Abstract
A microorganism isolated and identified as Acinetobacter courvalinii was found to be able to perform sequential free cyanide (CN−) degradation, simultaneous nitrification and aerobic denitrification (SNaD); this ability was associated with the multiphase growth profile of the microorganism when provided with multiple nitrogenous sources. The effect of CN− on SNaD including enzyme expression, activity and protein functionality of Acinetobacter courvalinii was investigated. It was found that CN− concentration of 1.9 to 5.8 mg CN−/L did not affect the growth of Acinetobacter courvalinii. Furthermore, the degradation rates of CN− and ammonium-nitrogen (NH4-N) were found to be 2.2 mg CN−/L/h and 0.40 mg NH4-N/L/h, respectively. Moreover, five models’ (Monod, Moser, Generic Rate law, Haldane, and Andrews) ability to predict SNaD under CN− conditions, indicated that, only the Rate law, Haldane and Andrew’s models, were suited to predict both SNaD and CN− degradation. The effect of CN− on NH4-N, nitrate-nitrogen (NO3−) and nitrite-nitrogen (NO2−) oxidizing enzymes indicated that the CN− did not affect the expression and activity of ammonia monooxygenase (AMO); albeit, reduced the expression and activity of nitrate reductase (NaR) and nitrite reductase (NiR). Nevertheless, a slow decrease in NO2− was observed after the supplementation of CN− to the cultures, thus confirming the activity of NaR and the activation of the denitrification pathway by the CN−. These special characteristics of the Acinetobacter courvalinii isolate, suggests its suitability for the treatment of wastewater containing multiple nitrogenous compounds in which CN− is present.
1. Introduction
Untreated total nitrogen (TN)-containing wastewater in the environment can result in eutrophication, thus the treatment of such wastewater containing TN is crucial in wastewater treatment plants (WWTPs) [1]. This type of wastewater may be treated by either physicochemical or biological methods. Most municipal and industrial wastewater treatment plants use biological methods such as simultaneous nitrification and aerobic denitrification (SNaD) for the treatment of contaminated water due to the cost-effectiveness of such processes [2,3].
However, SNaD is sensitive to toxicant loading [4,5,6], particularly free cyanide (CN−) [5]. Physical and chemical methods have been used to reduce CN− effects on SNaD; however, these methods are relatively expensive, complex and produce undesirable by-products such as hypochlorite, and toxicant loaded sludge [7]. Hence, biological removal of CN− prior to SNaD has recently received the most attention. Biological CN− removal has been proven to be an ideal option because of its cost effectiveness and environmental benignity [8]. Although, biological CN− removal reduces the production of undesirable by-products, conducting CN− degradation in a separate reactor with SNaD in sequential two stage reactor systems, may result in the escalation of costs associated with the operation of the multiple independent reactors dedicated to the biodegradation of CN− and TN. Han et al. [9] proposed that CN− degrading and resistant microorganisms can be used for SNaD in order to lower the inhibition effect of CN− towards SNaD in a single reactor system.
Mekuto et al. [10] reported that Bacillus species were capable of degrading up to 300 mg CN−/L and subsequently utilize the by-products which were NH4-N, NO3-N and NO2-N. Additionally, Mpongwana et al. [11] isolated a CN− tolerant strain capable of simultaneous CN− degradation and SNaD. However, the applicability of CN− degrading and resistant microorganisms is still restricted due to the lack of information necessary to adequately control SNaD under high CN− loading conditions. Hence, this paper reports on the isolation of an organism that simultaneously degrades CN− while performing SNaD, with the phenomena of simultaneous CN− degradation and NH4-N removal being modeled by the determination of kinetic parameters using numerous decay models. The mechanism by which CN− affects the nitrification and denitrification enzymes is still not documented, thus making it difficult to apply CN− resistant microorganisms in SNaD systems, in which CN− laden wastewater is being treated. To also address this research gap, the expression and activity of ammonia monooxygenase (AMO), nitrate reductase (NaR) and nitrite reductase (NiR) present in the CN− resistant isolates’ supernatant, deemed suitable for this research, was also conducted.
2. Materials and Methods
2.1. Isolation and Identification of the Bacterial Isolate of Interest
The isolate used in this study was isolated from CN− containing waste at the Bioresource Engineering Research Group (BioERG) laboratory, Cape Peninsula University of Technology, Cape Town, South Africa. This organism was sub-cultured to obtain pure colonies and subjected to toxicant (CN−) resistance testing to determine the highest CN− concentration it can withstand subsequent to gram staining. The genomic DNA was extracted from the pure cultures using the Quick-DNA™, Fungal/Bacterial Miniprep kit, to target the 16S rRNA gene (Zymo Research, Irvine, CA, USA, Catalogue No. D6005). This region was amplified using the OneTaq® Quick-Load® 2X Master Mix (Zymo Research, Catalogue No. M0486) and the primers which were used were the 16S-27F and 16S-1492R with sequence (5’ to 3’) AGAGTTTGATCMTGGCTCAG, and CGGTTACCTTGTTACGACTT, respectively.
The PCR products were run on an agarose gel and the gel fragments were extracted using the Zymoclean™ Gel DNA Recovery Kit (Zymo Research, Catalogue No. D4001). The extracted fragments were sequenced in the forward and reverse direction using the Nimagen BrilliantDye™ Terminator Cycle Sequencing kit v3.1 and further purified using the Zymo Research, ZR-96 DNA Sequencing Clean-up kit™. The purified fragments were analyzed using the ABI 3500XLGenetic Analyzer (Applied Biosystems, ThermoFisher Scientific, Waltham, MA, USA). CLC Bio Main Workbench v7.6 was used to analyze the files generated by the ABI 3500XL Genetic Analyzer and results were obtained through a BLAST search (NCBI).
2.2. Batch Culture Experiments
A basal medium was used for SNaD and CN− degradation studies. Erlenmeyer flasks (250 mL) were used with a working volume of 100 mL of basal media. The basal media was composed of: 1.5 g KH2PO4, 7.9 g Na2HPO4, 0.5 g MgSO4.7H2O, and 1 mL of trace element solution per liter, was used. The trace element solution contained (per liter): 50 g EDTA, 2.2 g ZnSO4.7H2O, 5.5 g CaCl2, 5.06 g MnCl2.4H2O, 5.0 g FeSO4.7H2O, 1.1 g (NH4)6Mo7O2.4H2O, 1.57 g CuSO4.5H2O, 1.61 g CoCl2.6H2O. The media was inoculated with a loop full of an overnight agar grown bacterial culture. The inoculated media was initially grown for 72 h without NH4-N and CN−, thereafter; 20 mgNH4-N /L and 20 mg CN−/L were added to the inoculated basal medium whereby samples were collected on a 24 h interval for 168 h to analyze CN−, NH4-N, NO2−, NO3− and microbial growth at OD660 nm. The biomass concentration at OD660 nm was calculated by Equation (1) (from calibrated values in triplicate):
OD660 = 0.0089 (CFU) − 0.3097
2.3. Enzyme Extraction
To analyze the activities of the enzymes, i.e., AMO, NaR and NiR, cold acetone was added to the collected samples, forming a pellet that was separated from the supernatant by centrifuging each sample at 5000 g for 15 min. The pellet was lysed using a sonicator and the supernatant was collected by centrifugation at 16000 rpm for 5 min. Thereafter, the precipitate was washed three times and resuspended in phosphate (360 mg/L) buffer solution (pH 7.4). NH4-N and CN CN− solutions were prepared and the enzyme extracts were added into the NH4-N and CN− solutions, while the changes in CN− (09701), NH4-N (00683), NO2-N (110057) and NO3-N (14773) were monitored and analyzed using the Merck test kits (Merck Millipore., Burlington, USA).
The activity of the NH4-N oxidizing enzyme was monitored by measuring the changes in NH4-N after the addition of the enzyme extracts into the NH4-N solution. The formation of NO3-N from NO2-N and the disappearance of NO3-N were also monitored in order to determine the activity of both NO3-N and NO2-N oxidizing enzymes, i.e., NaR and NiR. The CN− degrading enzymes were monitored with the changes in the concentration of CN−.
2.4. Analytical Procedure(s)
All the test kits used for the analyses of the samples in this study were obtained from Merck SA a subsidiary of Merck Millipore., Burlington, USA. Furthermore, the Merck Spectroquant Nova 60 instrument was used to quantify the residual CN−, NH4-N, NO2-N, and NO3− concentrations. The tests were conducted as per the manufacturer’s instructions.
2.5. Kinetics Model Developed
During CN− biodegradation, microorganisms convert CN− to NH4-N which was then used as a nitrogenous source. Mekuto et al. [10] reported the ability of cyanide degrading bacteria to degrade CN− and subsequently utilize NH4-N, with a further indication that some microorganisms are able to use NH4-N and CN− as nitrogenous sources simultaneously. It has been shown by Mpongwana et al. [11] that when a cyanide degrading microorganism is used for nitrification and denitrification under cyanide conditions, the microorganism undergoes, multiple growth phases, i.e., mostly diauxic growth; hence, growth models that describe diauxic growth must be considered when evaluating growth kinetic parameters of such microorganisms when removing TN. Multiphase growth rate was first described by Monod [12] as a sequential utilization of substrates accompanied by distinct exponential growth phases during a period in which the individual substrates were utilized.
In this study, Monod’s and other models were also used to describe simultaneous CN− degradation and SNaD, with all the experiments being carried out in a closed system (1L batch culture). The rate of toxicant pollutant degradation can be described as in Equation (2):
where ra is the rate of pollutant degradation, Ki is the saturation constant, Ca is the concentration of the pollutant, −dCa/dt is the disappearance rate of the pollutant, and n is the model fitting constant.
When TN is the sole nitrogenous source, the system is considered to be a non-inhibited system. As such, Monods’ proposed kinetic model (Equation (3)) for non-inhibited systems, is deemed suitable to model such systems:
where Vm, is the maximum specific degradation rate of the pollutant.
Haldane [13] also represented pollutant degradation kinetics using a model as shown in Equation (4):
Due to the possibility of inhibition of SNaD by CN−, a model (Andrew’s model) representing the kinetics of a primary pollutants’ degradation in which a secondary inhibitory pollutant is present, was deemed necessary to use, as presented in Equation (5) [13]:
where Ks is a secondary pollutants’ inhibition constant.
2.6. Regression of Experimental Data and Estimation of Model Kinetic Parameters
Mathematical models describing CN− and NH4-N accumulation and degradation were fitted to the experimental data generated. Additionally, the models describing NH4-N degradation with pollutant inhibition were also fitted into NH4-N accumulation and accumulation experimental data. A nonlinear regression function in the Polymath 6.0 software was used for simulations, thus the generation of simulation data (using estimated kinetic parameters) that was compared to experimental data.
2.7. Data Handling and Kinetic Parameters
The data were computed and analyzed using Microsoft Excel v2016. The mean was determined using Equation (6); Moreover, Polymath 6.0 was used to estimate kinetic parameters in Equations (1), (4) and (6). The data obtained from polymath was plotted using Microsoft Excel v2016:
where is the sum of the data points, while n is the number of experimental experiments conducted.
3. Results and Discussion
3.1. Identification of Bacterial Isolate
The CN− resistant bacterial strain was characterized by sequencing 16sRNA using the universal primers 27F and 1492R to amplify the target region. The results were obtained by a BLAST search. The sequence on NCBI indicated that the isolated bacteria was Acinetobacter courvalinii, accession number AB602910.1/NR_148843.1.
3.2. Degradation Kinetics of Cyanide and NH4-N
An experiment was conducted to investigate the growth of A. courvalinii under CN− conditions, subsequent to CN− and TN biodegradation studies by the isolate. The growth of A. courvalinii was shown not to be affected by CN− concentration of up to 20 mg CN−/L; therefore, it was selected for the assessment of its ability to perform SNaD under cyanogenic conditions. The isolate was found to possess a special ability of sequential utilization of CN− and TN by exploiting its multi phased growth attributes, by initially utilizing of CN− and subsequently, NH4-N, which was evident by the appearance of a secondary logarithmic phase as seen in Figure 1A.
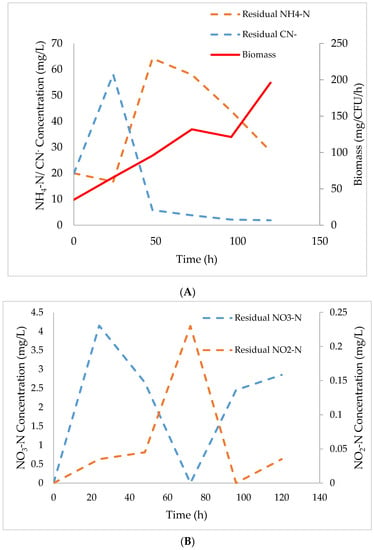
Figure 1.
Biodegradation kinetics of NH4-N and CN− by Acinetobacter courvalinii. (A): Growth of Acinetobacter courvalinii, sequential degradation of CN− and NH4-N. (B): NO2-N and NO3-N accumulation and degradation.
Figure 1A, demonstrates that A. courvalinii utilized CN− primarily, from a concentration of 20 mg CN−/L to 5.8 mg CN−/L followed by NH4-N. The rate of degradation for both CN− and NH4-N was 2.2 mg CN−/L/h and 0.40 mg NH4-N/L/h, respectively. Moreover, a cyanide-degrading consortium was also assessed (Figure S1 Supplementary file A) as it was having similar attributes to the isolate used in this study; albeit, due to a large percentage of organisms within the consortium being identified as unknown (Figure S2 Supplementary file A), the results were not included nor discussed herein. Overall, an increase of CN− in the first 24 h was observed, with a subsequent CN− decrease from 58.1 mg CN−/L to 5.8 mg CN−/L, being observed. The initial increase in CN− was associated with the activation of the ANR cascade which result in the production of HCN and denitrification enzymes (NAR, NIR, NOR, and N2OR) Figure 1A.
Arai et al. [14] reported the regulation of ANR cascade for transcription of denitrification enzymes resulting in the production of HCN by Pseudomonas aeruginosa. Moreover, the Pseudomonadaceae have been reported to possess the aerobic denitrification capability needed in the aiding and furtherance of SNaD [15]. A courvalinii exibited simillar charecteristics to those of Pseudomonas sp. Mpongwana et al. [10] also reported an increase in CN− concentration during the expression of denitrification enzymes. For this study, a decrease in NH4-N from 64.2 to 28.4 mgNH4-N/L was observed, indicating an initiation of nitrification from the threshold limit of 5.8 mg CN−/L Figure 1A. This indicated that a concentration of 5.8 mg CN−/L did not completely inhibit nitrification; albeit, this concentration is above the threshold concentration of CN− known to completely inhibit nitrification reaction [6]. Kim et al. [16] reported the complete inhibition of nitrification by 1 mg CN−/L. The inhibition is largely associated with enzyme redundancy, non-production and deactivation.
The NAR which is activated during the regulation of ANR is a significant enzyme that aid in aerobic respiration; thus, converting NO3-N back into NO2-N during aerobic denitrification [17]. The increase of CN− during the experiment, confirmed the ability of the A courvalinii to induce ANR advocated for its ability to carry out SNaD. Minute accumulation of NO2-N was observed compared to NO3-N which was produced in significant quantities in the first 24 h of the experiments (Figure 1B). This was deemed normal as some studies have reported that NO2-N accumulate in insignificant quantities during SNaD [18]. The NO2-N increase between 48–72 h, was associated with the conversion of NO3-N to NO2-N catalised by NAR during aerobic respiration culminating in the distinct feature of SNaD, a phenomena also supported by the observed decrease in NO3-N between 24 and 72 h which corresponded with the increase of NO2-N, confirming the ability of A courvalinii to carry-out aerobic respiration thus performing SNaD.
A multi-phased growth, as observed in Figure 1A, confirmed multiple distinct dual lag phases with a stationery phase being observed between 72–96 h. This stationery phase was associated with biomass switching utilization from one nitrogenous source to another, i.e., from using CN− to NH4-N. The first lag phase from 0–72 h was linked with the isolates’ growth as a result of CN− presence, using it as a source of nitrogen, while the second lag phase from 96–120 h was linked with the growth of A courvalinii as a result of NH4-N consumption and/or removal, with growth rate being 1.35 and 3.12 h−1 for first and second growth phases, respectively. This indicated that A courvalinii grows well when supported by less toxic nitrogenous source (NH4-N) than when growing in a inhibitory toxicant (CN−) even if it’s a nitrogenous source which can be utilized by the organism. However, the rate of NH4-N removal was low thus, optimization studies are needed, in order to increase the nitrification efficiency, which is the initial step in SNaD. Subsequent to the monitoring of TN and CN− removal, five models were used to assess their predictive ability of SNaD under CN− conditions, with kinetic parameters in each model being determined.
3.3. Model Parameter Determination and Statistical Analysis
The predictive capability of the three models on NH4-N degradation was analyzed. This was done by simulating the predicted model data in comparison to experimental data and also by plotting parity plots to assess the relationship between predicted data with the observed experimental values. The selected models included the Rate law, Haldane and Andrew’s models. The experimental data was fitted into the models to estimate kinetic constant in each model, using Polymath 6.0 software. The Rate law and Haldane models adequately described the experimental data of the nitrification step with a determination coefficient (R2) of 0.91 and 0.92, respectively (Table 1).

Table 1.
Estimated kinetic parameter values for the models for NH4-N degradation, a rate limiting step in nitrification.
Furthermore, the adjusted determination coefficient (Adj R2) was 0.89 and 0.89 respectively, with such a high Adj R2 signifying a high significance, thus suitability of the estimated model kinetic constants. Furthermore, the variance was 0.015 and 0.016 for the generic rate law and the Haldane model, respectively. This variance is very low and is evidence that the predicted (modelled) values generated using estimated kinetic parameters did not differ much from experimental data. Similarly, Andrews model generated data with substrate inhibition was also compared to experimental data. The model gave a better fit with a correlation coefficient of 0.94 and a linear correlation efficient of 0.98 and an Adj R2 0.90.
The average standard deviations were compared for the selection of the better predicting model for NH4-N degradation as a limiting step, with standard deviation of Haldane, Rate law and Andrew’s model being 0.041, 0.043, and 0.036, respectively. However, for the selection of the model that is adequate in predicting multiple nitrogenous pollutants (CN−, NH4-N, NO2-N and NO3-N) removal, the prediction ability of the model towards CN− degradation needed to be considered. The parity plot indicated a satisfactory correlation between the experimental rate of NH4-N degradation and the predicted rates with R2 being 0.96, 0.92, and 0.94 for Rate law, Haldane and Andrew’s model respectively.
Moreover, the variance for Andrews’s model also proved the significance of the model with variance being 0.015. This variance was minute which indicated that there was a negligible difference between the model and experimental data (see Figure 2 and Table 2). Monod and Moser models were also assessed for their predictive ability of NH4-N (Figure S3 and Table S1, Supplementary file A) and CN− (Figure S4 and Table S2, Supplementary file A) degradation rates; however, they did not fit well into the experimental data.
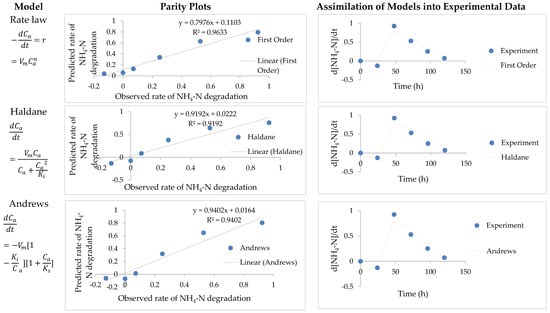
Figure 2.
Parity plots of predicted values versus experimental values. Assimilations of the model data into NH4-N degradation experimental data (Rate law, Haldane, and Andrew).

Table 2.
Estimated values of kinetic parameters for the models for CN− degradation.
Overall, the models used and reported in this research also fitted the CN− degradation data well (Figure 3). The models that gave a better fit were the rate law (R2 = 0.92, Adj R2 = 0.90, variance = 0.052 and standard deviation = 0.076) and Haldane model (R2 = 0.95, adj R2 = 0.94, variance = 0.034 and standard deviation= 0.061)—see Table 3. Andrew models’ had a poor prediction of CN− degradation with (R2 = −0.99, adj R2 = −2.31, variance = 1.75, and standard deviation = 0.38). This indicated that CN− degradation was not inhibited by the presence of NH4-N, as some organism would rather alternatively use the less inhibitory pollutant; although, inhibitory compound biodegradation might be a favorable alternative. Andrew’s model was developed to predict substrate utilization in systems with substrate inhibition, thus the models’ inability to model CN− removal.
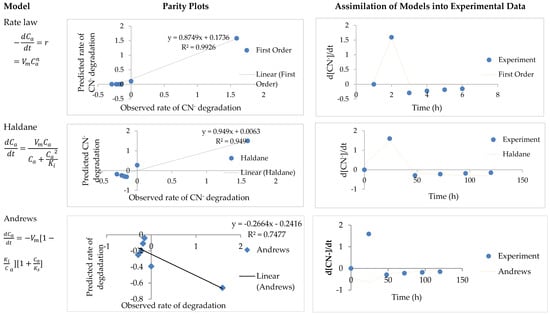
Figure 3.
Parity plots of predicted values versus experimental values. Assimilations of the model data into CN− degradation experimental data (Rate law, Haldane, and Andrews).

Table 3.
Kinetic parameters obtained from different studies assessing nitrification and aerobic denitrification.
Ge et al. [19] studied the kinetics of SNaD under high phenol concentrations using bacterial strains capable of phenol degradation, heterotrophic nitrification, and aerobic denitrification. Additionally, Vasiliadou et al. [20] also studied the kinetics of denitrification with nitrate inhibition. Additionally, Li et al. [21] also determined that the Haldane model predicted CN− well with R2 being 0.99 These studies indicated the feasibility of SNaD, even under inhibitory pollutant loading. Moreover, the kinetic parameters obtained were comparable to those obtained for this study (Table 3).
Overall, the variance and standard deviation of the model showed that all three models predicted NH4-N degradation better as opposed to CN− degradation. This was shown with the higher variances and standard deviation of CN− degradation than those of NH4-N degradation. Although, all three models adequately represented NH4-N removal with high R2 and adj R2, the rate law and Andrews models had higher average standard deviation and variance for predicted rate of CN− degradation in comparison to actual experimental data (Figure 3); thus rendering unusable as candidate prediction models for removal systems with multiple nitrogenous pollutants.
Moreover, Haldanes’ model was selected as a better predictor of NH4-N removal under CN− conditions. This decision was based on its standard deviation and the variance being the lowest for both NH4-N and CN− degradation, indicating that the predicted rate of NH4-N and CN− by Haldane had a minute deviation from the experimental data thus qualifying it as a better model to be used for multiple nitrogenous pollutants systems, especially, SNaD.
The result obtained it this study were consistent with those that were previously reported by Pradhan et al. [22] describing dual substrate kinetics of ammonia oxidation by Haldane model, with R2 > 0.98 and adj R2 > 0.98 and low standard errors (RMSE) < 0.61. Additionally, Wang et al. [23] reported a high correlation between Haldane prediction and experimental data with adjusted R2 (0.995), Ks (2.997 ± 0.041 mg/L) and Ki (64.736 ± 0.023 mg/L).
3.4. Effect of Cyanide on AMO, NaR, and NiR
Enzyme inhibition can occur in different ways such as the binding of an inhibitor onto an active site or for an example a site of the AMO responsible for NH4-N oxidation. The second inhibition mechanism can involve the removal of the Cu co-factor of AMO which in turn affects the activity of AMO [24]. To assess the effect of CN− towards SNaD, enzyme extracts from A. courvalinii cultivated in batch cultures were used. The effect of CN− towards the expression of NH4-N (AMO), NO3-N (NaR) and NO2-N (NiR) oxidizing enzymes was evaluated (Figure 4A).
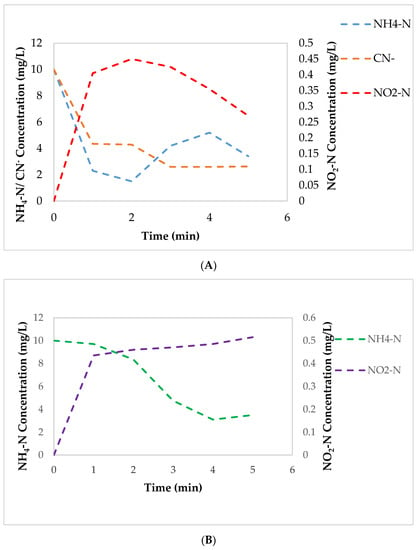
Figure 4.
The activity of NH4-N, NO3-N and NO2-N oxidizing enzymes and CN− degrading enzyme. (A): Effect of CN− on the induction of NH4-N, NO3-N and NO2-N oxidizing enzymes by A. courvalinii. (B): Effect of CN− on free cell NH4-N, NO3-N and NO2-N oxidizing enzymes.
The activity and presence of AMO, NaR, and NiR was conducted using cell-free extracts by the addition of the enzyme solution into solutions of NH4-N. The results revealed that A. courvalinii was able to express AMO, and not NaR and NiR; this was shown by the decrease of NH4-N and the accumulation of NO2-N; however, there was minimal NO2-N oxidation observed thus minimal NO3-N formation and accumulation detected (see Figure 4B). This observation led to a hypothesis that CN− inhibited the expression of NO3-N and NO2-N oxidizing enzymes or results in the excretion of non-active NO3-N and NO2-N oxidizing enzymes.
In addition, the competitive inhibition by the addition of CN− to free cell extract was examined. This experiment aimed to determine the effect of CN− on the expressed NH4-N, NO3-N, and NO2-N oxidizing enzymes, of which the outcomes indicated that the presence of CN− did not have an effect on the NH4-N reducing enzyme; albeit, a slow decrease of NO2-N was observed which suggested the activation of denitrification. Additionally, no NO3-N accumulation was observed which meant that A. courvalinii converted NO2-N directly to N2.
The activation of NO2-N genes when CN− was added to bacterial cultures, could be a confirmation that A. courvalinii expresses an ANR protein in response to the presence of CN− as a protective mechanism for the cyanide-resistant bacteria used in this research in order to mitigate against the toxicity of the CN−. Furthermore, the activation of this protein can lead to the induction of NAR, NIR, NOR and N2OR although this was not confirmed in this study [1].
4. Conclusions
The growth of Acinetobacter courvalinii accession number AB602910.1/NR_148843.1 was not affected by CN−. Furthermore, this microorganism was shown to carry-out SNaD under higher CN− concentration than the threshold concentration which is known to completely inhibit SNaD. The predictive ability of the rate law, Haldane, and Andrew’s models was assessed with results indicating that the rate law, Haldane, and Andrew’s models were better models to predict NH4-N removal as the initial step in SNaD, with high R2 and adj R2; thus, the evaluation of models ability to also predict CN− degradation, with the standard deviation and variance being a criterion for selection of the better predicting model. The Haldane model was found to have the lowest standard deviation for both CN− and NH4-N removal among all models evaluated. Thus it’s selection as a model that is suitable to predict the detoxification of TN in wastewater, using SNaD even under CN− laden conditions.
The influence of CN− on AMO, NaR, and NiR also showed that CN− did not affect the expression and activity of NH4-N oxidizing enzymes and the assumption was made that non-viable NO3-N and NO2-N reducing enzymes were expressed. This hypothesis comes from the observation of the slow decrease in NO2-N after the addition of CN− on crude enzyme extract assays, confirming the activity of NO3-N reducing enzyme and activation of the denitrification pathway by the CN−. Although A. courvalinii have been shown to be able to detoxify TN under CN− conditions, its physico-chemical parameters still need to be investigated in order to optimize the degradation efficiency of isolate, A. courvalinii
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3417/10/14/4823/s1, Figure S1: Simultaneous nitrification and aerobic denitrification performed by cyanide degrading consortia under high cyanide conditions. Figure S2: Metagenomics report for the consortium. A: Kingdom classification. A: Phylum classification. C: Class classification. D; Order classification. E: Family classification. Figure S3: Parity plots of predicted values versus experimental values. Assimilations of the model data into NH4-N degradation experimental data (Monod and Moser). Figure S4: Parity plots of predicted values versus experimental values. Assimilations of the model data into CN- degradation experimental data (Monod, Moser and Andrews). Table S1: Estimated values of kinetic parameters for the models for NH4-N degradation, a limiting step in nitrification. Table S2: Estimated values of kinetic parameters for the models for CN- degradation, a limiting step in nitrification.
Author Contributions
Conceptualization, N.M.; Formal analysis, N.M., E.I.O., S.K.O.N., L.C.R. and B.S.C.; Methodology and investigation, N.M., C.D. and M.R.M.; Funding acquisition, S.K.O.N.; Investigation, N.M.; Project administration, S.K.O.N., B.S.C., L.C.R. and E.I.O.; Resources, S.K.O.N.; Supervision, S.K.O.N., B.S.C. and E.I.O.; Writing–original draft, N.M.; Data interpretation, review & editing, N.M., S.K.O.N., E.I.O., L.C.R. and B.S.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research and the APC were funded by [Cape Peninsula University of Technology], grant number [URF RK16)].
Acknowledgments
The authors would like to thank the postgraduate students of Bioresource Engineering Research Group (BioERG), Faculty of Applied Sciences, CPUT for their technical support.
Conflicts of Interest
The authors declare that there is no conflict of interest related to this study.
References
- Duan, J.; Fang, H.; Su, B.; Chen, J.; Lin, J. Characterization of a halophilic heterotrophic nitrification–aerobic denitrification bacterium and its application on treatment of saline wastewater. Bioresour. Technol. 2015, 179, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Wang, S.; Yang, X.; Qiu, S.; Li, B.; Peng, Y. Detection of nitrifiers and evaluation of partial nitrification for wastewater treatment: A review. Chemosphere 2015, 140, 85–98. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Li, Z.; Sun, Q.; Xu, Y.; Ye, Q. Heterotrophic nitrification and aerobic denitrification by Pseudomonas tolaasii Y-11 without nitrite accumulation during nitrogen conversion. Bioresour. Technol. 2016, 200, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Daims, H.; Lebedeva, E.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A.; et al. Complete nitrification by Nitrospira bacteria. Nature 2015, 528, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Puyol, D.; Carvajal-Arroyo, J.; Sierra-Alvarez, R.; Field, J. Inhibition of anaerobic ammonium oxidation by heavy metals. J. Chem. Technol. Biotechnol. 2014, 90, 830–837. [Google Scholar] [CrossRef]
- Kim, Y.; Park, D.; Lee, D.; Park, J. Inhibitory effects of toxic compounds on nitrification process for cokes wastewater treatment. J. Hazard. Mater. 2008, 152, 915–921. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, V.; Li, X.; Elk, M.; Chandran, K.; Impellitteri, C.; Santo Domingo, J. Impact of Heavy Metals on Transcriptional and Physiological Activity of Nitrifying Bacteria. Environ. Sci. Technol. 2015, 49, 13454–13462. [Google Scholar] [CrossRef]
- Inglezakis, V.; Malamis, S.; Omirkhan, A.; Nauruzbayeva, J.; Makhtayeva, Z.; Seidakhmetov, T.; Kudarova, A. Investigating the inhibitory effect of cyanide, phenol and 4-nitrophenol on the activated sludge process employed for the treatment of petroleum wastewater. J. Environ. Manag. 2017, 203, 825–830. [Google Scholar] [CrossRef]
- Han, Y.; Jin, X.; Wang, Y.; Liu, Y.; Chen, X. Inhibitory effect of cyanide on nitrification process and its eliminating method in a suspended activated sludge process. Environ. Sci. Pollut. Res. Int. 2014, 21, 2706–2713. [Google Scholar] [CrossRef]
- Mekuto, L.; Ntwampe, S.K.O.; Jackson, V.A. Biodegradation of free cyanide and subsequent utilisation of biodegradation by-products by Bacillus consortia: Optimisation using response surface methodology. Environ. Sci. Pollut. Res. Int. 2015, 22, 10434–10443. [Google Scholar] [CrossRef]
- Mpongwana, N.; Ntwampe, S.; Mekuto, L.; Akinpelu, E.; Dyantyi, S.; Mpentshu, Y. Isolation of high-salinity-tolerant bacterial strains, Enterobacter sp., Serratia sp., Yersinia sp., for nitrification and aerobic denitrification under cyanogenic conditions. Water Sci. Technol. 2016, 73, 2168–2175. [Google Scholar] [CrossRef] [PubMed]
- Monod, J. The growth of bacterial cultures. Annu. Rev. Microbiol. 1942, 3, 371–394. [Google Scholar] [CrossRef]
- Annuar, M.S.M.; Tan, I.K.P.; Ibrahim, S.; Ramachandran, K.B. A kinetic model for growth and biosynthesis of medium-chain-length poly-(3-hydroxyalkanoates) in Pseudomonas putida. Braz. J. Chem. Eng. 2008, 25, 217–228. [Google Scholar] [CrossRef]
- Arai, H.; Kodama, T.; Igarashi, Y. Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa. Mol. Microbiol. 1997, 25, 1141–1148. [Google Scholar] [CrossRef] [PubMed]
- Yunjie, R.; Mohammad, J.T.; Dedong, K.; Huifeng, L.; Heping, Z.; Xiangyang, X.; Yu, L.; Lei, C. Nitrogen Removal Performance and Metabolic Pathways Analysis of a Novel Aerobic Denitrifying Halotolerant Pseudomonas balearica strain RAD-17. Microorganism 2020, 8, 72. [Google Scholar]
- Kim, Y.; Park, D.; Lee, D.; Park, J. Instability of biological nitrogen removal in a cokes wastewater treatment facility during summer. J. Hazard. Mater. 2007, 141, 27–32. [Google Scholar] [CrossRef]
- Mpongwana, N.; Ntwampe, S.K.O.; Omodanisi, E.I.; Chidi, B.S.; Razanamahandry, L.C. Sustainable Approach to Eradicate the Inhibitory Effect of Free-Cyanide on Simultaneous Nitrification and Aerobic Denitrification during Wastewater Treatment. Sustainability 2019, 11, 6180. [Google Scholar] [CrossRef]
- Sin, G.; Kaelin, D.; Kampschreur, M.J.; Takacs, I.; Wett, B.; Gernaey, K.V.; Rieger, L.; Siegrist, H.; van Loosdrecht, M. Modelling nitrite in wastewater treatment systems: A discussion of different modelling concepts. Water Sci. Technol. 2008, 58, 1155–1171. [Google Scholar] [CrossRef]
- Ge, Q.; Yue, X.; Wang, G. Simultaneous heterotrophic nitrification and aerobic denitrification at high initial phenol concentration by isolated bacterium Diaphorobacter sp. PD-7. Chin. J. Chem. Eng. 2015, 23, 835–841. [Google Scholar] [CrossRef]
- Vasiliadou, I.A.; Pavlou, S.; Vayenas, D.V. A kinetic study of hydrogenotrophic denitrification. Process Biochem. 2006, 41, 1401–1408. [Google Scholar] [CrossRef]
- Li, Q.; Lu, H.; Yin, Y.; Qin, Y.; Tang, A.; Liu, H.; Liu, Y. Synergic effect of adsorption and biodegradation enhance cyanide removal by immobilized Alcaligenes sp. strain DN25. J. Hazard. Mater. 2019, 364, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, N.; Thi, S.S.; Wuertz, S. Inhibition factors and kinetic model for anaerobic ammonia oxidation in a granular sludge bioreactor with Candidatus Brocadia. Chem. Eng. J. 2019, 123618. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.; Zhang, Y.; Zhang, J.; Li, J.; Wang, S.; Chen, G. Isolation and characterization of Acinetobacter sp. JQ1004 and evaluation of its inhibitory kinetics by free ammonia. Desalin. Water Treat. 2019, 147, 316–325. [Google Scholar] [CrossRef]
- Ruser, R.; Schulz, R. The effect of nitrification inhibitors on the nitrous oxide (N2O) release from agricultural soils—A review. J. Plant Nutr. Soil Sci. 2015, 178, 171–188. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).