A Reproducible Method for Growing Biofilms on Polystyrene Surfaces: Biomass and Bacterial Viability Evolution of Pseudomonas fluorescens and Staphylococcus epidermidis
Abstract
1. Introduction
2. Materials and Methods
2.1. Pre-Culture Preparation
2.2. Levels of Inoculum Density and TSB Concentrations
2.3. Biofilms Growth on a Defined Area
2.4. Enumeration of the Viable Cells and Optical Density Measurments
2.4.1. Enumeration of Biofilm Population Density through Viable Cell Counts
2.4.2. Biofilm Biomass Quantification through Optical Density Using Crystal Violet Assay
2.5. Confocal Laser Scanning Microscope
2.6. Mathematical Modelling of Biofilm Growth
2.7. Method Validation on Staphylococcus Epidermidis
2.8. Statistical Analysis
3. Results
3.1. Deveplopment of Biofilms on Defined Circular Areas
3.2. Growth Medium Concentration
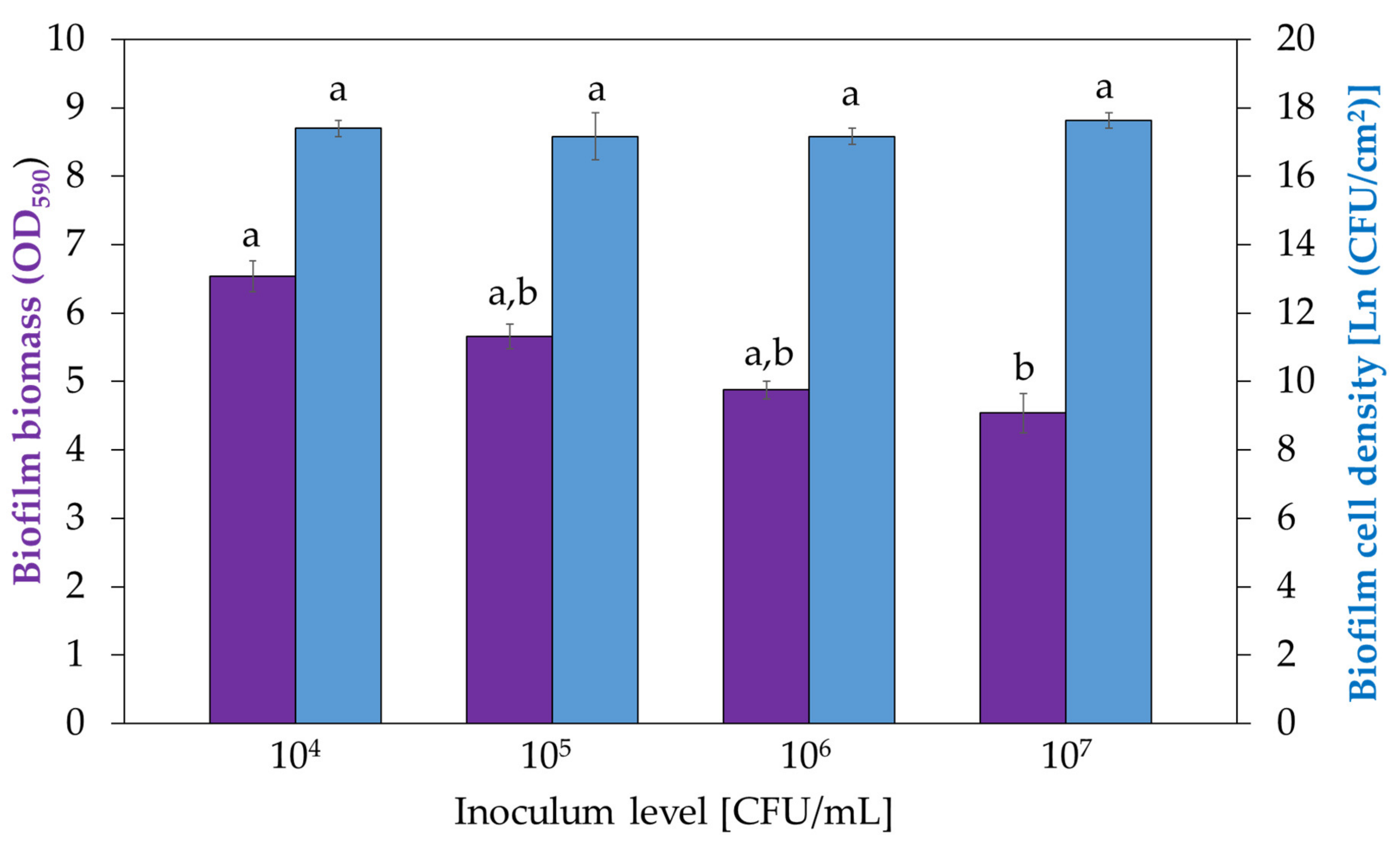
3.3. Influence of Inoculum Density on Biofilm Development
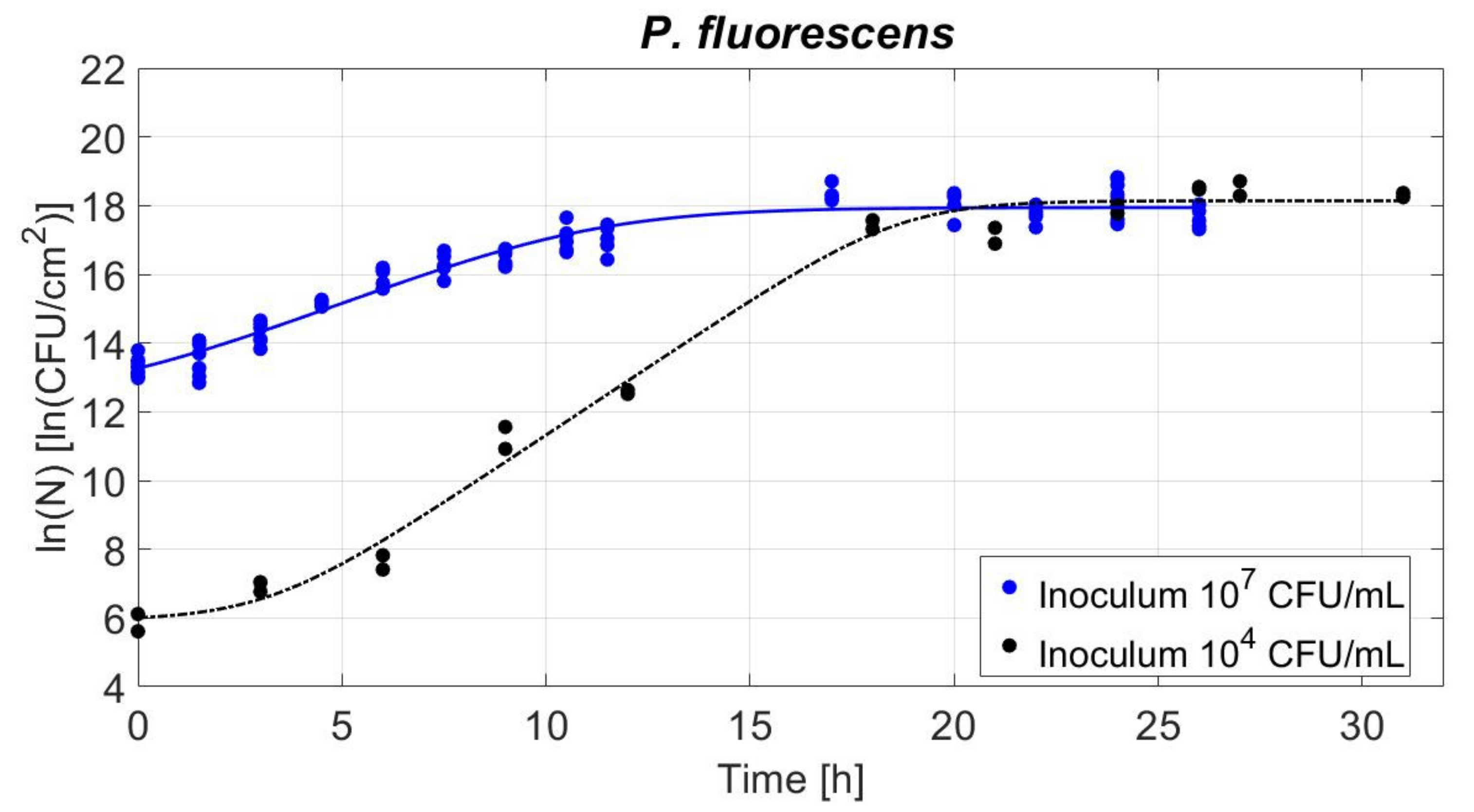
3.4. Biofilm Development in Time for Two Inoculum Levels
3.4.1. Study of Biofilm Population Density: Viable Cell Counts
3.4.2. Study of the Biofilm Biomass: Optical Density of CV Assay
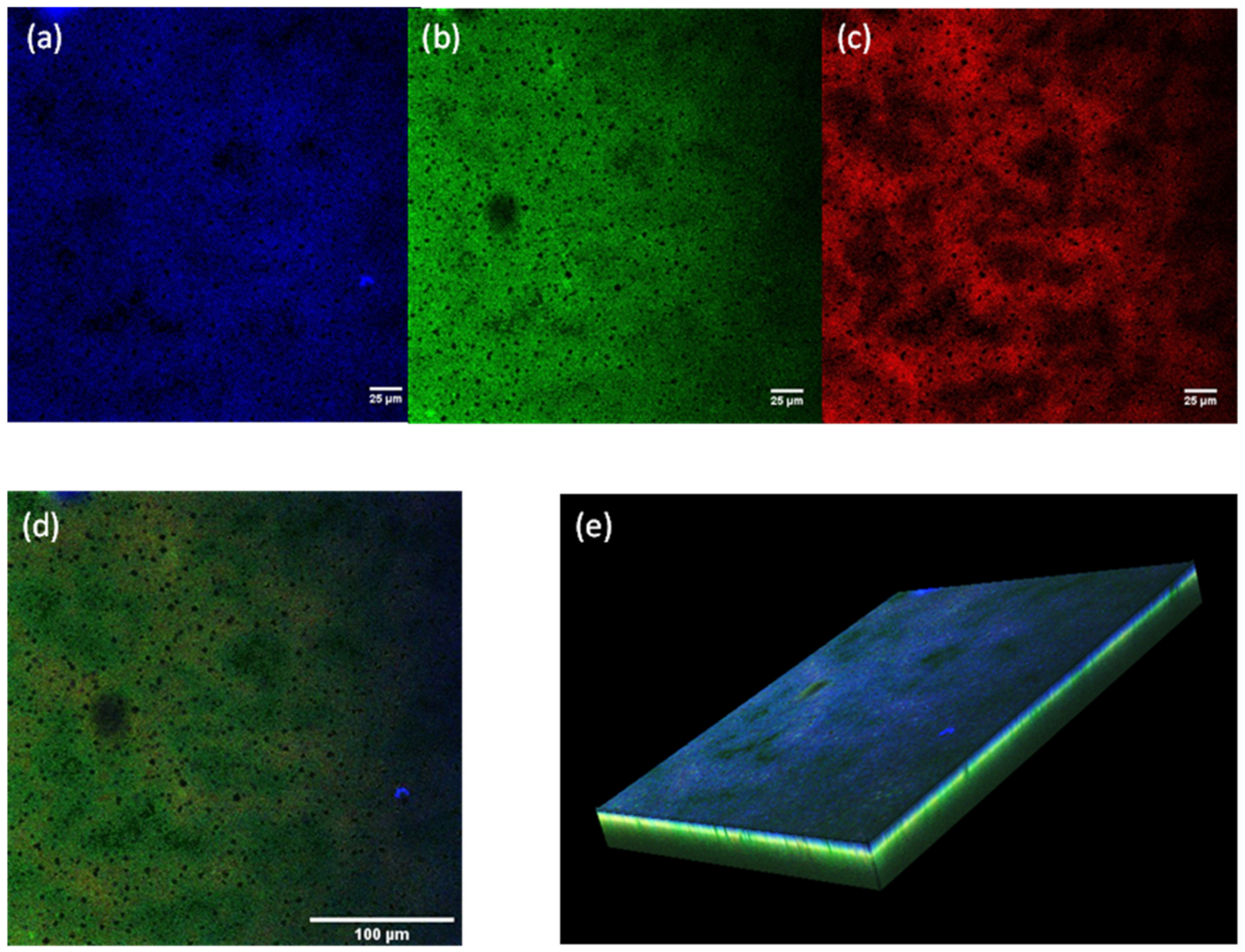
3.5. Reproducibility and Confocal Laser Scanning Microscopy Images
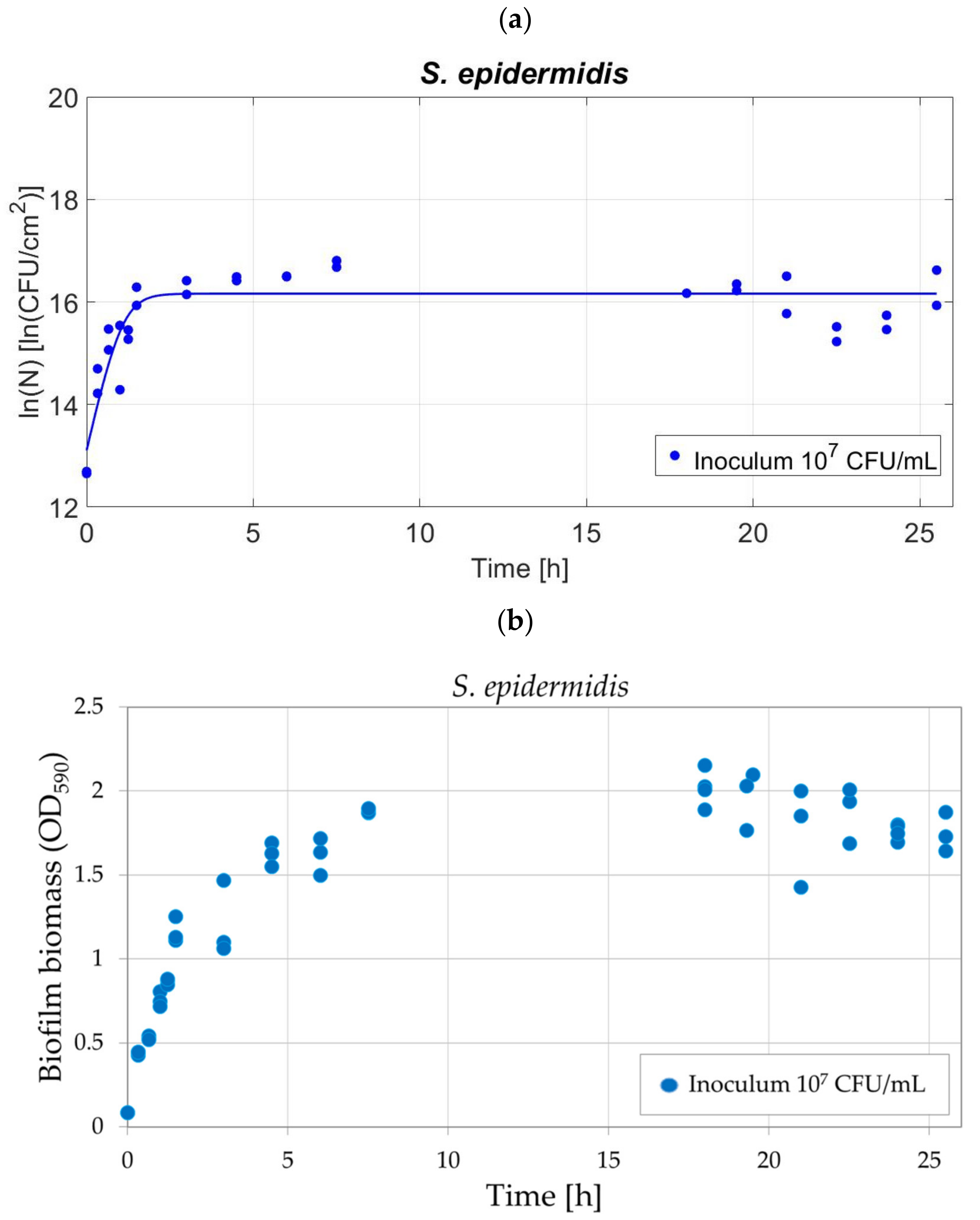
3.6. Method Validation on S. epidermidis
4. Discussion
4.1. Developing Strongly Attached Biofilms
4.2. Biofilm Dyamics for Two Inoculum Levels
4.2.1. Biofilm Population Density: Growth Parameters
4.2.2. Biofilm Biomass: Optical Density Evolution and Pellicles
4.3. Understanding Biofilm Development and Growth
4.4. Validation of the Protocol for Different Bacterial Strain: S. epidermidis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hentzer, M.; Teitzel, G.M.; Balzer, G.J.; Heydorn, A.; Molin, S.; Givskov, M.; Parsek, M.R. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 2001, 183, 5395–5401. [Google Scholar] [CrossRef] [PubMed]
- Baum, M.M.; Kainović, A.; O’Keeffe, T.; Pandita, R.; McDonald, K.; Wu, S.; Webster, P. Characterization of structures in biofilms formed by a Pseudomonas fluorescens isolated from soil. BMC Microbiol. 2009, 9, 103. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Hung, C.-C.; Santschi, P.H.; Gillow, J.B. Isolation and characterization of extracellular polysaccharides produced by Pseudomonas fluorescens Biovar II. Carbohydr. Polym. 2005, 61, 141–147. [Google Scholar] [CrossRef]
- Philips, J.; Rabaey, K.; Lovley, D.R.; Vargas, M. Biofilm formation by Clostridium ijungdahlii is induced by sodium chloride stress: Experimental evaluation and transcriptome analysis. PLoS ONE 2017, 12, e0170406. [Google Scholar] [CrossRef]
- Parsek, M.R.; Val, D.L.; Hanzelka, B.L.; Cronan, J.J.E.; Greenberg, E.P. Acyl homoserine-lactone quorum-sensing signal generation. PNAS 1999, 96, 4360–4365. [Google Scholar] [CrossRef]
- Dewanti, R.; Wong, A.C.L. Influence of culture conditions on biofilm frmation by Escherichia coli O157:H7. Int. J. Food Microbiol. 1995, 26, 147–164. [Google Scholar] [CrossRef]
- Speranza, B.; Corbo, M.R.; Sinigaglia, M. Effects of nutritional and environmental conditions on Salmonella sp. biofilm formation. J. Food Sci. 2011, 1, 12–16. [Google Scholar] [CrossRef]
- Bonin, E.; dos Santos, A.R.; Fiori da Silva, A.; Favero, M.E.; Campanerut-Sá, P.A.Z.; de Freitas, C.F.; Caetano, W.; Hioka, N.; Mikcha, J.M.G. Photodynamic inactivation of foodborne bacteria by eosin Y. J. Appl. Micorbiol. 2018, 124, 1617–1628. [Google Scholar] [CrossRef]
- Bonneville, L.; Ortiz, S.; Maia, V.; Brito, L.; Martínez-Suárez, J.V. Strain and Growth Conditions may Regulate Resistance of Listeria monocytogenes Biofilms to Benzalkonium Chloride. Appl. Sci. 2020, 10, 988. [Google Scholar] [CrossRef]
- Galdiero, E.; Di Onofrio, V.; Maione, A.; Gambino, E.; Gesuele, R.; Menale, B.; Ciaravolo, M.; Carraturo, M.; Guida, M. Allium ursinum and Allium oschaninii against Klebsiella pneumoniae and Candida albicans Mono- and Polymicrobic Biofilms in In Vitro Static and Dynamic Models. Microorganisms 2020, 8, 336. [Google Scholar] [CrossRef] [PubMed]
- Vikram, A.; Bomberger, J.M.; Bibby, K.J. Efflux as a glutaraldehyde resistance mechanism in Pseudomonas fluorescens and Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2015, 59, 3433–3440. [Google Scholar] [CrossRef] [PubMed]
- Allen, A.; Habimana, O.; Casey, E. The effects of extrinsic factors on the structural and mechanical properties of Pseudomonas fluorescens biofilms: A combined study of nutrient concentrations and shear conditions. Colloids Surf. B Biointerfaces 2018, 165, 127–134. [Google Scholar] [CrossRef]
- Skowron, K.; Wiktorczyk, N.; Kwiecińska-Piróg, J.; Sękowska, A.; Wałecka-Zacharska, E.; Gospodarek-Komkowska, E. Elimination of Klebsiella pneumoniae NDM from the air and selected surfaces in hospital using radiant catalytic ionization. Lett. Appl. Microbiol. 2019, 69, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Cirković, I.; Ranin, L.; Svabić-Vlahović, M. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett. Appl. Microbiol. 2004, 38, 428–432. [Google Scholar] [CrossRef]
- Pousti, M.; Zarabadi, M.P.; Amirdehi, M.A.; Paquet-Mercier, F.; Greener, J. Microfluidic bioanalytical flow cells for biofilm studies: A review. Analyst 2019, 144, 68–86. [Google Scholar] [CrossRef]
- Passerini, D.; Fécamp, F.; Marchand, L.; Kolypczuk, L.; Bonnetot, S.; Sinquin, C.; Verrez-Bagnis, V.; Hervio-Heath, D.; Colliec-Jouault, S.; Delbarre-Ladrat, C. Characterization of biofilm extracts from two marine bacteria. Appl. Sci. 2019, 9, 4971. [Google Scholar] [CrossRef]
- Lorenzo, F.; Sanz-Puig, M.; Bertó, R.; Orihuel, E. Assessment of performance of two rapid methods for on-site control of microbial and biofilm contamination. Appl. Sci. 2020, 10, 744. [Google Scholar] [CrossRef]
- Duddridge, J.; Kent, C.; Laws, J. Effect of surface shear stress on the attachment of Pseudomonas fluorescens to stainless steel under defined flow conditions. Biotechnol. Bioeng. 1982, 24, 153–164. [Google Scholar] [CrossRef]
- Assanta, M.A.; Roy, D.; Montpetit, D. Adhesion of Aeromonas hydrophila to water distribution system pipes after different contact times. J. Food Prot. 1998, 61, 1321–1329. [Google Scholar] [CrossRef]
- Chen, M.J.; Zhang, Z.; Bott, T.R. Effects of operating conditions on the adhesive strength of Pseudomonas fluorescens biofilms in tubes. Colloids Surf. B Biointerfaces 2005, 43, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Baranyi, J.; Roberts, T.A. Mathematics of predictive food microbiology. Int. J. Food Microbiol. 1994, 95, 199–218. [Google Scholar] [CrossRef]
- Kozajda, A.; Jeżak, K.; Kapsa, A. Airborne Staphylococcus aureus in different environments—A review. Environ. Sci. Pollut. Res. 2019, 26, 34741–34753. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Stanley, N.R.; Lazazzera, B.A. Environmental signals and regulatory pathways that influence biofilm formation. Mol. Microbiol. 2004, 52, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Schellenberg, J.; Leder, H.-J. Syndiotactic polystyrene: Process and applications. Adv. Polym. Technol. 2006, 25, 141–151. [Google Scholar] [CrossRef]
- Ho, B.T.; Roberts, T.K.; Lucas, S. An overview on biodegradation of polystyrene and modified polystyrene: The microbial approach. Crit. Rev. Biotechnol. 2018, 38, 308–320. [Google Scholar] [CrossRef]
- Guruvenket, S.; Rao, G.M.; Komath, M.; Raichurc, A.M. Plasma surface modification of polystyrene and polyethylene. Appl. Surf. Sci. 2004, 236, 278–284. [Google Scholar] [CrossRef]
- Lerman, M.J.; Lembong, J.; Muramoto, S.; Gillen, G.; Fisher, J.P. The evolution of polystyrene as a cell culture material. Tissue Eng. Part B Rev. 2018, 24, 359–372. [Google Scholar] [CrossRef]
- Janek, T.; Czyżnikowska, Ż.; Łukaszewicz, M.; Gałęzowska, J. The effect of Pseudomonas fluorescens biosurfactant pseudofactin II on the conformational changes of bovine serum albumin: Pharmacheutical and biomedical applications. J. Mol. Liquids 2019, 288, 111001. [Google Scholar] [CrossRef]
- Ude, S.; Arnold, D.L.; Moon, C.D.; Timms-Wilson, T.; Spiers, A.J. Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ. Microbiol. 2006, 8, 1997–2011. [Google Scholar] [CrossRef] [PubMed]
- Scale, B.S.; Dickson, R.P.; LiPuma, J.J.; Huffnagle, G.B. Microbiology, genomics, and clinical significance of the Pseudomonas fluorescens scpecies complex, an unappreciated colonizer of humans. Clin. Microbiol. Rev. 2014, 27, 927–948. [Google Scholar] [CrossRef] [PubMed]
- Banin, E.; Vasil, M.L.; Greenberg, E. Iron and Pseudomonas aeruginosa biofilm formation. PNAS USA 2005, 102, 11076–11081. [Google Scholar] [CrossRef]
- Pang, X.; Yuk, H.-G. Effects of the colonization sequence of Listeria monocytogenes and Pseudomonas fluorescens on survival of biofilm cells under food-related stresses and transfer to salmon. Food Microbiol. 2019, 82, 142–150. [Google Scholar] [CrossRef]
- Wei, H.; Zhang, L. Quorum-sensing system influences root colonization and biological control ability in Pseudomonas fluorescens 2P24. Antonie Van Leeuwenhoek 2006, 89, 267–280. [Google Scholar] [CrossRef]
- Quintieri, L.; Fanelli, F.; Caputo, L. Antibiotic resistant Pseudomonas spp. Spoilers in fresh dairy products: An underestimated risk and the control strategies. Foods 2019, 8, 372. [Google Scholar] [CrossRef]
- Bekker, A.; Steyn, L.; Charimba, G.; Jooste, P.; Hugo, C. Comparison of the growth kinetics and proteolytic activities of Chryseobacterium species and Pseudomonas fluorescens. Can. J. Microbiol. 2015, 61, 977–982. [Google Scholar] [CrossRef]
- Gazzola, G.; Habimana, O.; Quinn, L.; Casey, E.; Murphy, C.D. Population dynamics of a dual Pseudomonas putida-Pseudomonas fluorescens biofilm in a capillary bioreactor. Biofouling 2019, 35, 299–307. [Google Scholar] [CrossRef]
- Cattò, C.; Cappitelli, F. Testing anti-biofilm polymeric surfaces: Where to start? Int. J. Mol. Sci. 2019, 20, 3794. [Google Scholar] [CrossRef]
- Jalvo, B.; Faraldos, M.; Bahamonde, A.; Rosal, R. Antimicrobial and antibiofilm efficacy of self-cleaning surfaces functionalized by TiO2 photocatalytic nanoparticles against Staphylococcus aureus and Pseudomonas putida. J. Hazard Mater. 2017, 340, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Angarano, V.; Smet, C.; Akkermans, S.; Watt, C.; Chieffi, A.; Van Impe, J.F.M. Visible light as an antimicrobial strategy for inactivation of Pseudomonas fluorescens and Staphylococcus epidermidis biofilms. Antibiotics 2020, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Herigstad, B.; Hamilton, M.; Heersink, J. How to optimize the drop plate method for enumerating bacteria. J. Microbiol. Methods 2001, 44, 121–129. [Google Scholar] [CrossRef]
- Govaert, M.; Smet, C.; Baka, M.; Janssens, T.; Van Impe, J.F.M. Influence of incubation conditions on the formation of model biofilms by Listeria monocytogenes and Salmonella Typhimurium on abiotic surfaces. J. Appl. Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Achinas, S.; Charalampogiannis, N.; Euverink, G.J.W. A Brief Recap of Microbial Adhesion and Biofilms. Appl. Sci. 2019, 9, 2801. [Google Scholar] [CrossRef]
- Herald, P.J.; Zottola, E.A. Attachment of Listeria monocytogenes to stainless steel surfaces at various temperatures and pH values. J. Food Sci. 1988, 53, 1549–1562. [Google Scholar] [CrossRef]
- Shi, X.; Zhu, X. Biofilm formation and food safety in food industries. Trends Food Sci. Technol. 2009, 20, 407–413. [Google Scholar] [CrossRef]
- Gerstel, U.; Römling, U. Oxygen tension and nutrient starvation are major signals that regulate agfD promoter activity and expression of the multicellular morphotype in Salmonella Typhimurium. Environ. Microbiol. 2001, 3, 638–648. [Google Scholar] [CrossRef]
- Xu, K.D.; Stewart, P.S.; Fuhu, X.; Huang, C.-T.; McFeters, G.A. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 1998, 64, 4035–4039. [Google Scholar] [CrossRef]
- Lewandowski, Z.; Altobelli, S.A.; Fukushima, E. NMR and microelectrode studies of hydrodynamics and kinetics in biofilms. Biotechnol. Prog. 1993, 9, 40–45. [Google Scholar] [CrossRef]
- Borer, B.; Tecon, R.; Or, D. Spatial organization of bacterial populations in response to oxygen and carbon counter-gradient in pore networks. Nat. Commun. 2018, 22, 769. [Google Scholar] [CrossRef]
- Ausbacher, D.; Lorenz, L.; Pitts, B.; Stewart, P.S.; Goeres, D.M. Paired methods to measure biofilm killing and removal: A case study with Penicillin G treatment of Staphylococcus aureus biofilm. Lett. Appl. Microbiol. 2018, 66, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Espada, R.; Liu, X.; Goh, X.S.; Sharo, X. Antimicrobial blue light inactivation of polymicrobial biofilms. Front. Microbiol. 2019, 10, 721. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, F.; Peng, W.; Yan, K.; Zhao, H.; Liu, T.; Cheng, H.; Chang, P.; Yuan, F.; Chen, H.; et al. The CpxA/CpxR two-component system Affects biofilm formation and virulence in Actinobacillus pleuropneumoniae. Front. Cell. Infect. Microbiol. 2018, 8, 72. [Google Scholar] [CrossRef]
- Droste, R.L.; Andras, E.; Kennedy, K.J. Initial biofilm formation of acetoclastic methanogenic bacteria. Biofouling J. Bioadhes. Biofilm Res. 1990, 2, 191–210. [Google Scholar] [CrossRef]
- Cunault, C.; Faille, C.; Briandet, R.; Postollec, F.; Desriac, N.; Benezech, T. Pseudomonas sp. biofilm development on fresh-cut food equipment surfaces—A growth curve—Fitting approach to building a comprehensive tool for studying surface contamination dynamics. Food Bioprod. Process. 2018, 107, 70–87. [Google Scholar] [CrossRef]
- Neu, T.R. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol. Rev. 1996, 60, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari, A.; Dehghany, J.; Schwebs, T.; Müsken, M.; Häussler, S.; Meyer-Hermann, M. Inoculation density and nutrient level determine the formation of mushroom-shaped structures in Pseudomonas aeruginosa biofilms. Sci. Rep. 2016, 6, 32097. [Google Scholar] [CrossRef]
- Lange, R.; Hengge-Aronis, R. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor sigma S. J. Bacteriol. 1991, 173, 4474–4481. [Google Scholar] [CrossRef]
- Nguyen, H.D.N.; Yang, Y.S.; Yuk, H.G. Biofilm formation of Salmonella Typhimurium on stainless steel and acrylic surfaces as affected by temperature and pH level. LWT Food Sci. Technol. 2014, 55, 383–388. [Google Scholar] [CrossRef]
- Sabra, W.; Kim, E.; Zeng, A. Physiological responses of Pseudomonas aeruginosa PAO1 to oxidative stress in controlled microaerobic and aerobic cultures. Microbiology 2002, 148, 3195–3202. [Google Scholar] [CrossRef] [PubMed]
- Koza, A.; Hallett, P.D.; Moon, C.D.; Spiers, A.J. Characterization of a novel air-liquid interface biofilm of Pseudomonas fluorescens SBW25. Microbiology 2009, 155, 1397–1406. [Google Scholar] [CrossRef]
- Wang, S.; Liu, X.; Liu, H.; Zhang, L.; Guo, Y.; Yu, S.; Wozniak, D.J.; Ma, L.Z. The exopolysaccharide Psl–eDNA interaction enables the formation of a biofilm skeleton in Pseudomonas aeruginosa. Environ. Microbiol. Rep. 2015, 7, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Marsden, A.E.; Grudzinski, K.; Ondrey, J.M.; Deloney-Marino, C.R.; Visick, K.L. Impact of Salt and Nutrient Content on Biofilm Formation by Vibrio fischeri. PLoS ONE 2017, 12, e0169521. [Google Scholar] [CrossRef] [PubMed]
- Sinibaldi, G.; Iebba, V.; Chinappi, M. Swimming and rafting of E.coli microcolonies at air–liquid interfaces. MicrobiologyOpen 2018, 7. [Google Scholar] [CrossRef]
- Abee, T.; Kovács, A.T.; Kuipers, O.P.; van der Veen, S. Biofilm formation and dispersal in Gram-positive bacteria. Curr. Opin. Biotechnol. 2011, 22, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Lemon, K.P.; Higgins, D.E.; Kolter, R. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J. Bacteriol. 2007, 189, 4418–4424. [Google Scholar] [CrossRef]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998, 30, 285–293. [Google Scholar] [CrossRef]
- Kirov, S.M.; Castrisios, M.; Shaw, J.G. Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect. Immun. 2004, 72, 1939–1945. [Google Scholar] [CrossRef]
- Meier, M.J.; Subasinghe, R.M.; Beaudette, L.A. Draft genome sequence of the industrially significant bacterium Pseudomonas fluorescens ATCC 13525. Microbiol. Resour. Announc. 2018, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Houry, A.; Briandet, R.; Aymerich, S.; Gohar, M. Involvement of motility and flagella in Bacillus cereus biofilm formation. Microbiology 2010, 156, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Culibrk, B.; Kalab, M.; Schubert, P.; Yi, Q.-L.; Goodrich, R.; Ramirez-Arcos, S. Efficiency of riboflavin and ultraviolet light treatment against high levels of biofilm-derived Staphylococcus epidermidis in buffy coat platelet concentrates. Vox Sang. 2017, 112, 408–416. [Google Scholar] [CrossRef] [PubMed]
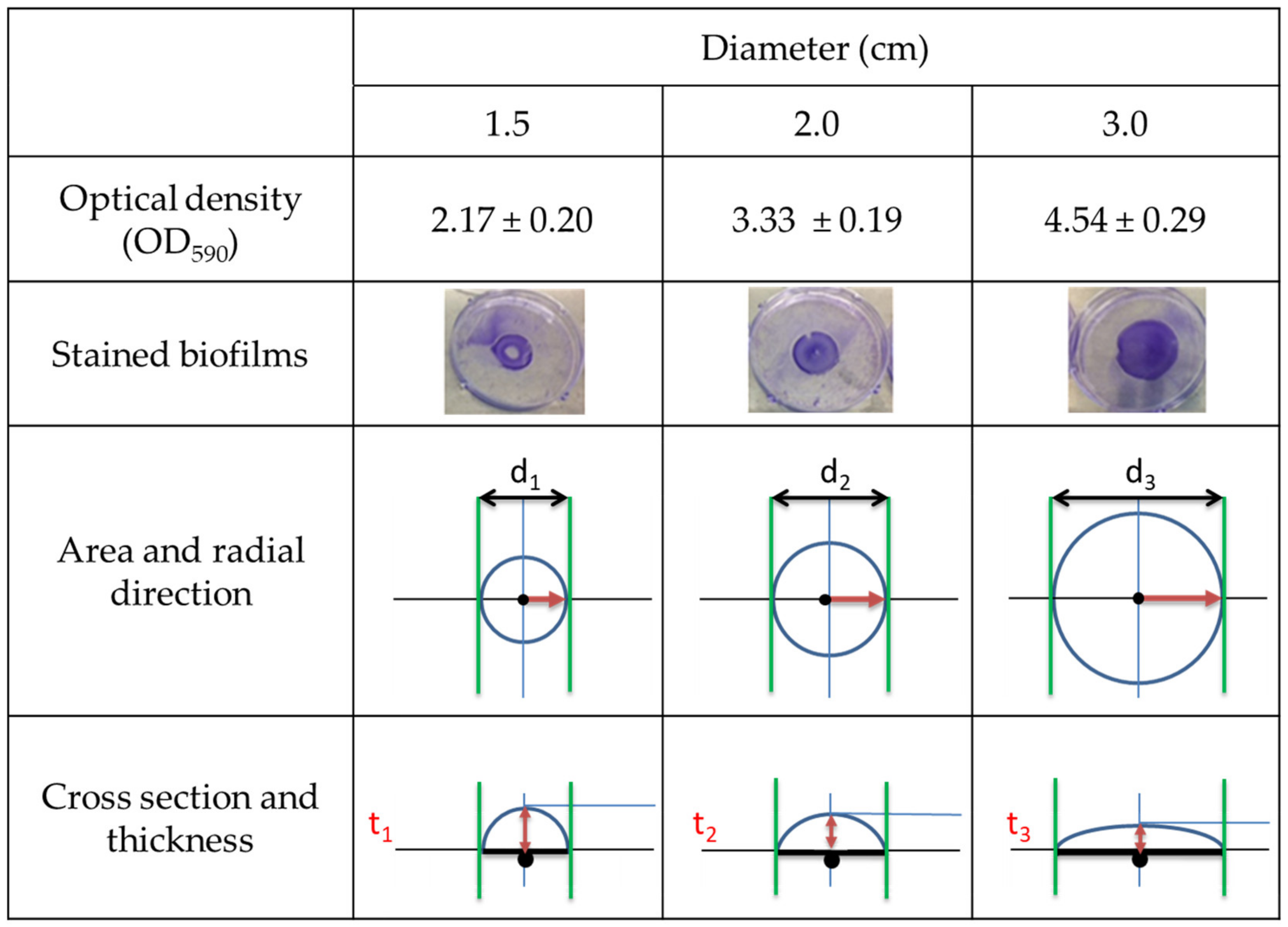
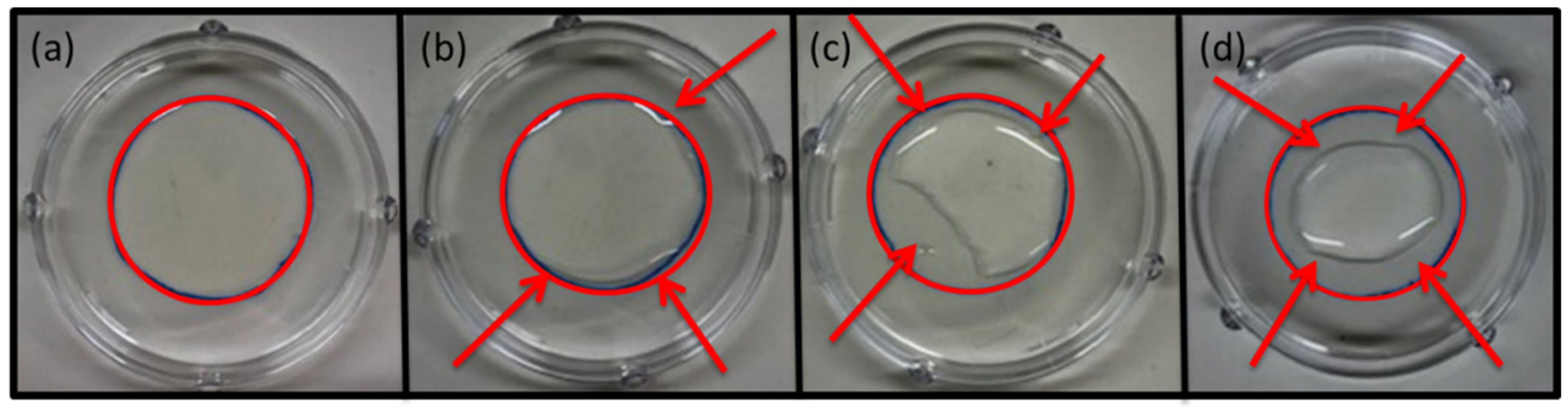

| Parameter | Inoculum Level [CFU/mL] | |
|---|---|---|
| Estimated Value | 104 | 107 |
| λ [h] | 3.28 ± 0.80 b | 1.00 ± 0.65 a |
| µmax [1/h] | 0.791 ± 0.057 b | 0.472 ± 0.040 a |
| ln(No) [ln(CFU/cm2)] | 6.00 ± 0.35 b | 13.26 ± 0.11 a |
| ln(Nmax) [ln(CFU/cm2)] | 18.14 ± 0.18 a | 17.94 ± 0.07 a |
| Population increase [ln(CFU/cm2)] | 12.14 ± 0.39 b | 4.68 ± 0.13 a |
| RMSE | 0.5479 | 0.1570 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angarano, V.; Smet, C.; Akkermans, S.; Akritidou, T.; Huyck, B.; Chieffi, A.; Impe, J.F.M.V. A Reproducible Method for Growing Biofilms on Polystyrene Surfaces: Biomass and Bacterial Viability Evolution of Pseudomonas fluorescens and Staphylococcus epidermidis. Appl. Sci. 2020, 10, 4544. https://doi.org/10.3390/app10134544
Angarano V, Smet C, Akkermans S, Akritidou T, Huyck B, Chieffi A, Impe JFMV. A Reproducible Method for Growing Biofilms on Polystyrene Surfaces: Biomass and Bacterial Viability Evolution of Pseudomonas fluorescens and Staphylococcus epidermidis. Applied Sciences. 2020; 10(13):4544. https://doi.org/10.3390/app10134544
Chicago/Turabian StyleAngarano, Valeria, Cindy Smet, Simen Akkermans, Theodora Akritidou, Bart Huyck, Andre Chieffi, and Jan F. M. Van Impe. 2020. "A Reproducible Method for Growing Biofilms on Polystyrene Surfaces: Biomass and Bacterial Viability Evolution of Pseudomonas fluorescens and Staphylococcus epidermidis" Applied Sciences 10, no. 13: 4544. https://doi.org/10.3390/app10134544
APA StyleAngarano, V., Smet, C., Akkermans, S., Akritidou, T., Huyck, B., Chieffi, A., & Impe, J. F. M. V. (2020). A Reproducible Method for Growing Biofilms on Polystyrene Surfaces: Biomass and Bacterial Viability Evolution of Pseudomonas fluorescens and Staphylococcus epidermidis. Applied Sciences, 10(13), 4544. https://doi.org/10.3390/app10134544





