Abstract
Automated medical diagnosis is one of the important machine learning applications in the domain of healthcare. In this regard, most of the approaches primarily focus on optimizing the accuracy of classification models. In this research, we argue that, unlike general-purpose classification problems, medical applications, such as chronic kidney disease (CKD) diagnosis, require special treatment. In the case of CKD, apart from model performance, other factors such as the cost of data acquisition may also be taken into account to enhance the applicability of the automated diagnosis system. In this research, we proposed two techniques for cost-sensitive feature ranking. An ensemble of decision tree models is employed in both the techniques for computing the worth of a feature in the CKD dataset. An automatic threshold selection heuristic is also introduced which is based on the intersection of features’ worth and their accumulated cost. A set of experiments are conducted to evaluate the efficacy of the proposed techniques on both tree-based and non tree-based classification models. The proposed approaches were also evaluated against several comparative techniques. Furthermore, it is demonstrated that the proposed techniques select around 1/4th of the original CKD features while reducing the cost by a factor of 7.42 of the original feature set. Based on the extensive experimentation, it is concluded that the proposed techniques employing feature-cost interaction heuristic tend to select feature subsets that are both useful and cost-effective.
1. Introduction
Chronic kidney disease (CKD) is an ailment that affects the functionality of a kidney in the body. Generally, CKD is divided into multiple stages in which the later stages are denoted as a renal failure when the kidney is unable to perform its functions of blood purification and balancing minerals in the body [1]. In the case of end-stage renal failure, hemodialysis is performed to supplant the kidney function. This intervention provides a temporary solution to the problem. Hence, it is of paramount importance that the CKD is detected at earlier stages where it can be addressed through medication and lifestyle changes [2]. CKD is a highly prevalent disease, according to an estimate one in nine Korean adults suffer from kidney disease [3]. Likewise, around 2.5–11.2% of the adult population in Europe also suffer from it, while around 59% of all the American adult population is at a high risk of developing kidney disease at some point [4,5]. The high incidence and prevalence of CKD are attributed to its late diagnosis, especially in developing countries [6].
In the domain of medical data mining, several intelligent clinical decision support systems are designed which tend to automate the diagnosis process [6,7]. These decision systems employ machine learning techniques that assist physicians in the diagnosis and treatment of CKD in an efficient manner [6,8,9]. Based on a number of important indicators such as blood pressure, albumin levels, blood and urea tests, potassium, and other comorbidities, e.g., diabetes, cardiovascular disease, etc., a patient is comprehensively assessed for CKD and its progression. As the earlier diagnosis of the disease onset can improve the chances of patients to favorably respond to treatment, therefore, most of the automated systems are optimized for enhancing the overall accuracy of the model [8,10]. It is noted by Itani et al. [7] that medical decision systems that solely focus on predictive performance are far from the field reality and hence are not unanimously approved by physicians. In this regard, the interpretability of the classification model is stipulated as one of the important requirements among others for a successful medical decision system [7,10]. Similarly, the cost factor as a practicability concern for medical decision systems recently gained traction in the medical data mining community [6,11,12,13]. Therefore, one of the key research directions pursued by the research community is to design decision systems that are accurate, interpretable, and cost-effective.
In a number of studies performed on CKD diagnosis, decision tree models consistently produced results with high predictive accuracy [8,9,12]. Hence, the main impetus for using tree-based models in an ensemble technique is two-fold. Firstly, tree models are easy to interpret by the domain experts, therefore, in domains such as medical diagnosis, it is desirable to assess the validity of the classification model through visual inspection [7,10,14]. Secondly, tree-models that are based on bagging and boosting techniques tend to produce highly accurate classifiers on small to medium datasets [6,8,9,15]. Hence, tree models are suitable approaches for considering in an ensemble for a CKD dataset, as they can cater to both types of requirements i.e., interpretability and accuracy.
Moreover, feature selection is becoming an essential task in building classification models where the objective is to select a subset of useful features [6,8,15,16,17]. The notion of usefulness is based on the worth of a feature in a dataset in terms of its relevancy and redundancy. There are generally three approaches for feature selection i.e., filter-based approach, wrapper-based approach, and embedded approach [16,18,19]. In the case of filter approaches, the worth of a feature is evaluated through univariate statistical approaches such as Chi-Square, Gini index, information gain, etc. Therefore, feature ranking techniques fall into the filter category. On the other hand, wrapper approaches generally, construct a set of candidate feature subsets that are evaluated on a classifier [19]. Embedded techniques are implicitly used by some of the classifiers, such as decision trees, while constructing a model.
A number of studies demonstrated that ensemble-based feature selection techniques generally perform better than non-ensemble techniques [8,16,17,20,21]. Ensemble feature selection approaches are composed of multiple evaluation functions for quantifying the worth of a feature or a subset of features. In this regard, multiple types of feature evaluation functions can be used such as univariate techniques, classification models, or a set of mixed techniques from the aforementioned categories [8]. Ensembles can be comprised of both homogenous and heterogeneous configurations. In this regard, for a homogenous configuration, a dataset is horizontally partitioned into multiple subsets where a single type of the feature evaluation function is executed on each partition [16,21]. On the other hand, for a heterogeneous configuration, multiple evaluation functions are executed on the dataset in parallel, and later their results are combined [16,17,21].
Similarly, ensemble feature ranking approaches can be arranged in either a homogenous configuration or a heterogeneous manner. In both cases, a global ranked list of features is obtained based on multiple feature lists produced by the individual feature ranking functions. One key challenge in this regard is to select a threshold value which divides the global ranked list into a set of retained and removed features [20].
Most of the studies in the CKD domain assume that the cost of data acquisition is symmetric i.e., having the same cost albeit not necessarily zero; therefore, the cost factor associated with each feature is generally ignored [6,12,13]. However, this assumption may not hold in many real-world medical applications where a patient is required to undergo multiple tests such as urine analysis, electrocardiogram, blood culture, etc., and the tests may vary in terms of incurred cost. Therefore, feature selection methods for CKD diagnosis applications may take into account the cost factor as well.
In Figure 1a,b a scenario is depicted in which features are listed in descending order of their importance. In Figure 1a, a feature merit calculation is performed based on the weightage of a feature and feature’s associated cost. Both these terms are combined using a trade-off constant factor, α, pre-specified by the user. As it can be seen in the implicit case that although different ‘α’ values may have the same overall trend, it is still not clear which value of a threshold to select among a set of candidate values. Furthermore, after feature number 13, the feature ranking is not consistent with different ‘α’ values. Therefore, the implicit case is not only sensitive to the pre-specified ‘α’, but it also makes it subjective to select a threshold value to retain a set of features. On the other hand, in Figure 1b the blue line denotes feature weightage (FW) while the orange line represents an accumulated cost of selecting a set of features. In the explicit case, a set of features can be easily distinguished at the point of intersection which is comparatively cost-effective and useful than those which are below the intersection point. Hence, in this study, the main question under investigation is this that can the point of feature-cost intersection be used for selecting a subset of features that are both accurate and cost-effective in the CKD diagnosis problem?
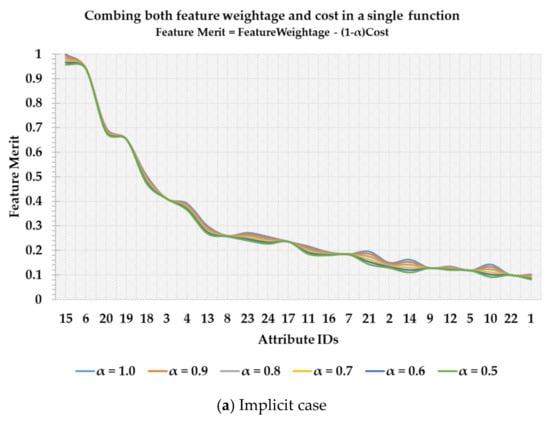
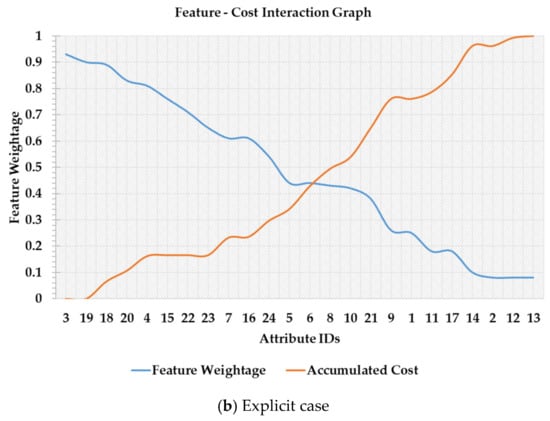
Figure 1.
Cost-sensitive feature ranking and threshold selection.
Recent studies reported significant scholarly work on developing chronic kidney disease diagnosis and management systems [6,8,9,11,15,18,22,23,24,25,26,27,28]. In this regard, this study is continuation of research performed on the CKD diagnosis problem. The study addresses the problem of cost-sensitive feature selection for building decision tree models for the CKD diagnosis problem. The main objective of the study is to demonstrate that economic considerations can be effectively taken into account along with retaining the overall performance of the CKD diagnosis systems.
The proposed approaches are based on ensemble ranking techniques with a cost-sensitive threshold selection. The proposed heuristic rule for threshold selection takes into account both worth of a set of features and the overall incurred cost concerning the selected features. To the best of our knowledge, it is the first study that addressed the notion of data acquisition cost within the framework of cost-sensitive ensemble feature ranking. We proposed two ensemble ranking techniques that use multiple decision tree-based classifiers as heterogeneous scoring functions. A schematic diagram for an ensemble feature ranker is shown in Figure 2. The proposed techniques differ in terms of the application of threshold operation. Ensemble-1 combines all the scores and thereafter automatically selects a threshold value whereas ensemble-2 applies a threshold to individual ranks, and subsequently a set of feature subsets are generated which are later combined into a consolidated feature subset. The multiple feature subsets are combined using the majority voting scheme which is also adopted by several studies [8,17,29].
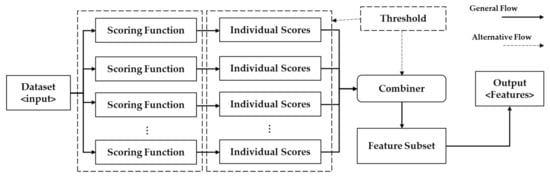
Figure 2.
Schematic diagram of an ensemble feature ranker.
The major contributions of this study are as follows:
- We propose an automatic cost-sensitive threshold selection heuristic that takes into account both the worth of a set of features and the overall accumulated cost.
- Decision tree-based classifiers performed favorably on the resulting features on a benchmark CKD dataset.
- The features could also yield higher average accuracy with more sophisticated classification structures, which shows that these features present high generalizability.
The rest of the paper is organized as follows: Section 2 deals with the literature review on the subject. The proposed methodology is discussed in Section 3 in which the proposed ensemble techniques are elaborated. Section 4 deals with the experimentation and the case study results, in which we provide a detailed treatment of both the proposed approaches along with their comparison with other related techniques. The conclusion of the study is provided in Section 5, along with a set of future directions for extending this research.
2. Literature Review
A number of studies shown that feature selection improves the generalization capabilities of the classification models [6,8,9,16,18,20]. Feature selection is similar to dimensionality reduction whereas the objective of the former is to retain the semantics present in the original dataset while the latter transforms the data in such a manner as the overall dimensions of the data are reduced [21]. Feature selection techniques are generally grouped into three broad categories, i.e., filter-techniques, wrapper-techniques, and embedded-techniques, as mentioned in the preceding section. Filter techniques score features based on the general characteristics of the dataset [18]. In this regard, most of the filter approaches are based on evaluating the correlation between features and the class label. Hence, features having a high correlation with the target concept are regarded as useful features. Feature ranking approaches are generally based on filter methods, but ranking can also be produced by employing a classification model which in turn evaluates a subset of features [16]. Wrapper approaches involve classification algorithms in the process of evaluating a subset of features. In this regard, a feature subset generation step is followed by an evaluation step [19]. The main objective of the wrapper approach is to find a subset of features that are neither irrelevant nor redundant. Filter methods are generally employed when the number of features is very large as these methods are computationally fast and do not get bogged down in a pairwise comparison of the candidate feature sets. Wrapper methods generally produce results that are relatively more optimized and accurate than that of the filter methods whereas the latter produces the result in relatively less time [18]. Embedded methods select a subset of features as an integral part of the process of building a classifier such as a decision tree algorithm that selects the most appropriate feature as it grows the tree [30]. A high-level summary (adapted from [31]) regarding the merits and demerits of feature selection techniques is given in Table 1.

Table 1.
Comparison among different feature subset selection techniques.
This research focuses on the application feature selection and classification for the CKD diagnosis problem. In this section, we discuss some of the representative works in which feature selection techniques are used for the CKD diagnosis. Salekin and Stankovic [11] proposed a wrapper based feature selection approach which reduces the overfitting of the Random Forest (RF) classification model on the CKD dataset. The reported resultant F1-measure of the model on top 5 features is 99.80%. Furthermore, the authors also reported promising results in terms of reduced cost. Chen et al. employed three models, i.e., K-Nearest Neighbor (KNN), Support Vector Machine (SVM), and Soft Independent Modeling of Class Analogy for decision modeling of the CKD patients. The reported approach achieved an accuracy of 93%. It is also reported that SVM was more robust in dealing with noisy data as compared to other models and hence achieved an accuracy of 99% [23]. Serpen [24] used the C4.5 decision tree model on the CKD dataset. The resultant tree model produced 8 production rules of the form IF <condition> THEN <conclusion> and achieved an accuracy of 98.25%, whereas, Al-Taee et al. [26] reported lower accuracy on the same dataset. Furthermore, the authors also identified 5 salient features in their study. In another study, the same framework as reported in [26] is used, in which they used three classifiers on a CKD dataset which was acquired from Prince Hamza Hospital, Jordan. The study reported that the decision tree model performed reasonably well on a number of performance metrics [27]. Tazin et al. [27] used several classification models such as SVM, Naive Bayes, KNN, and decision tree on the CKD dataset. Subsequently, a feature ranking is generated from which the top 10 features were selected. It is reported that the decision tree algorithm produced a model yielding accuracy of 99.75%. Polat et al. [18] proposed a feature selection technique for SVM based classification model. The authors used a hybrid feature selection by leveraging both filter and wrapper methods. They reported an accuracy rate of 98.50% on SVM using ‘Best First’ search technique using 11 attributes. Likewise, Ogunleye and Wang [8] selected the top 13 features for feature selection based on an ensemble of feature selection techniques. Afterward, the authors performed classification using an optimized RF classifier for the CKD dataset. The reported accuracy of 100% was over the reduced CKD dataset. In Ref. [22], the authors experimented with SVM and Artificial Neural Network (ANN) on the CKD dataset. They reported that ANN produced a comparatively higher accuracy model as that of SVM. All the experiments are performed on the top 12 features. Apart from feature selection, the data discretization process is reported to have a favorable effect on the decision tree-based model construction [28]. Qin et al. [15] experimented with several different data imputation configurations on a set of multiple classifiers. They reported that RF achieved the highest accuracy of 99.75% for the CKD diagnosis, while logistic regression (LG) was able to produce an accuracy of 98.95%. Afterward, the authors proposed an integrated model that employed both the aforementioned classifiers along with the perceptron and subsequently produced an accuracy of 99.83% using the integrated approach. Sobrindo et al. [6] performed a comprehensive study on CKD diagnosis using various machine learning algorithms. The authors reported the highest accuracy achieved by decision tree-based models in the pool of candidate models which included Naïve Bayes (NB), SVM, ANN, KNN.
We provided a general overview of the feature selection techniques and classification algorithms applied to CKD diagnosis and it can be observed that decision tree-based models are one of the popular modeling approaches for the CKD diagnosis. Our proposed approach is based on feature ranking therefore we herein mention a few studies which addressed the problem of automatically selecting an appropriate threshold value using heuristics. Most of the studies opt for a fixed threshold value for retaining a set of top features [32,33,34]. But as it is observed that a fixed threshold value may over-select or under-select an appropriate number of features [20,29,35,36]. Authors in [37] used data complexity measures for selecting a threshold value while authors in [38] used a minimum union method to combine multiple rankings and produced promising results on high dimensional datasets [39]. Tsai and Hsiao [17] performed a detailed study regarding combining multiple feature selection methods for stock prediction problem. Authors reported higher predictive accuracy over the ANN classifier based on the multi-point interaction among Principal Component Analysis (PCA), Genetic Algorithm (GA), and Decision Tree (DT) feature sets, among other combination strategies. Osanaiye et al. [40] proposed an ensemble feature selection technique in which the authors combine partial results from multiple filter measures. Subsequently, 1/3rd of features from each feature selection method are retained. Consolidated feature subset is obtained through the intersection of the candidate feature subsets.
Several threshold selection approaches are reported in the literature that are primarily geared towards optimizing the accuracy of the classification task. But the aforementioned approaches cannot take into account valuable meta-information associated with the feature set, such as cost of data acquisition. It is important to note that the cost-free feature selection approaches may also result in cost reduction but such a cost reduction would be effectively unintentional since the cost-free feature selection methods are oblivious to the cost meta-data associated with the features. Our proposed approaches are based on explicitly taking into account the cost factor along with the feature worth. In this regard, we show that an automatic threshold value can be selected based on the feature-cost interaction curve, as shown in Figure 1b, which results in a feature set that is both useful and cost-effective. In the following section, we elaborate on the proposed methodology for computing feature weightage along with the application of a threshold value in arriving at a final feature set.
3. Proposed Methodology
In this section, we elaborate on the underpinnings of our proposed approach for cost-sensitive feature ranking. In most of the feature ranking techniques, a feature weightage is produced which in turn is used for the feature ranking. Afterward, a threshold value is used to filter-out undesirable features while the retained features are fed to a data classification model. One of the major challenges in this regard is to find an appropriate threshold value as shown in Figure 1a. Furthermore, it is also important to select the feature scoring function which is not biased towards any particular data characteristics, e.g., information gain tends to favor attributes that take on a large number of distinct values [41]. Filter-based feature weighting measures such as the Gini index do not account for feature interaction and hence the measure may not be comprehensive enough to capture the complementary feature interaction i.e., a set of features that may not be highly relevant but when considered collectively, they enhance the overall model’s performance. In this regard, the feature ranking approach depends on the comprehensiveness of the weighting function. Therefore, in this research, we use three decision tree-based classification models, Classification and Regression Tree (CART), RF and, Gradient Boosting Trees (GBT), which evaluate both the relevance and redundancy of a feature set. It is important to note that the model-based scoring functions can be executed in parallel, therefore the running time of the ensemble is proportional to the running time of its slowest classifier. In this regard, using concurrent processes we can execute the classifiers and afterward combine their results. The main objective of the feature weightage step is to score features based on their importance as well as their interaction with other features in the dataset. Once a reliable feature score is obtained, based on their weightage, features are ranked in descending order of their importance. Subsequently, an averaging operation is performed on the obtained feature weightage and a final feature score is generated.
In this research, we propose two approaches for combining features i.e., ensemble-1 is based on combining individual scores, whereas ensemble-2 generates three partial solutions in the form of three subsets obtained from the scoring functions. Afterward, the partial solutions are combined using the majority vote. In ensemble-1, a threshold value based on the feature-cost intersection is selected after combining multiple lists, whereas, in ensemble-2, the threshold is applied on an individual list which is obtained from the scoring function and a resultant feature subset is generated. Each feature subset is treated as a partial solution. A final solution is obtained by applying the majority vote scheme to the partial solutions. We normalized the cost values; hence, the overall cost of the entire feature set adds-up to 1. The proposed ensemble-1 is depicted in Figure 3, while Figure 4 shows ensemble-2.
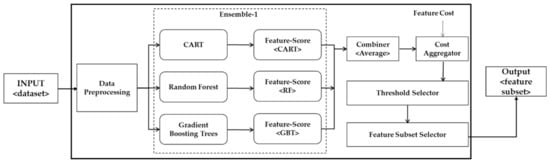
Figure 3.
The architecture of the proposed ensemble-1.
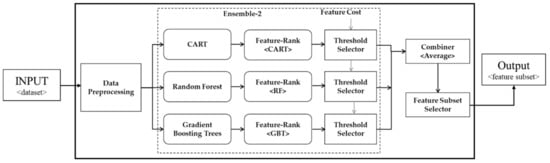
Figure 4.
The architecture for the proposed ensemble-2.
3.1. Data Preprocessing
Data preprocessing is a pre-requisite task in a number of data-driven applications. The main objective of this operation is to ensure that the data are of high quality before commencing the model construction process. The major operations performed by this component are missing values imputation and feature id-ness detection and removal. For this study, we used a k-nearest neighbor to impute missing values, where k = 3. This technique selects a set of records from the dataset which are similar to the missing value record and subsequently imputes the missing value based on the local information of the selected instances. Furthermore, we used Euclidean distance for computing the similarity of numerical attributes. Numerical values are replaced with taking the mean value of the selected attribute while mode value is used for nominal features. The chronic kidney disease dataset, discussed in the subsequent section, contains 242 records with missing values while the complete dataset is composed of 400 records. Therefore, around 60% of the records contained one or more missing values.
3.2. Classifier-Ensemble
The proposed approach uses decision tree-based classifiers for feature scoring. Following is a brief description of the classification models used in the ensemble:
Classification and Regression Tree: It is one of the popular decision tree induction algorithms. CART creates strictly binary decision trees by employing recursive partitioning to build the model [42]. CART selects a subset of features from the complete feature set for building a decision tree model. The choice of feature selection is based on the quantification of a feature’s worth in generating a homogenous data subset. Generally, the Gini index is used to calculate the importance of a feature at a specific level in the tree whereas information gain is also one of the popular choices for feature quantification. The main objective of the CART algorithm is to construct the model which can separate the data into homogenous subsets with respect to the class label. We used the Gini index to calculate the impurity of a feature subset. Feature weightage is obtained from CART by taking into account the effect of adding a feature to the decision tree and the subsequent decrease in the impurity in the model.
Random Forest: It is based on a decision tree model where a set of decision trees are generated. In this case, each tree is incrementally improved by partitioning the dataset into homogenous subsets [12]. RF creates a set of pre-defined decision trees where each tree is constructed from a bootstrapped dataset. RF generally performs well for small to medium-sized datasets and the resulting model is robust against overfitting, feature interaction, and spurious data patterns [43]. The key approach of this algorithm is to create a set of bootstrapped datasets through which a set of pre-defined randomly selected features are selected and a tree model is created. The standard RF model is a combination of binary decision trees. Unlike CART, RF is an ensemble tree model where multiple decision trees are created for introducing diversity in the overall model. Bagging is applied to the input dataset through which several data subsets are created. Generally, trees are constructed based on randomly-selected features but other feature selection schemes can also be used [12]. Finally, all the generated trees are integrated through a majority voting scheme for the classification problem. Although the resulting model is not fully interpretable as that of CART, the individual decision trees can be extracted. Similarly, feature weightage is computed by taking into account the average decrease in impurity over all the trees in an ensemble.
Gradient Boosted Trees: It is an ensemble modeling technique for either regression or classification problems. It is based on a forward-learning approach in which predictive results are incrementally improved through introducing weak models. A weak model is one that is slightly better than a random guess. GBT is based on a non-linear regression procedure that improves the accuracy of decision trees. Weak classification tree models are applied to different distributions of the dataset in a sequential manner where misclassified data points are given higher weightage. As weak models are added to the integrated model the error recorded through the loss function minimizes by applying the gradient descent approach. In the case of GBT, the feature weightage can be obtained by the sum of improvements for a given attribute at a node.
3.3. Combiner
It plays an important role in the overall proposed architecture.
- Ensemble-1 produces multiple feature weightages obtained from individual feature scoring functions. In this case, the task of combiner is to consolidate the individual feature weightages into a consolidated score. The final scores are obtained by taking the average across multiple scoring functions as shown in Equation (1):
The final score of a given feature is the average weightage across three independent scoring functions σ applied on a feature , where N is the total number of functions in the ensemble. IL is an intermediate object that contains feature weightage and the associated information for each scoring function. A feature list is denoted by an object L that stores a feature’s score, its ID and the accumulated cost for a given feature f. *L represents the sorted list based on features’ score. Please note that the scoring values are scaled between 0 and 1 before applying Equation (1).
- Ensemble-2 deals with multiple partial solutions in the form of feature subsets. In this case, each scoring function produces an independent ranked list. A threshold operation is applied to each list. Subsequently, three different subsets are produced. All the subsets are combined by taking the majority voting scheme. In our case, as the ensemble is comprised of 3 scoring functions, therefore the majority voting is effectively translated into the selection of a feature that is present into at least 2/3rd of the subsets.
Table 2 provides summary of the functions used in both Algorithms 1 and 2.
| Algorithm 1 Ensemble-1 |
| Input: Dataset D, Cost vector Cv, List of scoring functions N Output: Selected feature set S 1: Begin 2: foreach m in N do: 3: IL[m] = score_function(m, D) //N=3 i.e., DT, RF, GBT 4: endfor 5: L = average (IL) // using Equation (1) 6: *L = sort(L) // *L, is sorted in descending order 7: foreach f in *L do: 8: Acc_Cost (f) = Cv(f) //using Equation (2), cost accumulation based on all the elements in *L up to ‘f’ 9: *L.cost[f] = Acc_Cost (f) //accumulated cost assignment 10: endfor 11: T = intersection (*L) // where *L.score < *L.cost 12: S = retained (*L, T) // retained features in *L after applying T 13: return S 14: End |
| Algorithm 2 Ensemble-2 |
| Input: Dataset D, Cost vector Cv, List of scoring functions N Output: Selected feature set S 1: Begin 2: foreach m in N do: 3: L[m] = score_function(m, D) //same as Algorithm 1 4: *L[m] = sort(L[m]) //separate list for each scoring function ‘m’ 5: foreach f in *L[m] do: 6: Acc_Cost(f) = Cv(f) //same as Algorithm 1 7: *L[m].cost = Acc_Cost (f) //accumulated cost assignment for ‘m’ 8: endfor 9: T = intersection (*L) //same as Algorithm 1 10: Sm = retained (*L, T) //feature subset is obtained for ‘m’ 11: endfor 12: S = combine( //using majority vote scheme 13: return S 14: End |

Table 2.
Summary table for elaborating the proposed methods.
3.4. Feature Cost Aggregator
Feature cost aggregator is used for selecting a threshold value over a feature weightage curve. Both ensemble-1 and ensemble-2 approaches use a threshold to select a subset of features. Features are arranged in the descending order of their combined score. The individual cost of a feature is retrieved and accumulated in a top-down manner as given in Equation (2).
where i = 1, 2, 3, …, m. And denotes a cost vector; therefore, .
In this study, values of the cost feature are normalized between 0 and 1. Please note that, although the feature score curve is calculated only once, whereas, different cost factors associated with a feature may generate different accumulated cost (Cscore) curves. We investigated the economic cost perspective, whereas other cost factors such as data’s availability, risk, or the computational cost may also be taken into account.
3.5. Threshold and Feature Subset Selector
The purpose of the threshold value is to select a subset of features from the given feature list after incorporating the cost value. Ensemble-1 produces a feature list based on average scores. In this regard, we can find a point of intersection between feature weightage and the corresponding accumulated cost values. The point of intersection between ‘FW’ and accumulated cost score ‘Cscore’ can be found where . A sample graph based on the feature-cost intersection curve is depicted in Figure 1b. A threshold value is automatically selected based on the point of intersection, e.g., the point of intersection is at feature number 6 in Figure 1b. Hence, all the features starting from feature number 3 leading up to feature number 6 would be retained while the rest of the features would be discarded. The assumption taken in this regard is that the features over the interaction point are reasonably useful and cost-effective, whereas, the accumulated cost of features below the intersection point out weigh their importance.
In the case of ensemble-2, we consolidate individual feature subsets by accounting for the occurrence of a feature in multiple subsets and taking a majority vote among the partial solutions. For example, we have features such as α, β, and γ, placed at arbitrary positions in three separate subsets produced by three scoring functions, , , and . Then we compute the frequency of these features e.g., <α:3>, <β:2>, and <γ:1>. According to the aforementioned selection strategy, we select both α and β features based on our majority voting heuristic, i.e., 2/3rd of feature frequency, while discard γ and all other features which are having a frequency as that of γ or lower. The intuition between the second approach is that, if a feature appears more frequently in multiple subsets, then it is less likely due to any spurious patterns or any particular bias of the scoring function. Table 3 shows a sample scenario for the ensemble-2 approach. In this case, we have three different subsets. We generate an integrated subset by taking into account the frequency of a particular feature regardless of its position in the subset. The highest score of a feature is determined by the number of scoring functions in the ensemble. As we have three classifiers, therefore, the highest score a feature may get is 3. In Table 3, the selected features are denoted with boldface letters while the remaining features are discarded.

Table 3.
Ensemble-2 frequency-based feature ranking.
It is important to note that the subsets generated by scoring functions may not be of equal size. Since for each scoring function e.g., CART, features’ weightage is obtained and then subsequently based on the intersection of the FW and the accumulated cost i.e., Cscore, similar to the ensemble-1 approach, a subset of features is selected for each function. More details regarding this step are presented in Section 4.
4. Experimentation
This section deals with the experimentation details of the study. In this regard, a brief description of the dataset is provided along with a summarized analysis of the quality of the dataset. Afterward, we elaborate on the performance metrics used in this study and the interpretation of the results. Furthermore, we carried out two sets of experiments. Experiment 1 deals with evaluating the efficacy of ensemble-1 with that of the baseline models, while ensemble-2 is compared with baseline models in experiment 2. Once we establish the performance of both the proposed approaches, we then compare them with other similar methods mentioned in Section 2, over several performance metrics and incurred cost.
To demonstrate the efficacy and applicability of our proposed approach, we used a benchmark dataset from the University of California (UCI, Irvine, CA, USA) online repository [44]. The chronic kidney disease (CKD) is a real-world dataset acquired over a period of two months by Apollo Hospitals, Tamilnadu, India.
4.1. Dataset Description
The CKD dataset is composed of 400 instances where each instance is comprised of 24 attributes excluding the class attribute. There are 13 categorical attributes while 11 of the attributes have numerical values. The dataset is used to model a dichotomous decision variable i.e., 1 represents a given patient is diagnosed with the disease, while −1 denotes otherwise. The overall dataset contains 250 CKD patients while the rest of the patients have a non-CKD diagnosis. The acquired data are preprocessed to impute missing values and ID attributes. Table 4 provides a summary of the CKD dataset, along with the economic cost of acquiring data for a particular feature. The cost factor associated with each attribute is adapted from the work of Salekin and Standkovic [11].

Table 4.
Chronic Kidney Disease (CKD) dataset description.
The attribute importance in terms of its correlation with the class variable is shown in Figure 5. As it can be seen that several features have a high correlation with the target concept. The nature of correlated features and their treatment are generally domain-dependent and therefore, a decision-maker is generally involved to decide on either retaining or removing highly correlated features. In the absence of a domain expert, features having higher correlation are generally preferred over lower correlation features but in the case where the availability of such features is not certain at the time of decision making then it is recommended to remove such highly correlated features.
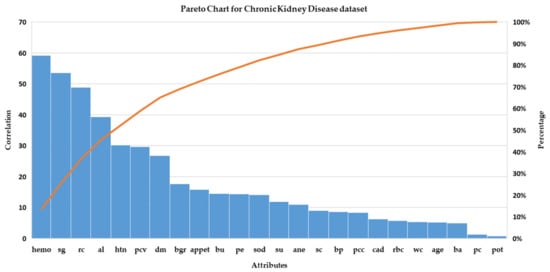
Figure 5.
Pareto-chart of features in the Chronic Kidney Disease (CKD) dataset.
4.2. Experimental Setup
In this study, we used seven classification algorithms for evaluating the performance of the proposed approaches. The selected classifiers are comprised of decision tree-based models as well as other well-known classification algorithms. Moreover, extensive experimentation is also performed in which a number of relevant techniques are compared with that of the proposed approaches. All experiments are performed on a system having processor AMD Ryzen 3 2200 G with 8 GB RAM and 64-bit Windows 10 Enterprise Edition. In this study, we used the RapidMiner Studio 9.6 version [45] for simulating the proposed approaches. Workflows related to generating feature weightage for different feature scoring functions are hosted at MyExperiment.org, which is a collaborative repository to share workflows and other associated files [46,47,48].
Table 5 shows the parameters selected against each classification model.

Table 5.
Classification models parameters.
To evaluate the efficacy of the proposed approaches, we used several evaluation metrics such as accuracy, precision, recall also known as sensitivity, specificity, F1-measure, and Area under Receiver Operating Characteristics Curve (AUC). The evaluation metrics are computed through the confusion matrix such as:
- True Positive (TP): denotes positive instances predicted as positive.
- True Negative (TN): denotes negative instances predicted as negative.
- False Positive (FP): denotes negative instances predicted as positive.
- False Negative (FN): denotes positive instances predicted as negative.
Based on the aforementioned definitions, the quality metrics of interest are calculated as follows:
The evaluation results are reported on 5 fold cross-validation in which the original dataset is horizontally partitioned into 5 partitions. In each iteration, four of the partitions are used for scoring features and obtaining a final feature subset. The remaining fifth partition is used for building classification models. In this manner, the reported result values for each classification model are averaged over different testing partitions.
5. Results and Analysis
5.1. Baseline Results
In this section, we report the results of the baseline models over the full CKD dataset without any feature selection, as shown in Table 6.

Table 6.
Evaluation results for baseline models.
5.2. Feature Weightage Calculation and Feature Subset Acquisition
As mentioned earlier, both ensemble-1 and ensemble-2 approaches require a robust feature scoring function. In this regard, three decision tree-based classifiers are used as the scoring functions to obtain a consolidated feature score. In the following section, we elaborate on the feature scores obtained from different functions along with the feature subsets selected. Please note that ensemble-1 combines individual weightage obtained from multiple scoring functions and afterward selects a threshold, whereas ensemble-2 is based on an eager approach in which threshold is applied to individual scoring functions and a set of feature subsets are obtained which are afterward combined into a consolidated feature set.
- Decision Tree Score: Features are scored through the CART decision tree classifier. The blue line in Figure 6 shows the feature weightage (FW) in the decreasing order of their importance while the orange line denotes accumulated cost (Cscore). Both values are normalized. The point of intersection between FW and Cscore is found around feature number 5 as shown in Figure 6.
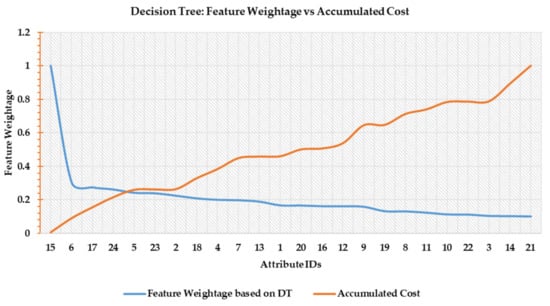 Figure 6. Decision tree-based feature scoring.
Figure 6. Decision tree-based feature scoring. - Random Forest Score: The second scoring function is based on random forest. The blue line in Figure 7 shows the feature weightage (FW) while the orange line denotes accumulated cost (Cscore). Both the values are normalized. The point of intersection between FW and Cscore can be observed around feature number 9 as shown in Figure 7.
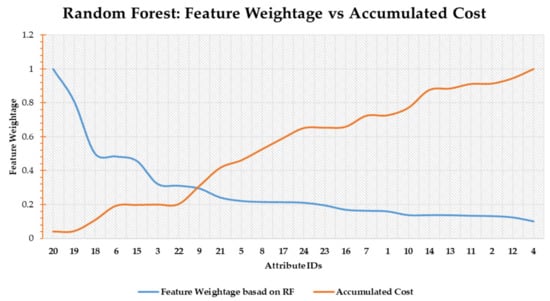 Figure 7. Random forest-based feature scoring.
Figure 7. Random forest-based feature scoring. - Gradient Boosted Trees Score: The last scoring function is based on GBT. The blue line in Figure 8 shows the feature weightage (FW) while the orange line denotes accumulated cost (Cscore). Both the values are normalized. The point of intersection between FW and Cscore is around feature number 3 as shown in Figure 8.
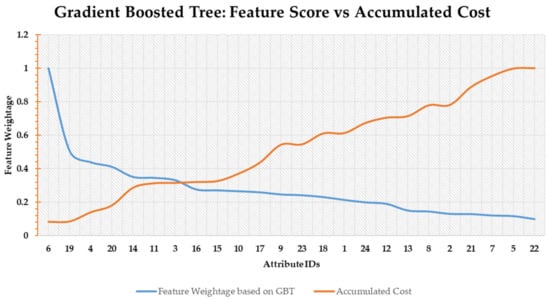 Figure 8. Gradient boosted trees based feature scoring.
Figure 8. Gradient boosted trees based feature scoring.
In this study, all the feature scoring functions are based on decision tree family models. Therefore, it would be interesting to investigate the correlation of generated ranking lists. We used the Kendall rank correlation coefficient [49] to compute the pair-wise correlation of lists produced by DT, RF, GBT as shown in Table 7.

Table 7.
Kendall rank correlation coefficient for different scoring functions.
As it can be observed that the correlation between different lists is closer to zero. The correlation results of the ranked lists support the null hypothesis of mutual independence. Therefore, we can conclude that there is statistically significant independence between the ranked lists. Furthermore, the lists generated by both GBT and DT are relatively in agreement with each other while the lists produced by DT and RF, and as well as RF and GBT show disagreement. In this regard, each scoring function account for important characteristics of the dataset while not diverging too much in their final results.
Furthermore, we can also look at different feature subsets obtained from the aforementioned scoring functions as reported in Table 8 and their Jaccard index value [50]. Figure 9 depicts a Venn diagram for all possible logical relations among different feature subsets.

Table 8.
Selected features by individual scoring functions.
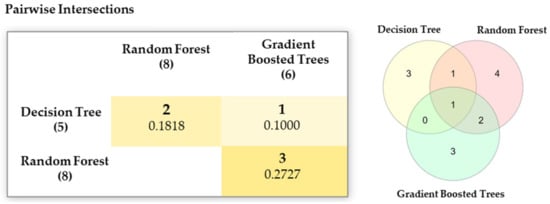
Figure 9.
Jaccard index for different feature subsets.
It is interesting to note that, although the ranked lists produced by GBT and DT have a higher correlation, whereas, the feature subsets obtained after applying a threshold value to the respective lists have a lower value on the Jaccard index i.e., 0.1000. On the other hand, a higher Jaccard index value is obtained between RF and GBT feature subsets while their ranked lists reported a negative correlation. Based on these observations, we can conclude that both the lists and the subsequent feature subsets obtained from the scoring functions are not redundant.
The resulting selected features collected in lists 1, list 2, and list 3 are based on the decision tree, random forest, and gradient boosted trees, respectively. As it can be seen that there are some variations in the selected features, which shows that each scoring function has its own inductive bias while constructing a model as shown in Table 8. A detailed study of the inductive bias of decision tree models is not within the scope of this study.
The averaged evaluation results of the aforementioned seven classification models are provided in Table 9. These results reflect the performance of classification models on reduced datasets acquired from each scoring function. In this regard, the decision tree classifier is constructed based on features present in list L1. Likewise, classifiers for random forest and gradient boosted trees are built on L2 and L3, respectively.

Table 9.
Averaged evaluation results for individual scoring functions on seven classification models.
The overall performance of the random forest and gradient boosted trees increased. Although the decision tree could not improve the accuracy over the full dataset, it slightly improved the sensitivity. As it can be seen that the automatic threshold selection successfully opted for threshold values which resulted in the selection of important features while also keeping the overall cost of the selected feature set low. Hence, the feature subsets produced by different scoring functions are both relevant in enhancing the accuracy of the classification models and distinct.
5.3. Ensemble-1 Results
Feature weightage obtained from individual scoring functions are consolidated through an averaging operation. The intuition behind the consolidation process is that the final scores will reflect both low-cost and high-accuracy characteristics of the individual functions. Figure 10 shows the feature-cost interaction graph for ensemble-1.
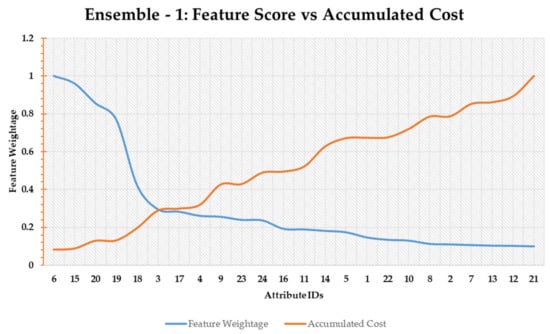
Figure 10.
Ensemble-1 based feature scoring.
As can be seen in Figure 10 shows that the point of intersection is around feature number 3. Therefore, all the features starting from feature number 6 up to feature number 3, i.e., 6, 15, 20, 19, 18, and 3, would be selected. In this regard, Table 10 shows the results of evaluation metrics for features selected by the ensemble-1 approach.

Table 10.
Ensemble-1 results based on selected features.
A cursory glance at Table 9 and Table 10 shows that the proposed ensemble-1 technique successfully reduced the overall cost while improving the key evaluation metrics over the individual scoring functions. Moreover, multiple combinations of feature scoring functions and their respective averaged results over seven classification models are shown in Table 11.

Table 11.
Multiple combinations of scoring functions and their respective results.
5.4. Ensemble-2 Results
As mentioned earlier in Section 3.5, the ensemble-2 is based on consolidating the feature subset acquired from different scoring functions in such a manner that the majority of the features are retained from the individual subsets while the rest of the features are discarded. Table 12 shows features subsets acquired from different scoring functions and the final consolidated solution based on ensemble-2.

Table 12.
Feature selected through ensemble-2.
In this regard, Table 13 shows the evaluation metrics applied to the classification models constructed from the ensemble-2 feature subset.

Table 13.
Ensemble-2 results based on selected features.
It is important to note that the ensemble-2 approach employs the majority vote among individual partial solutions. In the case of three scoring functions, the majority vote based selection heuristic can also be stated as a 2/3rd rule i.e., a feature would be selected if it is present in at least two partial solutions. The alternative options available are a union case in which all the distinct features obtained in partial solutions are selected, and in intersection case, only those features present in all partial solutions are admissible. In this regard, the majority vote can be seen as a multi-point intersection as discussed in [17]. Comparative results of the aforementioned subset combining cases are provided in Table 14.

Table 14.
Multiple combinations of scoring functions and their respective results.
In terms of comparative analysis between ensemble-1 and ensemble-2 it can be observed that for the CKD problem, the latter performs better in terms of accuracy, precision, specificity, F1-measure, the cardinality of selected features and the overall cost of the solution. It would be interesting to explore whether these results generalize to other cost-sensitive diagnosis problems or not?
5.5. Comparison with Other Similar Approaches
In this section, we compare our results with other feature selection approaches on the CKD dataset. All the experiments are performed on the aforementioned seven classifiers, and subsequently, the averaged results are reported in Table 15.

Table 15.
Multiple combinations of scoring functions and their respective results.
A detailed comparison based on similar techniques and proposed techniques is drawn in Table 16. The boldface values denote the best performance achieved under a specific criterion. As it can be seen that most of the comparative techniques are primarily optimized to produce models with higher accuracy. Therefore, in terms of accuracy, the difference between the competing approaches is not statistically significant as reported in Table 16. Furthermore, the entire dataset was used at the feature scoring stage and this would have introduced a potential source of over-estimation of the results. The results of the statistical test are consistent with the overall observation that the proposed approaches produce results for the CKD diagnosis problem that are comparable with other methods in terms of predictive accuracy while at the same time are relatively more cost-effective.

Table 16.
Comparison between proposed and other methods in terms of statistical difference (two-tailed unpaired student t-test, significant level 0.05).
Moreover, we ranked the comparative techniques in terms of predicted accuracy, precision, recall, specificity, F1-measure, AUC, feature set cardinality, and cost of the solution. Lower the rank, better the approach, as the best approach for each criterion is placed at rank 1. Although it is already established in the preceding sections the ensemble-2 approach outperformed all other comparative techniques but as it can be seen in Figure 11, ensemble-1 obtained a lower rank than ensemble-2. It is because ensemble-2 resulted in the lowest recall value among all the competing approaches, which effectively placed it at 9th position. Therefore, both the proposed approaches have their strengths and weaknesses. Ensemble-1 performed consistently over both performance and incurred-cost evaluation factors. On the other hand, ensemble-2 was able to outperform ensemble-1 in terms of incurred cost and on general accuracy metrics over the CKD dataset. It is also noted that, in terms of the recall factor, ensemble-2 lagged behind the other techniques mentioned in the comparative analysis. It can be seen that the cost difference between ensemble-1 and ensemble-2 is $31, while the recall difference is around 2.5%. In this regard, it might be worth spending an extra $31 to timely diagnosis patients. In this regard, both ensemble-1 and ensemble-2 can be treated as non-dominating solutions, i.e., the merit of one approach cannot be undermined by that of the another. In such cases, the final selection is left to the discretion of the decision maker.
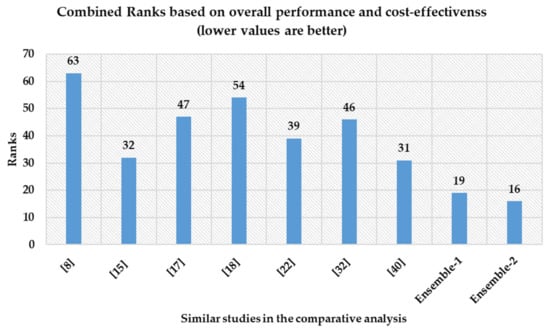
Figure 11.
Combined ranks of different approaches across performance and cost factors.
6. Conclusions
Cost-sensitive feature selection is one of the important areas of research where the cost of data acquisition plays an important role in the applicability of the solution. Generally, it is assumed that cost of data acquisition is the same, i.e., not necessarily zero. This assumption does not hold in certain application domains such as disease diagnosis. In this study, we used a well-known benchmarked dataset for disease diagnosis i.e., chronic kidney disease. A significant scholarly work is reported in designing algorithms and systems for chronic kidney disease efficiently. The proposed techniques for cost-sensitive feature selection reported in this study is in the continuation of CKD research, where the key objective is to enhance the performance of decision tree-based classification models.
Decision tree-based classification models shown a great promise in the domain of medical diagnosis, especially in dealing with structured heterogeneous datasets, e.g., electronic medical records for chronic kidney disease patients. This research deals with addressing the applicability concerns of decision tree models through ensemble feature ranking techniques. In the proposed techniques a set of multiple feature scoring functions are used which are based on the decision tree family. It is also demonstrated that the partial solutions obtained from these scoring functions are not redundant and hence are useful in creating an ensemble technique. Furthermore, a heuristic technique based on feature weightage and the accumulated cost is introduced to select a subset of features. It is also demonstrated that the features selected based on the threshold value are both useful and cost-effective.
The two proposed approaches for cost-sensitive ensemble feature selection primarily differ in the application of the threshold operation. Ensemble-1 combines multiple feature scores into a consolidated score and thereafter applies the threshold operation. In the case of ensemble-2, the threshold is applied to the individual lists obtained from multiple scoring functions. In this case, multiple feature subsets are produced as partial solutions. Afterward, all the solutions are combined using the majority voting scheme. Extensive experimentation is performed, in which it is demonstrated that although ensemble-2 is better in terms of general evaluation criteria for the CKD problem, ensemble-1 produces more consistent results. Both the aforementioned techniques are compared with other similar feature selection methods. It is demonstrated that cost-free feature selection techniques generally produce a solution with high accuracy but as the cost is not taken into account therefore, the resultant solutions are not cost-effective. Based on the comparative analysis, it can be seen that proposed techniques produce solutions for the CKD diagnosis problem which are accurate and cost-effective. The proposed approaches selected a final feature subset for the CKD dataset by retaining around 1/4th of the original features, decreasing the cost by a factor of 7.42 of the original feature set, and achieving comparable average accuracy as that of other methods in this study.
This research can be extended in a number of directions such as we used a classifier-ensemble to account for feature interaction. Although this approach provided promising feature weights but the overall running time of the scoring functions can be reduced by employing lightweight filter techniques. Furthermore, cost can be modeled as a multi-objective function along with the error rate and hence a number of candidate solutions can be generated for the decision maker for an informed decision making. Another important direction worth exploring is to treatment multiple features, collectively. In this study, each feature has its own cost, while, in the medical domain, it is generally the case that data acquired from multiple features are clubbed together under different medical tests.
Author Contributions
S.I.A. is principal researcher who conceptualized the idea, designed and simulated the methodology, performed experiments, and authored the manuscript. B.A. assisted in the manuscript preparation along with assisting in evaluating the methodology. J.H. assisted in simulating the methodology. M.H. contributed in comparing the proposed methodologies, F.A.S. assisted in reviewing the manuscript. G.H.P. and S.L. contributed in the funding acquisition, along with supervising the overall experiment design, providing valuable feedback and reviewed the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the MSIT (Ministry of Science and ICT), Korea, under the ITRC (Information Technology Research Center) support program (IITP-2017-0-01629) supervised by the IITP (Institute for Information & communications Technology Promotion), this work was supported by the Institute for Information & communications Technology Promotion (IITP) grant funded by the Korea government (MSIT) (No.2017-0-00655), this research was supported by the MSIT(Ministry of Science and ICT), Korea, under the Grand Information Technology Research Center support program(IITP-2020-0-01489) supervised by the IITP(Institute for Information & communications Technology Planning & Evaluation), NRF-2016K1A3A7A03951968 (EU), and NRF-2019R1A2C2090504.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| CKD | Chronic Kidney Disease |
| FW | Feature Weightage |
| CScore | Cost Score |
| KNN | K-Nearest Neighbor |
| SVM | Support Vector Machine |
| RF | Random Forest |
| ANN | Artificial Neural Network |
| PCA | Principal Component Analysis |
| GA | Genetic Algorithm |
| CART | Classification And Regression Trees |
| GBT | Gradient Boosted Trees |
| UCI | University of California, Irvine |
| AUC | Area under Receiver Operating Characteristics Curve |
| TP | True Positive |
| TN | True Negative |
| FP | False Positive |
| FN | False Negative |
References
- Kidney Disease: Improving Global Outcomes (KDIGO) Transplant Work Group. KDIGO clinical practice guideline for the care of kidney transplant recipients. Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transpl. Surg. 2009, 9, S1. [Google Scholar] [CrossRef] [PubMed]
- Kellum, J.A.; Lameire, N. KDIGO AKI Guideline Work Group Diagnosis, evaluation, and management of acute kidney injury: A KDIGO summary (Part 1). Crit. Care 2013, 17, 204. [Google Scholar] [CrossRef] [PubMed]
- Park, J.I.; Baek, H.; Jung, H.H. Prevalence of chronic kidney disease in korea: The korean national health and nutritional examination survey 2011–2013. J. Korean Med. Sci. 2016, 31, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-L.; Rothenbacher, D. Prevalence of chronic kidney disease in population-based studies: Systematic review. BMC Public Health 2008, 8, 117. [Google Scholar] [CrossRef]
- Moyer, V.A.; Force, U.P.S.T. Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann. Intern. Med. 2012, 157, 120–134. [Google Scholar] [CrossRef]
- Álvaro, S.; Queiroz, A.C.M.D.S.; Da Silva, L.D.; Costa, E.D.B.; Pinheiro, M.E.; Perkusich, A. Computer-Aided Diagnosis of Chronic Kidney Disease in Developing Countries: A Comparative Analysis of Machine Learning Techniques. IEEE Access 2020, 8, 25407–25419. [Google Scholar] [CrossRef]
- Itani, S.; Lecron, F.; Fortemps, P. Specifics of medical data mining for diagnosis aid: A survey. Expert Syst. Appl. 2019, 118, 300–314. [Google Scholar] [CrossRef]
- Ogunleye, A.A.; Qing-Guo, W. XGBoost Model for Chronic Kidney Disease Diagnosis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 1. [Google Scholar] [CrossRef]
- Khan, B.; Naseem, R.; Muhammad, F.; Abbas, G.; Kim, S. An Empirical Evaluation of Machine Learning Techniques for Chronic Kidney Disease Prophecy. IEEE Access 2020, 8, 55012–55022. [Google Scholar] [CrossRef]
- Cios, K.J.; Krawczyk, B.; Cios, J.; Staley, K.J. Uniqueness of Medical Data Mining: How the New Technologies and Data They Generate are Transforming Medicine. arXiv 2019, arXiv:1905.09203. [Google Scholar]
- Salekin, A.; Stankovic, J. Detection of Chronic Kidney Disease and Selecting Important Predictive Attributes. In Proceedings of the 2016 IEEE International Conference on Healthcare Informatics (ICHI), Chicago, IL, USA, 4–7 October 2016; pp. 262–270. [Google Scholar]
- Zhou, Q.; Zhou, H.; Li, T. Cost-sensitive feature selection using random forest: Selecting low-cost subsets of informative features. Knowl.-Based Syst. 2016, 95, 1–11. [Google Scholar] [CrossRef]
- Min, F.; Liu, Q. A hierarchical model for test-cost-sensitive decision systems. Inf. Sci. 2009, 179, 2442–2452. [Google Scholar] [CrossRef]
- Vasquez-Morales, G.R.; Martinez-Monterrubio, S.M.; Moreno-Ger, P.; Recio-Garcia, J.A. Explainable Prediction of Chronic Renal Disease in the Colombian Population Using Neural Networks and Case-Based Reasoning. IEEE Access 2019, 7, 152900–152910. [Google Scholar] [CrossRef]
- Qin, J.; Chen, L.; Liu, Y.; Liu, C.; Feng, C.; Chen, B. A Machine Learning Methodology for Diagnosing Chronic Kidney Disease. IEEE Access 2020, 8, 20991–21002. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Alonso-Betanzos, A. Ensembles for feature selection: A review and future trends. Inf. Fusion 2019, 52, 1–12. [Google Scholar] [CrossRef]
- Tsai, C.-F.; Hsiao, Y.-C. Combining multiple feature selection methods for stock prediction: Union, intersection, and multi-intersection approaches. Decis. Support Syst. 2010, 50, 258–269. [Google Scholar] [CrossRef]
- Polat, H.; Mehr, H.D.; Cetin, A. Diagnosis of Chronic Kidney Disease Based on Support Vector Machine by Feature Selection Methods. J. Med. Syst. 2017, 41, 55. [Google Scholar] [CrossRef]
- Taradeh, M.; Mafarja, M.; Heidari, A.A.; Faris, H.; Aljarah, I.; Mirjalili, S.; Fujita, H. An evolutionary gravitational search-based feature selection. Inf. Sci. 2019, 497, 219–239. [Google Scholar] [CrossRef]
- Seijo-Pardo, B.; Bolón-Canedo, V.; Alonso-Betanzos, A. On developing an automatic threshold applied to feature selection ensembles. Inf. Fusion 2019, 45, 227–245. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Sanchez-Marono, N.; Alonso-Betanzos, A. An ensemble of filters and classifiers for microarray data classification. Pattern Recognit. 2012, 45, 531–539. [Google Scholar] [CrossRef]
- Almansour, N.A.; Syed, H.F.; Khayat, N.R.; Altheeb, R.K.; Juri, R.E.; Alhiyafi, J.; Alrashed, S.; Olatunji, S.O. Neural network and support vector machine for the prediction of chronic kidney disease: A comparative study. Comput. Biol. Med. 2019, 109, 101–111. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, X.; Zhang, Z. Clinical risk assessment of patients with chronic kidney disease by using clinical data and multivariate models. Int. Urol. Nephrol. 2016, 48, 2069–2075. [Google Scholar] [CrossRef] [PubMed]
- Serpen, A.A. Diagnosis Rule Extraction from Patient Data for Chronic Kidney Disease Using Machine Learning. Int. J. Biomed. Clin. Eng. 2016, 5, 64–72. [Google Scholar] [CrossRef][Green Version]
- Al-Hyari, A.Y.; Al-Taee, A.M.; Al-Taee, M.A. Clinical decision support system for diagnosis and management of chronic renal failure. In Proceedings of the 2013 IEEE Jordan Conference on Applied Electrical Engineering and Computing Technologies, Amman, Jordan, 3–5 December 2013; pp. 1–6. [Google Scholar]
- Ani, R.; Sasi, G.; Sankar, U.R.; Deepa, O.S. Decision support system for diagnosis and prediction of chronic renal failure using random subspace classification. In Proceedings of the 2016 International Conference on Advances in Computing, Communications and Informatics, Jaipur, India, 21–24 September 2016; pp. 1287–1292. [Google Scholar]
- Tazin, N.; Sabab, S.A.; Chowdhury, M.T. Diagnosis of Chronic Kidney Disease using effective classification and feature selection technique. In Proceedings of the 2016 International Conference on Medical Engineering, Health Informatics and Technology, Dhaka, Bangladesh, 17–18 December 2016; pp. 1–6. [Google Scholar]
- Cahyani, N.; Muslim, M.A. Increasing Accuracy of C4. 5 Algorithm by Applying Discretization and Correlation-based Feature Selection for Chronic Kidney Disease Diagnosis. J. Telecommun. Electron. Comput. Eng. (JTEC) 2020, 12, 25–32. [Google Scholar]
- Seijo-Pardo, B.; Bolón-Canedo, V.; Alonso-Betanzos, A. Testing Different Ensemble Configurations for Feature Selection. Neural Process. Lett. 2017, 46, 857–880. [Google Scholar] [CrossRef]
- Lal, T.N.; Chapelle, O.; Weston, J.; Elisseeff, A. Embedded methods. In Feature Extraction; Springer: Berlin/Heidelberg, Germany, 2008; pp. 137–165. [Google Scholar]
- Jain, D.; Singh, V. Feature selection and classification systems for chronic disease prediction: A review. Egypt. Inform. J. 2018, 19, 179–189. [Google Scholar] [CrossRef]
- Ali, M.; Ali, S.I.; Kim, H.; Hur, T.; Bang, J.; Lee, S.; Kang, B.H.; Hussain, M. uEFS: An efficient and comprehensive ensemble-based feature selection methodology to select informative features. PLoS ONE 2018, 13, e0202705. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Sanchez-Marono, N.; Alonso-Betanzos, A. A review of feature selection methods on synthetic data. Knowl. Inf. Syst. 2012, 34, 483–519. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Sánchez-Maroño, N.; Alonso-Betanzos, A. Recent advances and emerging challenges of feature selection in the context of big data. Knowl.-Based Syst. 2015, 86, 33–45. [Google Scholar] [CrossRef]
- Khoshgoftaar, T.M.; Golawala, M.; Van Hulse, J. An empirical study of learning from imbalanced data using random forest. In Proceedings of the 19th IEEE International Conference on Tools with Artificial Intelligence, Patras, Greece, 29–31 October 2007; Volume 2, pp. 310–317. [Google Scholar]
- Mejia-Lavalle, M.; Sucar, E.; Arroyo, G. Feature selection with a perceptron neural net. In Proceedings of the International Workshop on Feature Selection for Data Mining, Bethesda, MA, USA, 22 April 2006; pp. 131–135. [Google Scholar]
- Seijo-Pardo, B.; Bolón-Canedo, V.; Alonso-Betanzos, A. Using data complexity measures for thresholding in feature selection rankers. In Proceedings of the Spanish Association for Artificial Intelligence, Salamanca, Spain, 14–16 September 2016; pp. 121–131. [Google Scholar]
- Willett, P. Combination of Similarity Rankings Using Data Fusion. J. Chem. Inf. Model. 2013, 53, 1–10. [Google Scholar] [CrossRef]
- Seijo-Pardo, B.; Bolón-Canedo, V.; Alonso-Betanzos, A. Using a feature selection ensemble on DNA microarray datasets. In Proceedings of the ESANN. European Symposium on Artificial Neural Networks, Bruges, Belgium, 27–29 April 2016. [Google Scholar]
- Osanaiye, O.; Cai, H.; Choo, K.-K.R.; Dehghantanaha, A.; Xu, Z.; Dlodlo, M.E. Ensemble-based multi-filter feature selection method for DDoS detection in cloud computing. EURASIP J. Wirel. Commun. Netw. 2016, 2016, 130. [Google Scholar] [CrossRef]
- Chiew, K.-L.; Tan, C.L.; Wong, K.; Yong, K.S.; Tiong, W.-K. A new hybrid ensemble feature selection framework for machine learning-based phishing detection system. Inf. Sci. 2019, 484, 153–166. [Google Scholar] [CrossRef]
- Kurt, I.; Türe, M.; Kurum, T. Comparing performances of logistic regression, classification and regression tree, and neural networks for predicting coronary artery disease. Expert Syst. Appl. 2008, 34, 366–374. [Google Scholar] [CrossRef]
- Sathe, S.; Aggarwal, C.C. Nearest Neighbor Classifiers Versus Random Forests and Support Vector Machines. In Proceedings of the 2019 IEEE International Conference on Data Mining (ICDM), Beijing, China, 8–11 November 2019; pp. 1300–1305. [Google Scholar]
- Dua, D.; Graff, C. UCI Machine Learning Repository; University of California, School of Information and Computer Science: Irvine, CA, USA, 2019; Available online: http://archive.ics.uci.edu/ml (accessed on 5 May 2020).
- Mierswa, I.; Klinkenberg, R. RapidMiner Studio (9.2) [Data Science, Machine Learning, Predictive Analytics]. 2018. Available online: https://rapidminer.com/ (accessed on 30 June 2020).
- CHART Based Feature Weightage. 2020. Available online: http://www.myexperiment.org/workflows/5148.html (accessed on 30 June 2020).
- Gradient Boosting Trees Based Feature Weightage. 2020. Available online: http://www.myexperiment.org/workflows/5149.html (accessed on 30 June 2020).
- Random Forest Based Feature Weightage. 2020. Available online: http://www.myexperiment.org/workflows/5150.html (accessed on 30 June 2020).
- Kendall, M.G. Rank Correlation Methods; American Psychological Association: Washington, DC, USA, 1948. [Google Scholar]
- Jaccard, P. Étude comparative de la distribution florale dans une portion des Alpes et des Jura. Bull. Soc. Vaud. Sci. Nat. 1901, 37, 547–579. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).