Study on the Effect of Contrast Agent on Biofilms and Their Visualization in Porous Substrate Using X-ray μCT
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Growth Medium for Biofilm Culture
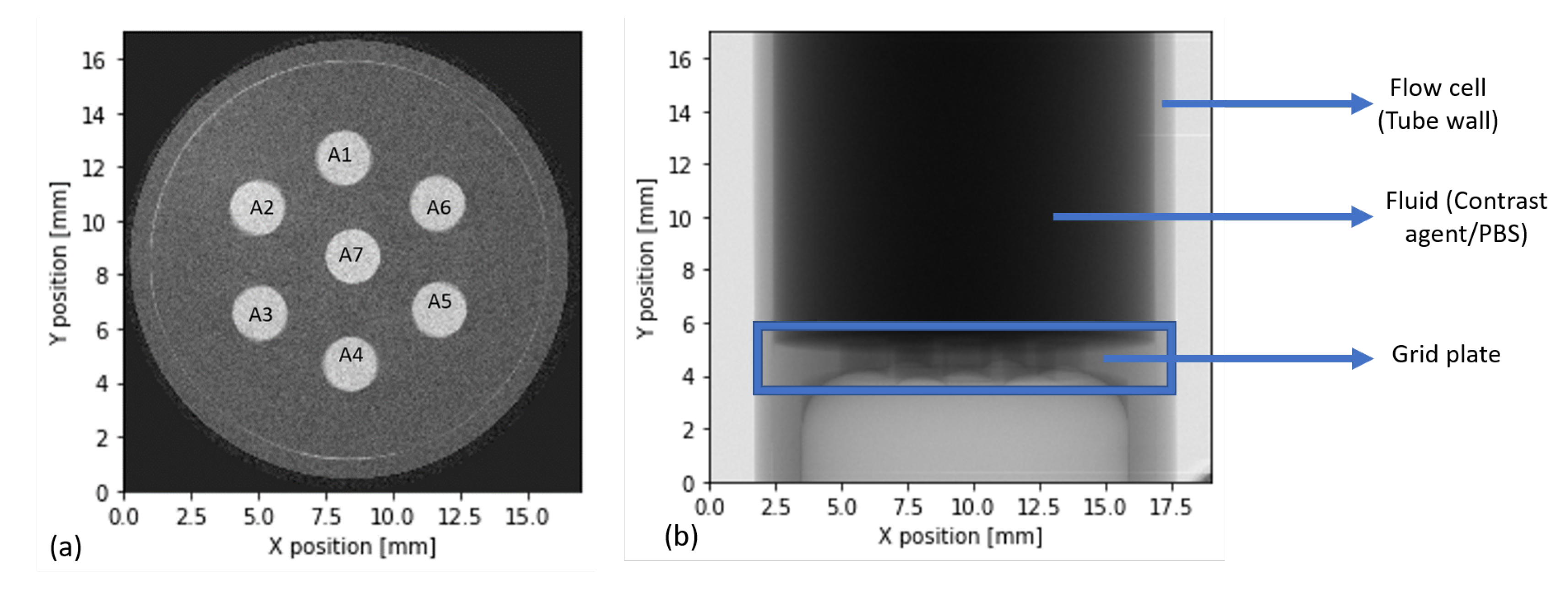
2.2. Porous Substrate and Flow Cell
2.3. Contrast Agents
2.4. Scanner System and Microscopy
2.4.1. X-ray CT Scanner Settings
2.5. Simulations of X-ray μCT Scan
2.6. Determination of Contrast Agent’s Toxicity
2.6.1. By Plate Counting Method
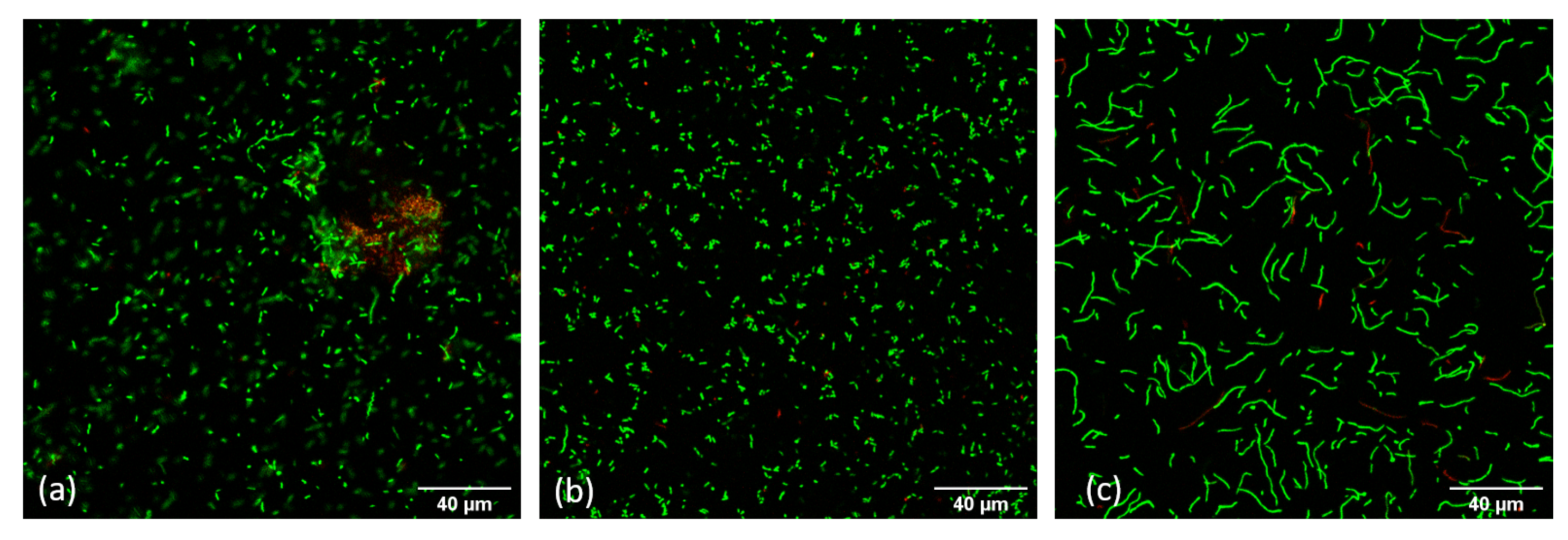
2.6.2. By Confocal Laser Scanning Microscope
2.7. Protocol for Bacterial Incubation in Flow Cell
2.8. Determination of Irradiation Effect of X-rays on Biofilms
2.9. Diffusion Experiment
2.10. In-Situ Experiments for Biofilm Visualization
Characterisation of Biofilm
3. Results
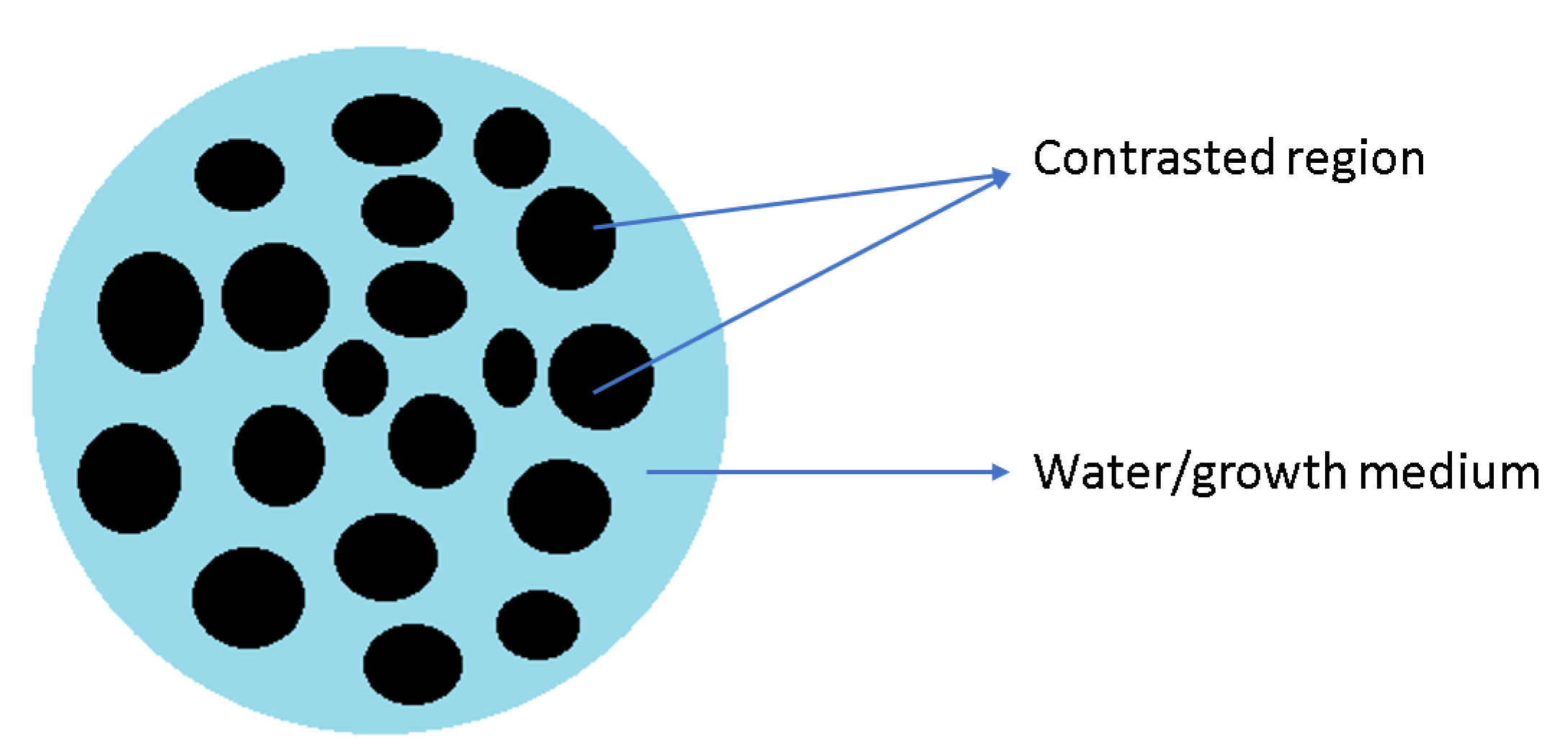
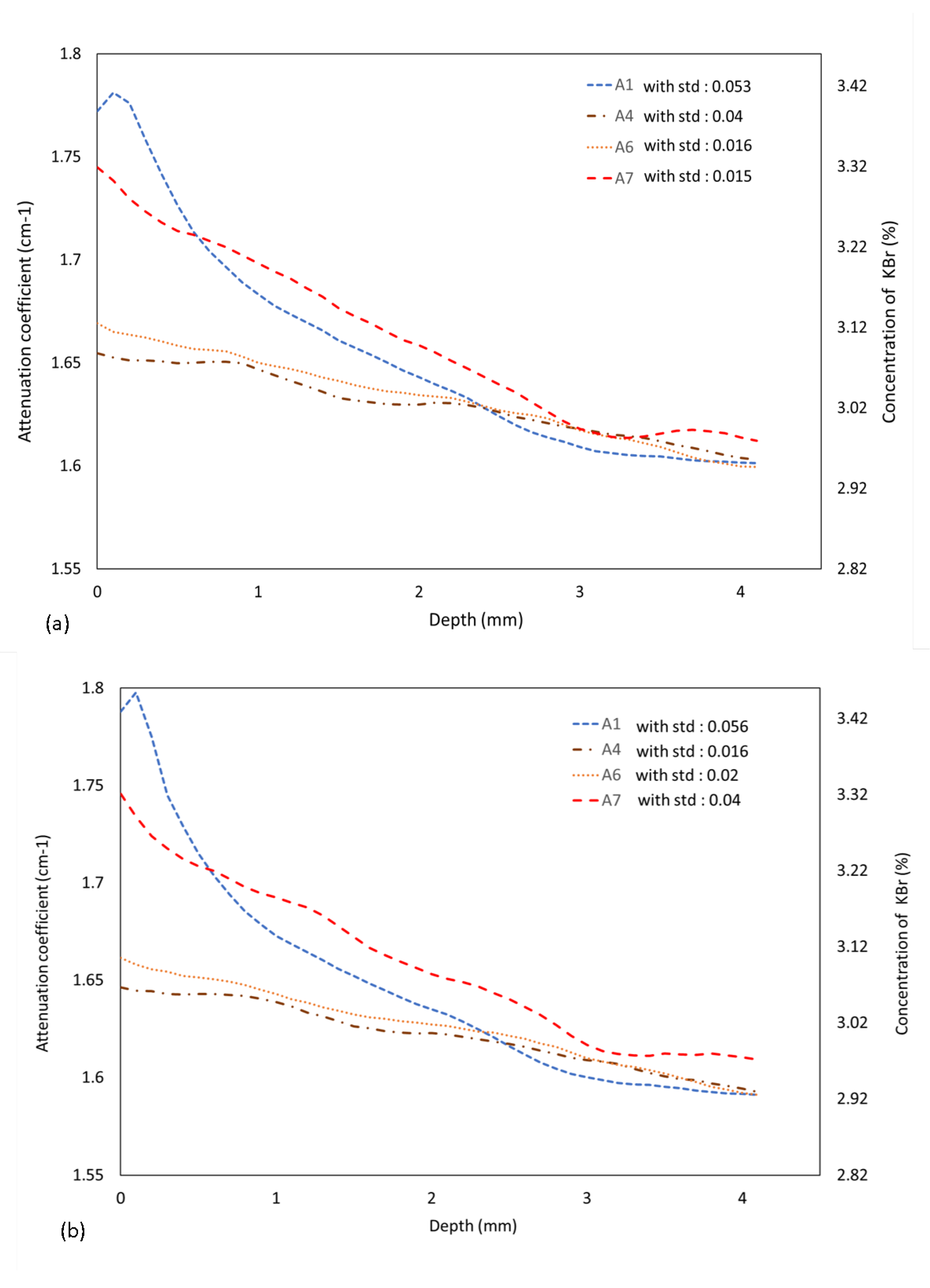
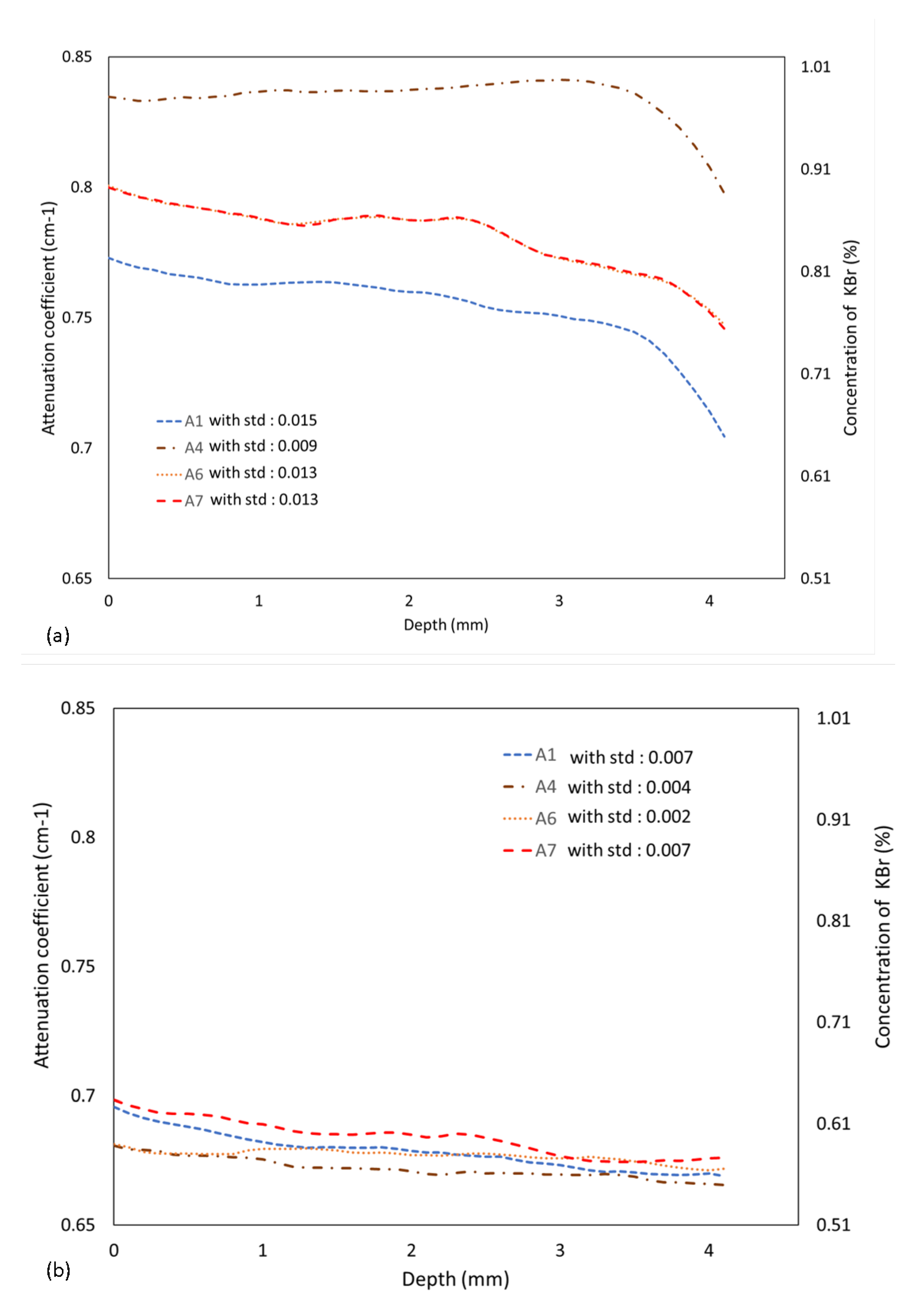
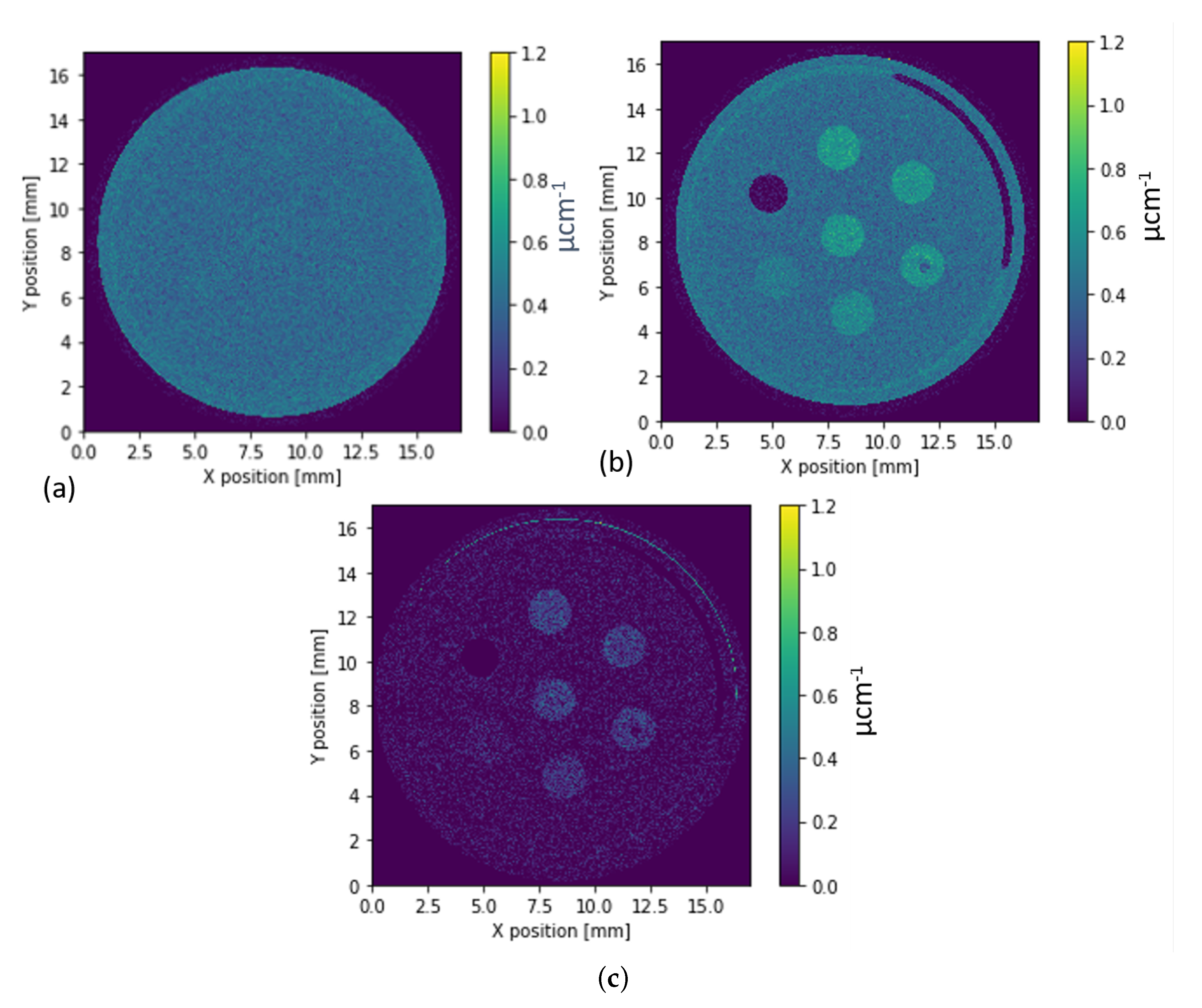
3.1. Simulated Data of the Virtual Sample with KBr
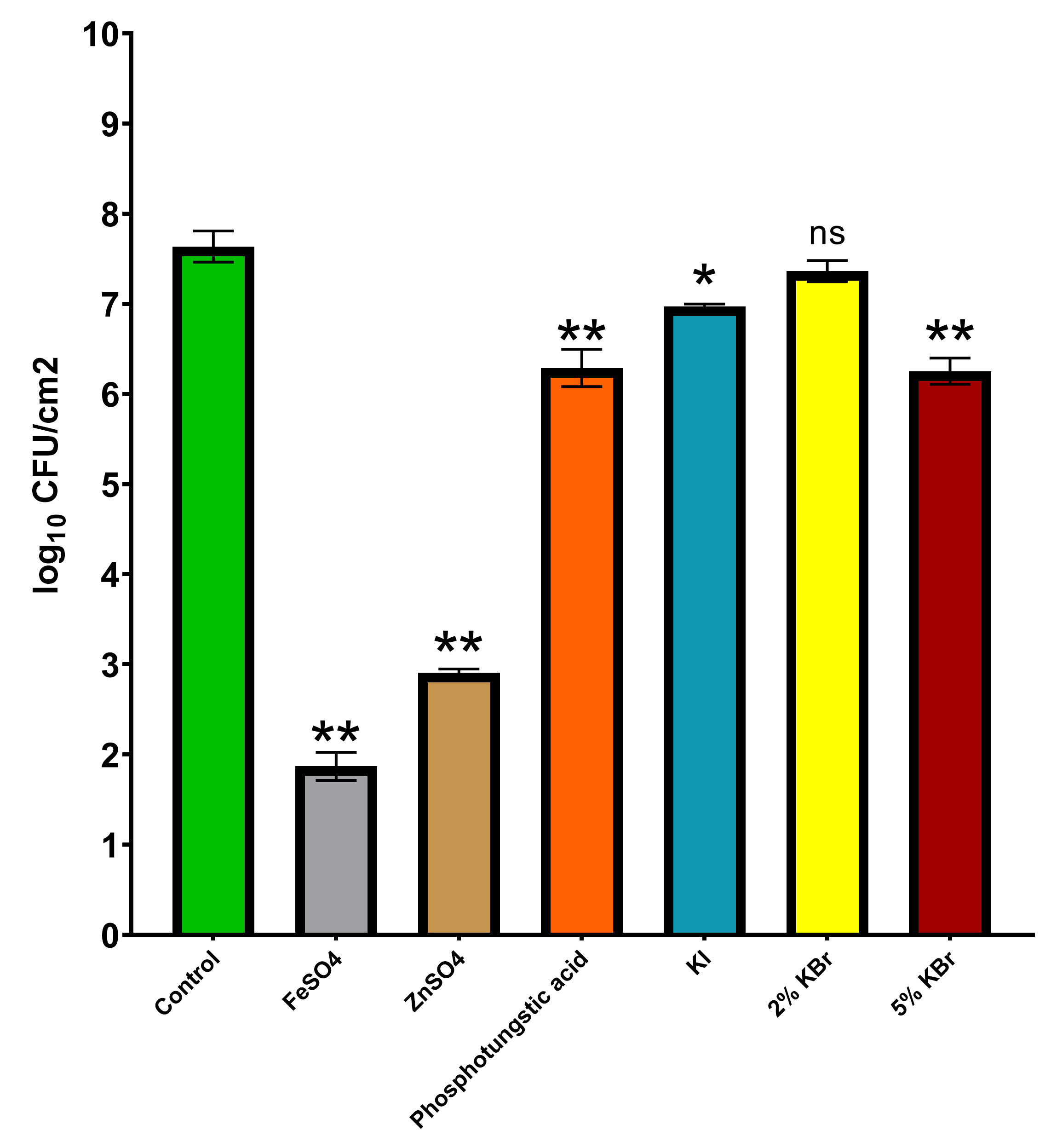
3.2. Long-Term Toxicity Test of Contrast Agents on P. Fluorescens Pf-01 Biofilms
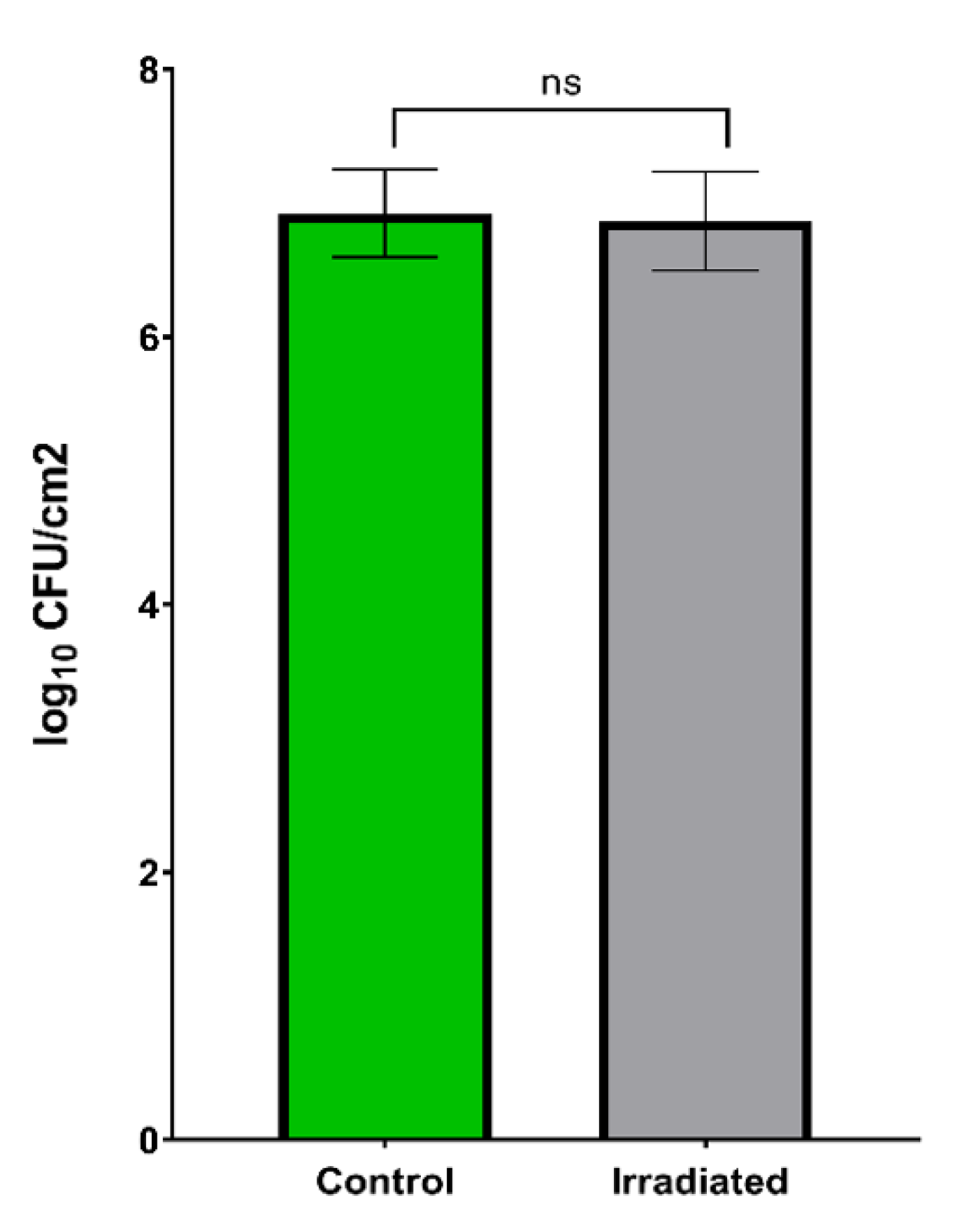
3.3. Irradiation Effect of X-rays on P. Fluorescens Pf-01 Biofilms
3.4. Diffusion Experiments
3.4.1. Contrast Agent Application
3.4.2. Phosphate Buffer Saline Application
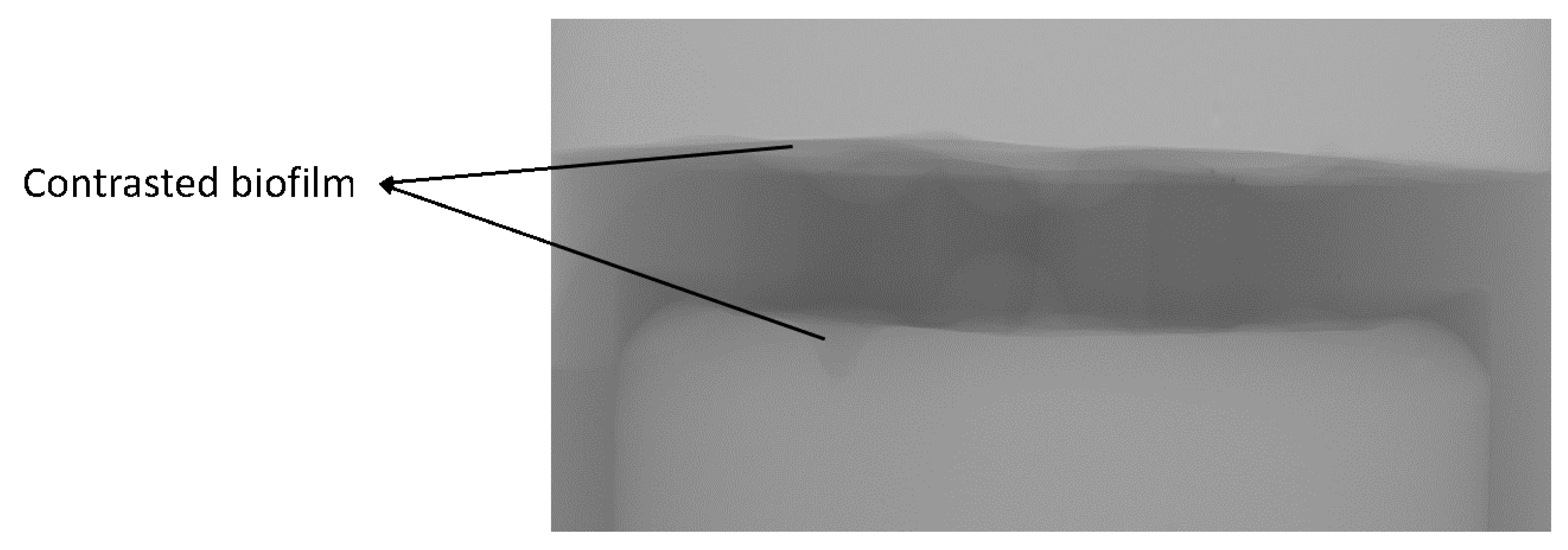
3.5. Visualization of Biofilms with Application of Contrast Agent
4. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CT | Computed Tomography |
| LB | Lysogeny broth |
| TSB | Tryptic soy broth |
| PBS | Phosphate buffer saline |
| w/v | weight/volume |
References
- López, D.; Vlamakis, H.; Kolter, R. Biofilms. Cold Spring Harbor Perspect. Biol. 2010, 2, a000398. [Google Scholar] [CrossRef] [PubMed]
- Boyle, K.E.; Heilmann, S.; van Ditmarsch, D.; Xavier, J.B. Exploiting social evolution in biofilms. Curr. Opin. Microbiol. 2013, 16, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Ghoul, M.; Mitri, S. The ecology and evolution of microbial competition. Trends Microbiol. 2016, 24, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar]
- Flemming, H.C.; Neu, T.R.; Wozniak, D.J. The EPS matrix: The “house of biofilm cells”. J. Bacteriol. 2007, 189, 7945–7947. [Google Scholar] [CrossRef]
- Whiteley, R.K.; Pajkos, A.; Vickery, K. Biofilms that impact on human health. J. Pharm. Pract. Res. 2002, 32, 153–158. [Google Scholar] [CrossRef]
- Crabbé, A.; Jensen, P.; Bjarnsholt, T.; Bjarnsholt, T.; Coenye, T. Antimicrobial tolerance and metabolic adaptations in microbial biofilms. Trends Microbiol. 2019, 27, 850–863. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Halan, B.; Buehler, K.; Schmid, A. Biofilms as living catalysts in continuous chemical syntheses. Trends Biotechnol. 2012, 30, 453–465. [Google Scholar] [CrossRef]
- Govender, S.; Pillay, V.; Odhav, B. Nutrient manipulation as a basis for enzyme production in a gradostat bioreactor. Enzym. Microb. Technol. 2010, 46, 603–609. [Google Scholar] [CrossRef]
- Volkmar, K.; Bremer, E. Effects of seed inoculation with a strain of Pseudomonas fluorescens on root growth and activity of wheat in well-watered and drought-stressed glass-fronted rhizotrons. Can. J. Plant Sci. 1998, 78, 545–551. [Google Scholar] [CrossRef]
- Suh, Y.J.; Park, J.M.; Yang, J.W. Biodegradation of cyanide compounds by Pseudomonas fluorescens immobilized on zeolite. Enzym. Microb. Technol. 1994, 16, 529–533. [Google Scholar] [CrossRef]
- Turhan, E.Ü.; Erginkaya, Z.; Korukluoğlu, M.; Konuray, G. Beneficial Biofilm Applications in Food and Agricultural Industry. In Health and Safety Aspects of Food Processing Technologies; Springer: Berlin/Heidelberg, Germany, 2019; pp. 445–469. [Google Scholar]
- Bridier, A.; Briandet, R.; Thomas, V.; Dubois-Brissonnet, F. Resistance of bacterial biofilms to disinfectants: A review. Biofouling 2011, 27, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.G.; Young, J.P.W.; Fitter, A.H. Interactions between Pseudomonas fluorescens biocontrol agents and Glomus mosseae, an arbuscular mycorrhizal fungus, within the rhizosphere. FEMS Microbiol. Lett. 1998, 166, 297–303. [Google Scholar] [CrossRef]
- Wang, H.; Cai, L.; Li, Y.; Xu, X.; Zhou, G. Biofilm formation by meat-borne Pseudomonas fluorescens on stainless steel and its resistance to disinfectants. Food Control 2018, 91, 397–403. [Google Scholar] [CrossRef]
- Vikram, A.; Bomberger, J.M.; Bibby, K.J. Efflux as a glutaraldehyde resistance mechanism in Pseudomonas fluorescens and Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2015, 59, 3433–3440. [Google Scholar] [CrossRef]
- Workentine, M.L.; Harrison, J.J.; Stenroos, P.U.; Ceri, H.; Turner, R.J. Pseudomonas fluorescens’ view of the periodic table. Environ. Microbiol. 2008, 10, 238–250. [Google Scholar] [CrossRef]
- Ge, Y.; Zhu, J.; Ye, X.; Yang, Y. Spoilage potential characterization of Shewanella and Pseudomonas isolated from spoiled large yellow croaker (Pseudosciaena crocea). Lett. Appl. Microbiol. 2017, 64, 86–93. [Google Scholar] [CrossRef]
- Nya, E. Biofilms and biofouling: Cost and effect on drinking water quality for human development. J. Glob. Biosci. 2015, 4, 1571–1578. [Google Scholar]
- Scales, B.S.; Dickson, R.P.; LiPuma, J.J.; Huffnagle, G.B. Microbiology, genomics, and clinical significance of the Pseudomonas fluorescens species complex, an unappreciated colonizer of humans. Clin. Microbiol. Rev. 2014, 27, 927–948. [Google Scholar] [CrossRef]
- Cortizo, M.C.; De Mele, M.F.L. Preliminary characterization of thin biofilms by optical microscopy. Biofouling 2000, 15, 253–260. [Google Scholar] [CrossRef][Green Version]
- Bogachev, M.I.; Volkov, V.Y.; Markelov, O.A.; Trizna, E.Y.; Baydamshina, D.R.; Melnikov, V.; Murtazina, R.R.; Zelenikhin, P.V.; Sharafutdinov, I.S.; Kayumov, A.R. Fast and simple tool for the quantification of biofilm-embedded cells sub-populations from fluorescent microscopic images. PLoS ONE 2018, 13, e0193267. [Google Scholar] [CrossRef] [PubMed]
- Thornton, R.B.; Rigby, P.J.; Wiertsema, S.P.; Filion, P.; Langlands, J.; Coates, H.L.; Vijayasekaran, S.; Keil, A.D.; Richmond, P.C. Multi-species bacterial biofilm and intracellular infection in otitis media. BMC Pediatrics 2011, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Reichhardt, C.; Parsek, M. Confocal laser scanning microscopy for analysis of Pseudomonas aeruginosa biofilm architecture and matrix localization. Front. Microbiol. 2019, 10, 677. [Google Scholar] [CrossRef] [PubMed]
- Beech, I.B.; Tapper, R.C.; Gubner, R.J. Microscopy methods for studying biofilms. In Biofilms: Recent Advances in Their Study and Control; CRC Press: Boca Raton, FL, USA, 2000; pp. 51–70. [Google Scholar]
- Cnudde, V.; Boone, M.N. High-resolution X-ray computed tomography in geosciences: A review of the current technology and applications. Earth-Sci. Rev. 2013, 123, 1–17. [Google Scholar] [CrossRef]
- Krummel, A.T.; Datta, S.S.; Münster, S.; Weitz, D.A. Visualizing multiphase flow and trapped fluid configurations in a model three-dimensional porous medium. AIChE J. 2013, 59, 1022–1029. [Google Scholar] [CrossRef]
- Long, H.; Swennen, R.; Foubert, A.; Dierick, M.; Jacobs, P. 3D quantification of mineral components and porosity distribution in Westphalian C sandstone by microfocus X-ray computed tomography. Sediment. Geol. 2009, 220, 116–125. [Google Scholar] [CrossRef]
- Valvatne, P.H. Predictive Pore-Scale Modelling of Multiphase Flow. Ph.D. Thesis, Department of Earth Science and Engineering, Imperial College London, London, UK, 2004. [Google Scholar]
- Zhou, M.; Lu, D.; Dunsmuir, J.; Thomann, H. Irreducible water distribution in sandstone rock: Two phase flow simulations in CT-based pore network. Phys. Chem. Earth Part A Solid Earth Geod. 2000, 25, 169–174. [Google Scholar] [CrossRef]
- Keren-Paz, A.; Brumfeld, V.; Oppenheimer-Shaanan, Y.; Kolodkin-Gal, I. Micro-CT X-ray imaging exposes structured diffusion barriers within biofilms. NPJ Biofilms Microbiomes 2018, 4, 1–4. [Google Scholar] [CrossRef]
- Vyas, N.; Walmsley, A.; Pecheva, E.; Dehghani, H.; Grover, L.; Sammons, R. Visualizing synthetic dental biofilm on teeth using micro computed tomography. In Proceedings of the Bruker Micro-CT User Meeting, University of Birmingham, Birmingham, UK, 29 April 2014; Volume 75. [Google Scholar]
- Davit, Y.; Iltis, G.; Debenest, G.; Veran-Tissoires, S.; Wildenschild, D.; Gérino, M.; Quintard, M. Imaging biofilm in porous media using X-ray computed microtomography. J. Microsc. 2011, 242, 15–25. [Google Scholar] [CrossRef]
- Carrel, M.; Beltran, M.A.; Morales, V.L.; Derlon, N.; Morgenroth, E.; Kaufmann, R.; Holzner, M. Biofilm imaging in porous media by laboratory X-ray tomography: Combining a non-destructive contrast agent with propagation-based phase-contrast imaging tools. PLoS ONE 2017, 12, e0180374. [Google Scholar] [CrossRef] [PubMed]
- Smułek, W.; Zdarta, A.; Pacholak, A.; Runka, T.; Kaczorek, E. Increased biological removal of 1-chloronaphthalene as a result of exposure: A study of bacterial adaptation strategies. Ecotoxicol. Environ. Saf. 2019, 185, 109707. [Google Scholar] [CrossRef] [PubMed]
- Rendleman, J. Metal-polysaccharide complexes—Part II. Food Chem. 1978, 3, 127–162. [Google Scholar] [CrossRef]
- Maleki, S.; Almaas, E.; Zotchev, S.; Valla, S.; Ertesvåg, H. Alginate biosynthesis factories in Pseudomonas fluorescens: Localization and correlation with alginate production level. Appl. Environ. Microbiol. 2016, 82, 1227–1236. [Google Scholar] [CrossRef]
- Kolar, M.H.; Tabarrini, O. Halogen Bonding in Nucleic Acid Complexes: Miniperspective. J. Med. Chem. 2017, 60, 8681–8690. [Google Scholar]
- Cullen, J.; Phillips, M.; Shipley, G. The effects of temperature on the composition and physical properties of the lipids of Pseudomonas fluorescens. Biochem. J. 1971, 125, 733–742. [Google Scholar] [CrossRef][Green Version]
- Luria, S.; Adams, J.; Ting, R. Transduction of lactose-utilizing ability among strains of E. coli and S. dysenteriae and the properties of the transducing phage particles. Virology 1960, 12, 348–390. [Google Scholar] [CrossRef]
- Dulbecco, R.; Vogt, M. Plaque formation and isolation of pure lines with poliomyelitis viruses. J. Exp. Med. 1954, 99, 167–182. [Google Scholar] [CrossRef]
- Ha, J.; Kim, J.; Jung, Y.; Yun, G.; Kim, D.N.; Kim, H.Y. Poro-elasto-capillary wicking of cellulose sponges. Sci. Adv. 2018, 4, eaao7051. [Google Scholar] [CrossRef]
- Dierick, M.; Van Loo, D.; Masschaele, B.; Van den Bulcke, J.; Van Acker, J.; Cnudde, V.; Van Hoorebeke, L. Recent micro-CT scanner developments at UGCT. Nucl. Instrum. Methods Phys. Res. Sec. B Beam Interact. Mater. At. 2014, 324, 35–40. [Google Scholar] [CrossRef]
- Vlassenbroeck, J.; Dierick, M.; Masschaele, B.; Cnudde, V.; Van Hoorebeke, L.; Jacobs, P. Software tools for quantification of X-ray microtomography at the UGCT. Nucl. Instrum. Methods Phys. Res. Sec. A Accel. Spectrometers Detect. Assoc. Equip. 2007, 580, 442–445. [Google Scholar] [CrossRef]
- Brabant, L.; Vlassenbroeck, J.; De Witte, Y.; Cnudde, V.; Boone, M.N.; Dewanckele, J.; Van Hoorebeke, L. Three-dimensional analysis of high-resolution X-ray computed tomography data with Morpho+. Microsc. Microanal. 2011, 17, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Dhaene, J.; Pauwels, E.; De Schryver, T.; De Muynck, A.; Dierick, M.; Van Hoorebeke, L. Arion: A realistic projection simulator for optimizing laboratory and industrial micro-CT. In Proceedings of the 2nd International conference on Tomography of Materials and Structures (ICTMS 2015), Québec, QB, Canada, 29 June 2015; pp. 60–64. [Google Scholar]
- Sutherland, I.W. The biofilm matrix–an immobilized but dynamic microbial environment. Trends Microbiol. 2001, 9, 222–227. [Google Scholar] [CrossRef]
- Zuberer, D.A. Recovery and enumeration of viable bacteria. Methods Soil Anal. Part 2 Microbiol. Biochem. Prop. 1994, 5, 119–144. [Google Scholar]
- Al-kafaween, M.A.; Hilmi, A.B.M.; Jaffar, N.; Al-Jamal, H.A.N.; Zahri, M.K. Determination of optimum incubation time for formation of Pseudomonas aeruginosa and Streptococcus pyogenes biofilms in microtiter plate. Bull. Natl. Res. Cent. 2019, 43, 100. [Google Scholar] [CrossRef]
- Turner, R.D.; Hurd, A.F.; Cadby, A.; Hobbs, J.K.; Foster, S.J. Cell wall elongation mode in Gram-negative bacteria is determined by peptidoglycan architecture. Nat. Commun. 2013, 4, 1–8. [Google Scholar] [CrossRef]
- Delhaye, A.; Collet, J.F.; Laloux, G. Fine-tuning of the Cpx envelope stress response is required for cell wall homeostasis in Escherichia coli. MBio 2016, 7, e00047-16. [Google Scholar] [CrossRef]
- Wang, G.; He, Y.; Wang, H.; Zhang, L.; Yu, Q.; Peng, S.; Wu, X.; Ren, T.; Zeng, Z.; Xue, Q. A cellulose sponge with robust superhydrophilicity and under-water superoleophobicity for highly effective oil/water separation. Green Chem. 2015, 17, 3093–3099. [Google Scholar] [CrossRef]
- Joshi, M.; Pant, H.; Tiwari, A.; Maharjan, B.; Liao, N.; kim, H.; Park, C.; Kim, C. Three-dimensional cellulose sponge: Fabrication, characterization, biomimetic mineralization, and in-vitro cell infiltration. Carbohydr. Polym. 2016, 136, 154–162. [Google Scholar] [CrossRef]
- Ye, N.; Jian, F.; Lai, W. Effect of voxel size and partial volume effect on accuracy of tooth volumetric measurements with cone beam CT. Dentomaxillofacial Radiol. 2013, 42, 20130070. [Google Scholar] [CrossRef]
- Epstein, W. The roles and regulation of potassium in bacteria. Prog. Nucleic Acid Res. Mol. Biol. 2003, 75, 293–320. [Google Scholar] [PubMed]
- Arney, D.R.; Phillips, C.J. The effects of changes in sodium and potassium concentration on the growth of mastitogenic bacteria in vitro. Int. J. Appl. Res. Vet. Med. 2005, 3, 242–248. [Google Scholar]
- Gründling, A. Potassium uptake systems in Staphylococcus aureus: New stories about ancient systems. MBio 2013, 4, e00784-13. [Google Scholar] [CrossRef] [PubMed]
- Robertson, M.; Hapca, S.M.; Moshynets, O.; Spiers, A.J. Air–liquid interface biofilm formation by psychrotrophic pseudomonads recovered from spoilt meat. Antonie Van Leeuwenhoek 2013, 103, 251–259. [Google Scholar] [CrossRef]
- Ghafoor, A.; Hay, I.D.; Rehm, B.H. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 2011, 77, 5238–5246. [Google Scholar] [CrossRef]
- Takahashi, Y. Binding properties of alginic acid and chitin. In Inclusion Phenomena in Inorganic, Organic, and Organometallic Hosts; Springer: Berlin/Heidelberg, Germany, 1987; pp. 417–426. [Google Scholar]


| List of Experiments | ON Culture of Bacteria at 30 C | Incubation of Bacteria in TSB 1/10 for 48 h | Replacement of TSB 1/10 and Further Incubation for 24 h | Application of 5% w/v of KBr to Fully Developed Biofilms | PBS Application * | PBS Wash ** | Serial Dilution and Plate Counting |
|---|---|---|---|---|---|---|---|
| Irradiation effect of X-rays on biofilms | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Diffusion experiment | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| In-situ Experiment for biofilm visualization | ✓ | ✓ | ✓ | ✓ | ✓ |
| Incubation Type | CFU/cm | Total Biomass (OD) | std | Biomass/Cell | std |
|---|---|---|---|---|---|
| Control | 3.62 × | 0.13 | 0.01 | 1.33 × | 2.03 × |
| 5% KBr | 2.25 × | 0.10 | 0.02 | 7.78 × | 6.02 × |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shastry, A.; Villanueva, X.; Steenackers, H.; Cnudde, V.; Robles, E.; Boone, M.N. Study on the Effect of Contrast Agent on Biofilms and Their Visualization in Porous Substrate Using X-ray μCT. Appl. Sci. 2020, 10, 5435. https://doi.org/10.3390/app10165435
Shastry A, Villanueva X, Steenackers H, Cnudde V, Robles E, Boone MN. Study on the Effect of Contrast Agent on Biofilms and Their Visualization in Porous Substrate Using X-ray μCT. Applied Sciences. 2020; 10(16):5435. https://doi.org/10.3390/app10165435
Chicago/Turabian StyleShastry, Abhishek, Xabier Villanueva, Hans Steenackers, Veerle Cnudde, Eric Robles, and Matthieu N. Boone. 2020. "Study on the Effect of Contrast Agent on Biofilms and Their Visualization in Porous Substrate Using X-ray μCT" Applied Sciences 10, no. 16: 5435. https://doi.org/10.3390/app10165435
APA StyleShastry, A., Villanueva, X., Steenackers, H., Cnudde, V., Robles, E., & Boone, M. N. (2020). Study on the Effect of Contrast Agent on Biofilms and Their Visualization in Porous Substrate Using X-ray μCT. Applied Sciences, 10(16), 5435. https://doi.org/10.3390/app10165435







