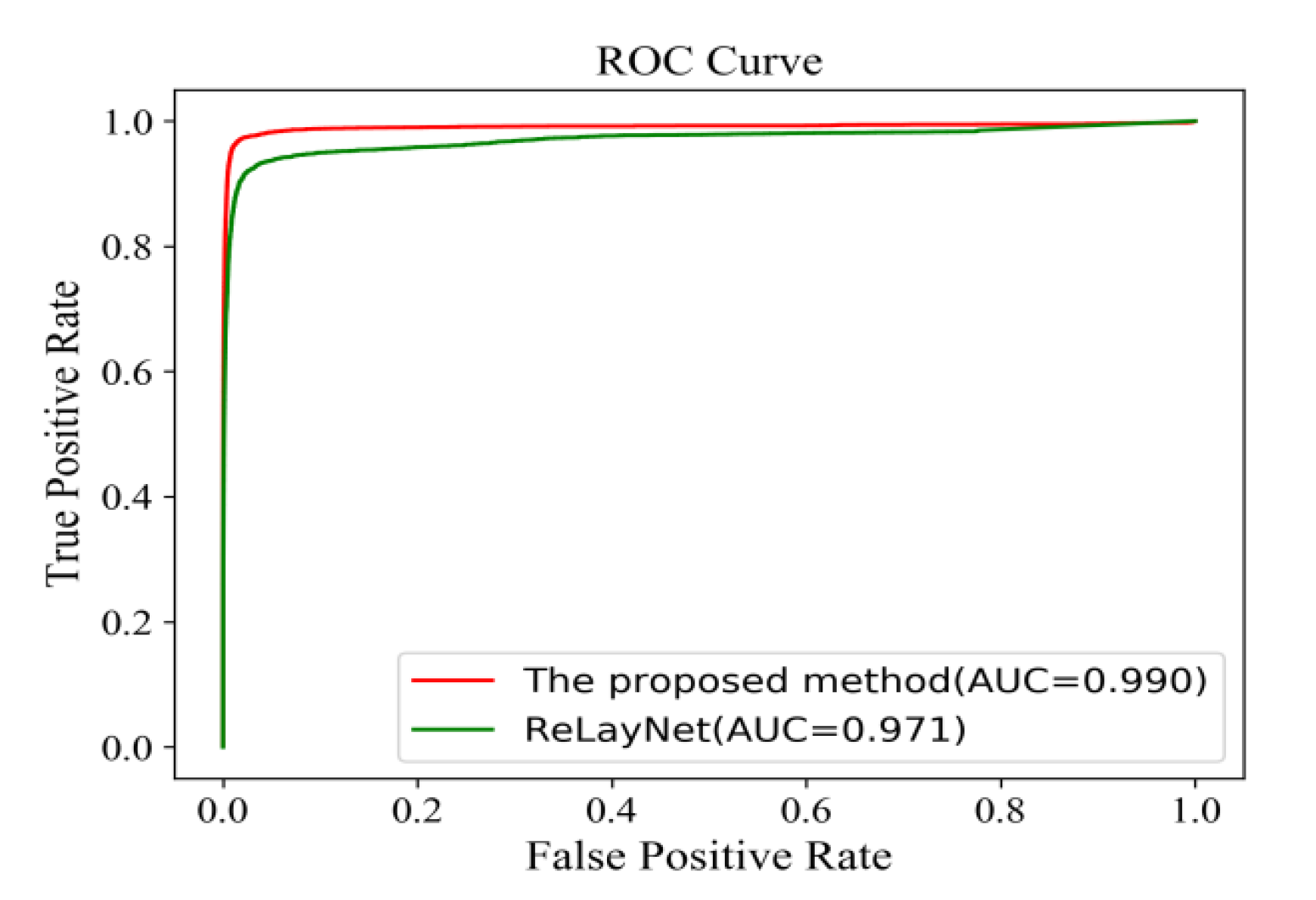
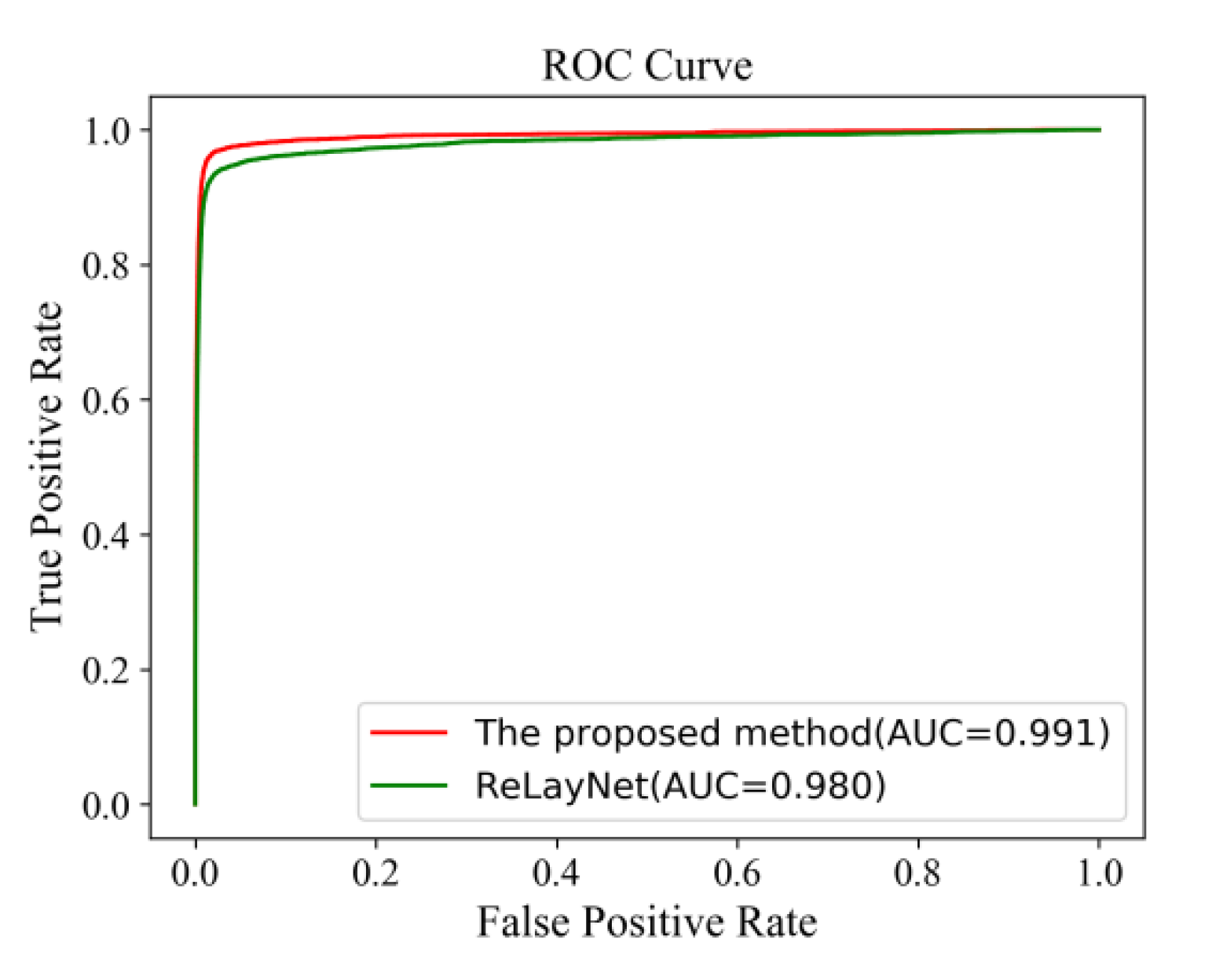
Figure 18 shows the comparison of ROC curves for the expert 2 annotations as a ground truth. The AUC value of 0.991 of the proposed method was 0.011 values higher than ReLayNet at 0.980.
Having expert 2 annotations as the ground truth,
Table 7 illustrates the comparison of quantitative results around fovea edema regions. Compared with ReLayNet, Dice, Iou, Recall, and precision were improved by 0.01, 0.01, 0.01, and 0.02 in the proposed method.
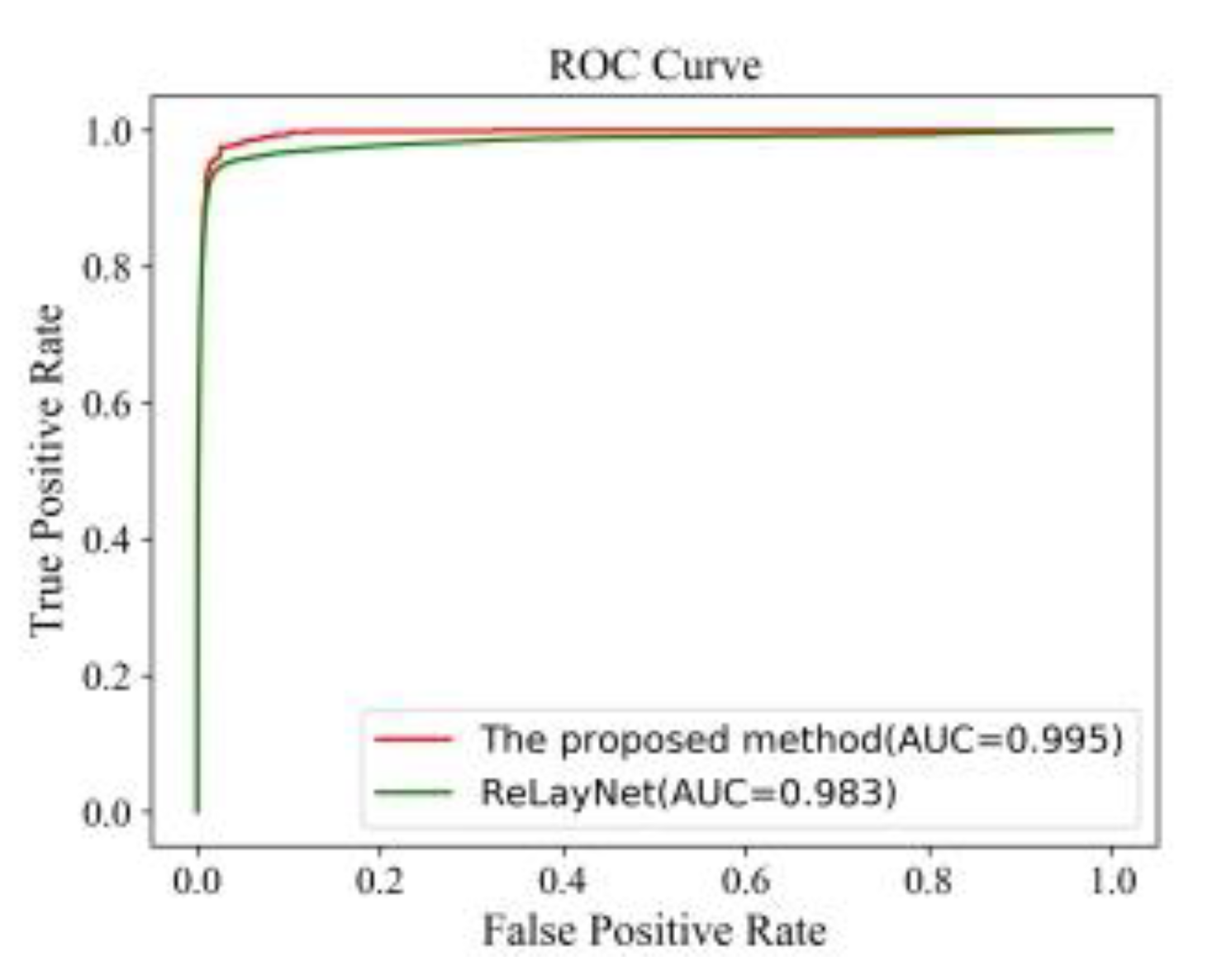
Figure 19 also shows the comparison of ROC curves. The AUC value of 0.995 of the proposed method was 0.012 values higher than the ReLayNet at 0.983.
Having expert 2 annotations as the ground truth,
Table 8 illustrates the comparison results of the diverse edemas in multi-regions. The proposed method also effectively upgraded to 0.77, 0.75, 0.80, and 0.77 in the four indicators, being much better than the ReLayNet method and expert 1 for diverse edemas in multi-regions.
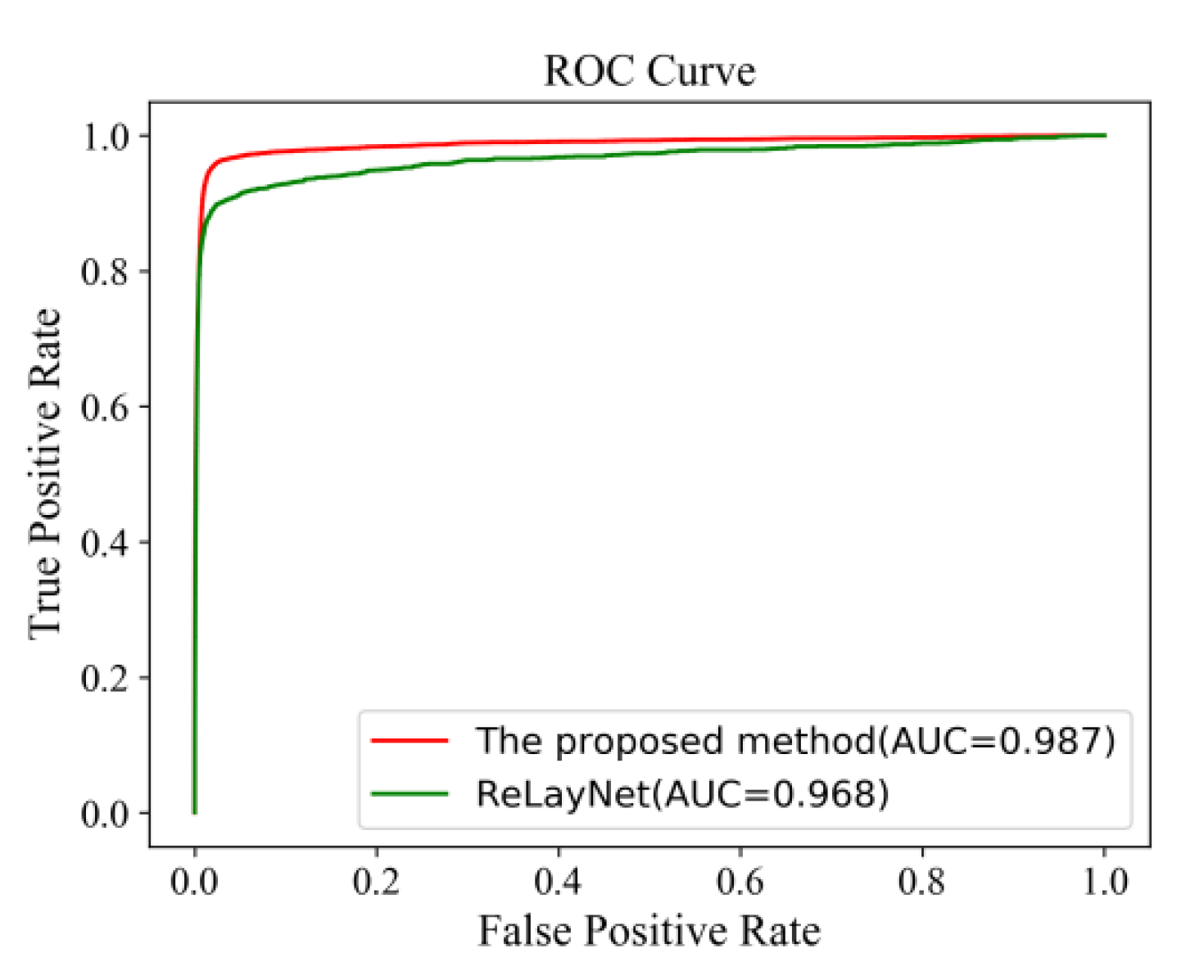
Figure 20 shows the comparison of ROC curves. The AUC value of 0.987 of the proposed method was 0.019 values higher than the ReLayNet at 0.968.