Accelerating Super-Resolution and Visual Task Analysis in Medical Images
Abstract
:1. Introduction
2. Related Work and Contributions
2.1. Super-Resolution
- Simplicity and Stability. Our customized SISR model, inspired by VDSR (20 conv. layers) [21], has a shallower structure (7 conv. layers) with a lower number of training parameters. In addition, our proposed SISR model is easy to train and has a single network contrary to GAN-based networks which are difficult to train and require both generator and discriminator networks. Further, our proposed model is more stable and less sensitive to hyper-parameters selection as compared to most GAN-based models. As large models with massive number of parameters are restricted to computing platforms with large memory banks and computing capability, developing smaller and stable networks without losing representative accuracy is important to reduce the number of parameters and the storage size of the networks. This would boost the usage of these networks in limited-resource settings and embedded healthcare systems.
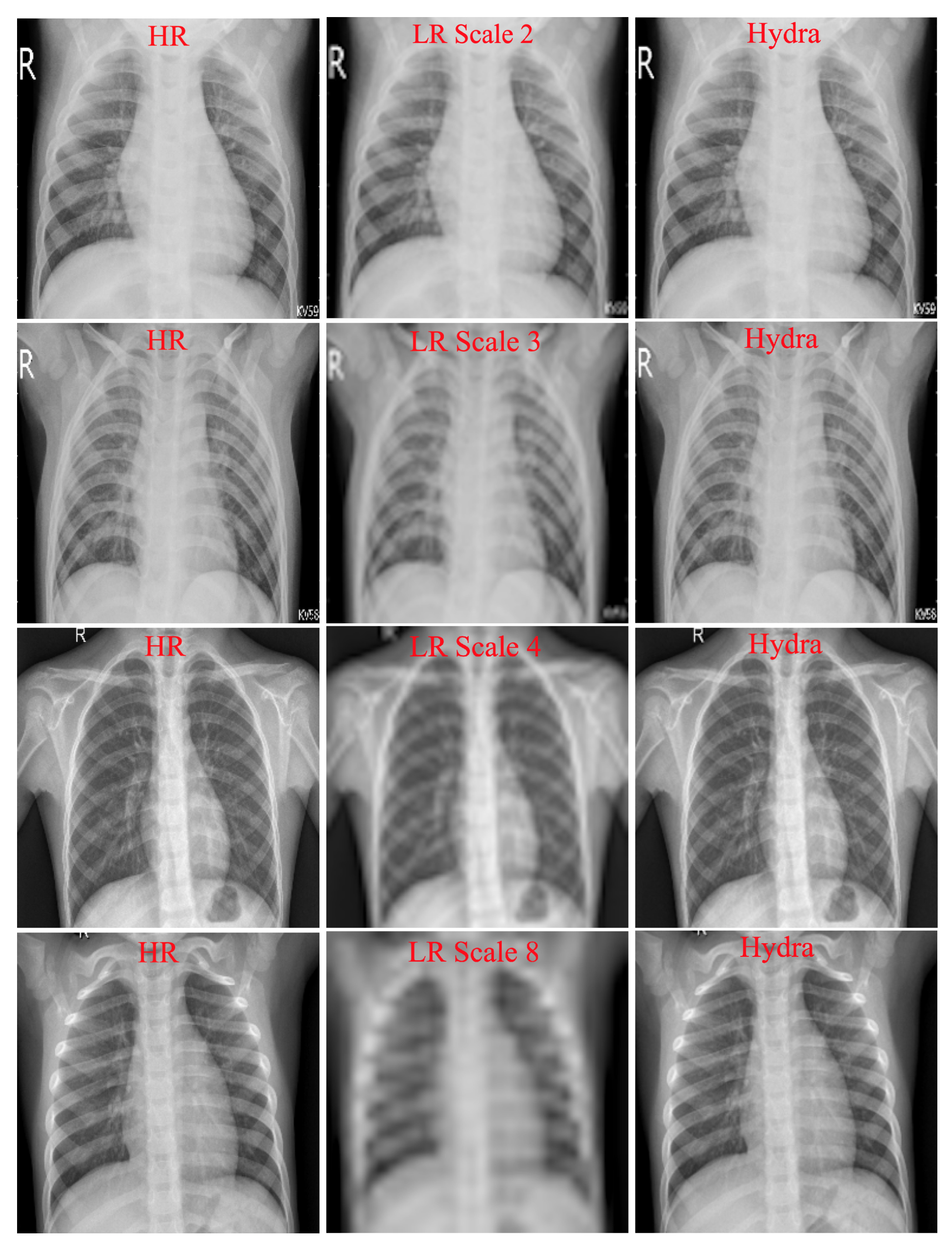
- Multiple Scales Training: Our SISR model is trained with different scale factors at once. The trained network can then be tested with any scale used during training. As discussed in [21], training a single model with multiple scale factors is more efficient, accurate, and practical as compared to training and storing several scale-dependent models.
- Context: We utilize information from the entire image region. Existing methods either rely on the context of small image regions (e.g., [20]) or large image regions (e.g., [21]), but not the entire image region. Our experimental results demonstrate that using the entire image region leads to better overall performance while decreasing computations.
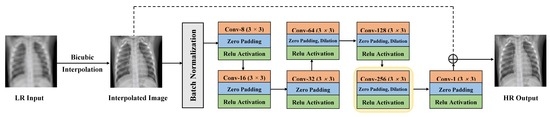
- Raw Image Channels: We propose to compute the residual image from the raw image (RGB or grayscale) directly instead of converting the images to a different color space (e.g., YCbCr [21]). The residual image is computed by subtracting the HR reference image from the LR image that has been upscaled using interpolation to match the size of the reference image. The computed residual image contains information of the image’s high-frequency details. The main benefit of directly working on the raw color space is that we decrease the total computational time by dropping two operations: (1) converting from raw color space to another color space (e.g., YCbCr) and (2) converting the image back to its original color space. Our customized SISR model computes the residual images directly from the original color space and learns to estimate these images. To construct an HR image, the estimated residual image is added to the upsampled LR image.
- Combined Learning Loss: We propose to train the proposed SISR model using a loss function that combines the advantages of the mean absolute error (MAE) and the Multi-scale Structural Similarity (MS-SSIM). Our experimental results show that MAE can better assess the average model performance as compared to other loss metrics. Also, our experimental results show that the MS-SSIM preserves the contrast in high-frequency regions better than other loss functions (e.g., SSIM). To capture the best characteristics of both loss functions, we propose to combine both loss terms (MAE + MS-SSIM).
2.2. Visual Task Analysis
2.3. Contributions
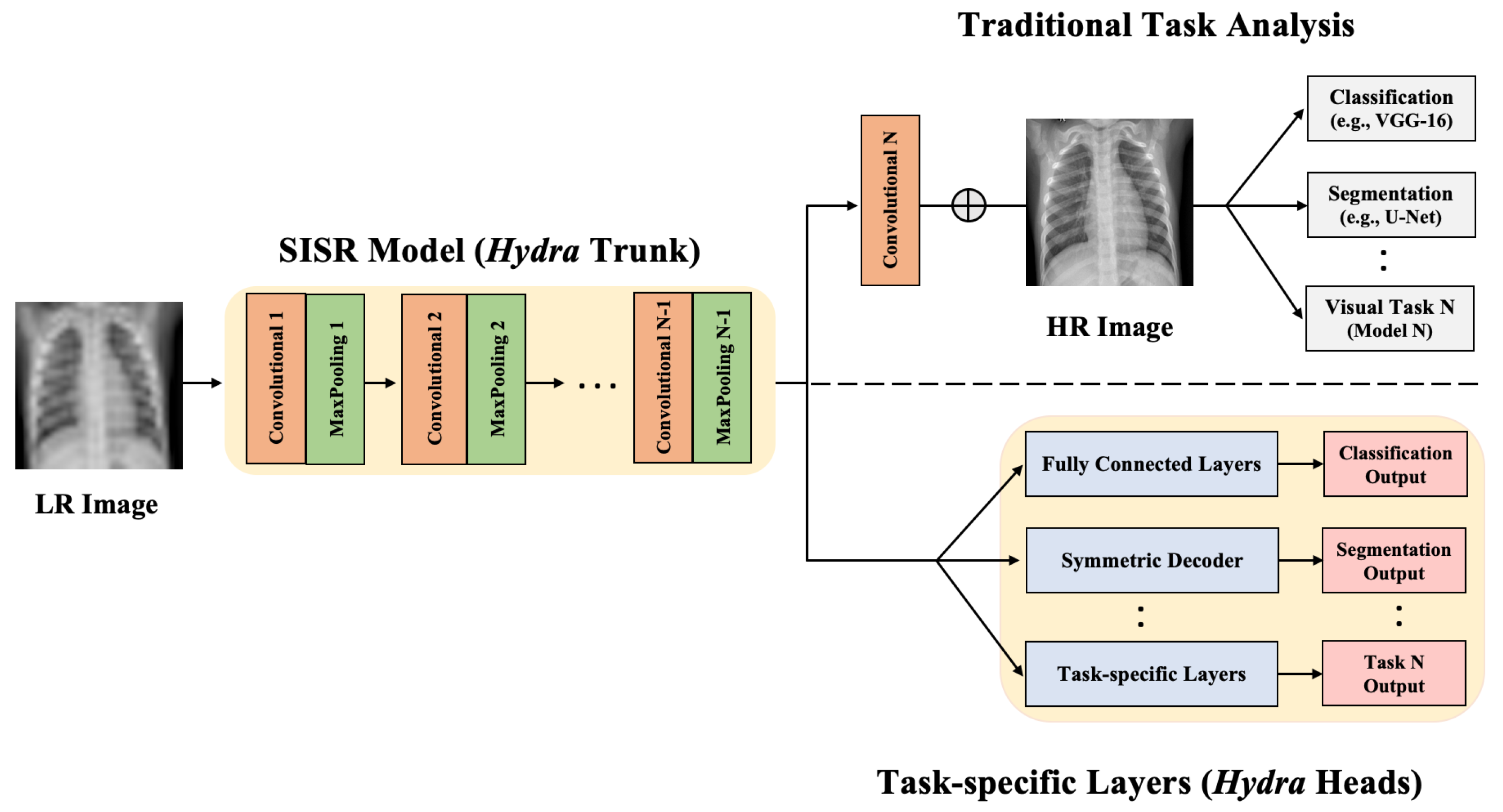
- We propose the Hydra approach for enhancing medical image resolution and visual task analysis. The Hydra consists of two components: a shared trunk and computing Heads.
- Hydra trunk is a proposed customized SISR model that learns the mapping from LR to HR. This SISR model has a simple architecture and is trained using the entire raw image and multiple scales at once to minimize a proposed loss function. Our experimental results show that the proposed SISR model, which has a markedly lower number of training time and parameters, achieves state-of-the-art performance.
- We propose to append the customized SISR trunk with multiple computing heads to learn different visual tasks in medical images. We evaluate our approach using CXR datasets to generate HR representation followed by jointly performing lung segmentation (visual task 1) and abnormality classification (visual task 2). We focus mainly on these two tasks because classification and segmentation are the key tasks in most medical image analysis applications.
- We empirically demonstrate the superiority and efficiency of our approach, in terms of performance and computation, for SR and medical image analysis as compared to the traditional approach.
3. Materials and Methods
3.1. Datasets for Training and Testing
3.1.1. RSNA CXR
3.1.2. Shenzhen CXR
3.1.3. Montgomery CXR
3.1.4. JSRT CXR
3.2. Proposed Approach: Hydra
3.2.1. Hydra SISR Trunk
3.2.2. Hydra Task-Specific Heads
4. Experiments and Results
4.1. Hydra Trunk Performance
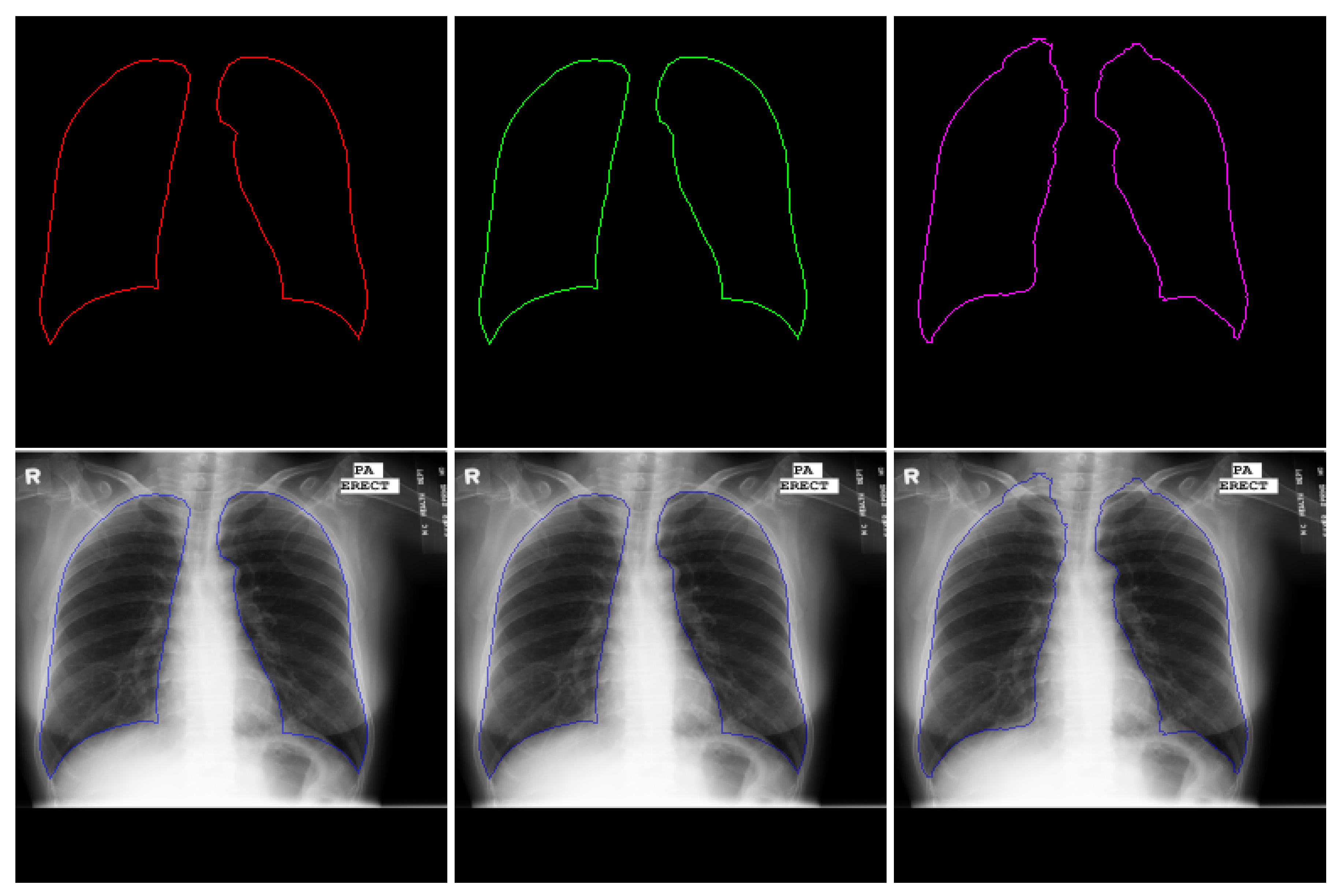
4.2. Hydra Heads Performance
4.2.1. Abnormality Classification
4.2.2. Lung Segmentation
4.3. Training Parameters and Time
5. Conclusions and Possible Extensions
Author Contributions
Funding
Conflicts of Interest
References
- Arai, K.; Kapoor, S. Advances in Computer Vision: Proceedings of the 2019 Computer Vision Conference (CVC); Springer: New York, NY, USA, 2019; Volume 2. [Google Scholar]
- Wang, Z.; Chen, J.; Hoi, S.C. Deep learning for image super-resolution: A survey. arXiv 2019, arXiv:1902.06068. [Google Scholar] [CrossRef] [Green Version]
- Giger, M.L. Machine learning in medical imaging. J. Am. Coll. Radiol. 2018, 15, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Tashiro, S. Estimation of the effects of medical diagnostic radiation exposure based on DNA damage. J. Radiat. Res. 2018, 59, ii121–ii129. [Google Scholar] [CrossRef]
- Li, X.; Fu, W. Regularized super-resolution restoration algorithm for single medical image based on fuzzy similarity fusion. EURASIP J. Image Video Process. 2019, 2019, 83. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Zeng, X.; Huang, Z.; Li, W.; Zhang, H. Low-dose chest X-ray image super-resolution using generative adversarial nets with spectral normalization. Biomed. Signal Process. Control 2020, 55, 101600. [Google Scholar] [CrossRef]
- Yun, S.J.; Ryu, C.W.; Choi, N.Y.; Kim, H.C.; Oh, J.Y.; Yang, D.M. Comparison of low-and standard-dose CT for the diagnosis of acute appendicitis: A meta-analysis. Am. J. Roentgenol. 2017, 208, W198–W207. [Google Scholar] [CrossRef] [PubMed]
- Mouton, A.; Breckon, T.P. On the relevance of denoising and artefact reduction in 3d segmentation and classification within complex computed tomography imagery. J. X-ray Sci. Technol. 2019, 27, 51–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sippola, S.; Virtanen, J.; Tammilehto, V.; Grönroos, J.; Hurme, S.; Niiniviita, H.; Lietzen, E.; Salminen, P. The accuracy of low-dose computed tomography protocol in patients with suspected acute appendicitis: The OPTICAP study. Ann. Surg. 2020, 271, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Rundo, L.; Tangherloni, A.; Cazzaniga, P.; Nobile, M.S.; Russo, G.; Gilardi, M.C.; Vitabile, S.; Mauri, G.; Besozzi, D.; Militello, C. A novel framework for MR image segmentation and quantification by using MedGA. Comput. Methods Programs Biomed. 2019, 176, 159–172. [Google Scholar] [CrossRef]
- Rundo, L.; Tangherloni, A.; Nobile, M.S.; Militello, C.; Besozzi, D.; Mauri, G.; Cazzaniga, P. MedGA: A novel evolutionary method for image enhancement in medical imaging systems. Expert Syst. Appl. 2019, 119, 387–399. [Google Scholar] [CrossRef]
- Lukin, A.; Krylov, A.S.; Nasonov, A. Image interpolation by super-resolution. Proc. GraphiCon 2006, 2006, 239–242. [Google Scholar]
- Keys, R. Cubic convolution interpolation for digital image processing. IEEE Trans. Acoust. 1981, 29, 1153–1160. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Orchard, M.T. New edge-directed interpolation. IEEE Trans. Image Process. 2001, 10, 1521–1527. [Google Scholar]
- Sun, J.; Xu, Z.; Shum, H.Y. Image super-resolution using gradient profile prior. In Proceedings of the 2008 IEEE Conference on Computer Vision and Pattern Recognition, Anchorage, AK, USA, 24–26 June 2008; pp. 1–8. [Google Scholar]
- Schulter, S.; Leistner, C.; Bischof, H. Fast and accurate image upscaling with super-resolution forests. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3791–3799. [Google Scholar]
- Li, H.; Lam, K.M.; Wang, M. Image super-resolution via feature-augmented random forest. Signal Process. Image Commun. 2019, 72, 25–34. [Google Scholar] [CrossRef] [Green Version]
- Gu, P.; Zheng, L. Fast low-dose Computed Tomography image Super-Resolution Reconstruction via Sparse coding and Random Forests. In Proceedings of the 2019 IEEE 8th Joint International Information Technology and Artificial Intelligence Conference (ITAIC), Chongqing, China, 24–26 May 2019; pp. 1400–1403. [Google Scholar]
- Freeman, W.T.; Jones, T.R.; Pasztor, E.C. Example-based super-resolution. IEEE Comput. Graph. Appl. 2002, 22, 56–65. [Google Scholar] [CrossRef] [Green Version]
- Dong, C.; Loy, C.C.; Tang, X. Accelerating the super-resolution convolutional neural network. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2016; pp. 391–407. [Google Scholar]
- Kim, J.; Kwon Lee, J.; Mu Lee, K. Accurate image super-resolution using very deep convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 1646–1654. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Mahapatra, D.; Bozorgtabar, B. Progressive Generative Adversarial Networks for Medical Image Super resolution. arXiv 2019, arXiv:1902.02144. [Google Scholar]
- Goodfellow, I. NIPS 2016 tutorial: Generative adversarial networks. arXiv 2016, arXiv:1701.00160. [Google Scholar]
- You, C.; Li, G.; Zhang, Y.; Zhang, X.; Shan, H.; Li, M.; Ju, S.; Zhao, Z.; Zhang, Z.; Cong, W.; et al. CT super-resolution GAN constrained by the identical, residual, and cycle learning ensemble (GAN-CIRCLE). IEEE Trans. Med. Imaging 2019, 39, 188–203. [Google Scholar] [CrossRef] [Green Version]
- Joshi, S.H.; Marquina, A.; Osher, S.J.; Dinov, I.; Darrell, J. Image Resolution Enhancement and Its Applications to Medical Image Processing; Laboratory of Neuroimaging University of of California: Los Angeles, CA, USA, 2008. [Google Scholar]
- Yang, W.; Zhang, X.; Tian, Y.; Wang, W.; Xue, J.H.; Liao, Q. Deep learning for single image super-resolution: A brief review. IEEE Trans. Multimed. 2019, 21, 3106–3121. [Google Scholar] [CrossRef] [Green Version]
- Gulati, T.; Sengupta, S.; Lakshminarayanan, V. Application of an enhanced deep super-resolution network in retinal image analysis. In Proceedings of the Ophthalmic Technologies XXX, San Francisco, CA, USA, 1–6 February 2020; Volume 11218, p. 112181K. [Google Scholar]
- Christodoulidis, A.; Hurtut, T.; Tahar, H.B.; Cheriet, F. A multi-scale tensor voting approach for small retinal vessel segmentation in high resolution fundus images. Comput. Med. Imaging Graph. 2016, 52, 28–43. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shen, L.; Xie, X.; Huang, S.; Xie, Z.; Hong, X.; Yu, J. Multi-resolution convolutional networks for chest X-ray radiograph based lung nodule detection. Artif. Intell. Med. 2019, 103, 101744. [Google Scholar] [CrossRef] [PubMed]
- Shih, G.; Wu, C.C.; Halabi, S.S.; Kohli, M.D.; Prevedello, L.M.; Cook, T.S.; Sharma, A.; Amorosa, J.K.; Arteaga, V.; Galperin-Aizenberg, M.; et al. Augmenting the National Institutes of Health chest radiograph dataset with expert annotations of possible pneumonia. Radiol. Artif. Intell. 2019, 1, e180041. [Google Scholar] [CrossRef]
- Jaeger, S.; Candemir, S.; Antani, S.; Wáng, Y.X.J.; Lu, P.X.; Thoma, G. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging. Med. Surg. 2014, 4, 475. [Google Scholar] [PubMed]
- Shiraishi, J.; Katsuragawa, S.; Ikezoe, J.; Matsumoto, T.; Kobayashi, T.; Komatsu, K.I.; Matsui, M.; Fujita, H.; Kodera, Y.; Doi, K. Development of a digital image database for chest radiographs with and without a lung nodule: Receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. AJR Am. J. Roentgenol. 2000, 174, 71–74. [Google Scholar] [CrossRef] [PubMed]
- SCR Reference Lung Boundaries. Available online: https://www.isi.uu.nl/Research/Databases/SCR/ (accessed on 24 May 2012).
- Wang, Z.; Simoncelli, E.P.; Bovik, A.C. Multiscale structural similarity for image quality assessment. In Proceedings of the Thrity-Seventh Asilomar Conference on Signals, Systems & Computers, Pacific Grove, CA, USA, 9–12 November 2003; Volume 2, pp. 1398–1402. [Google Scholar]
- Brunet, D.; Vrscay, E.R.; Wang, Z. On the mathematical properties of the structural similarity index. IEEE Trans. Image Process. 2011, 21, 1488–1499. [Google Scholar] [CrossRef]
- Chollet, F. Keras. Available online: https://github.com/fchollet/keras (accessed on 19 June 2015).
- Sharma, N.; Ray, A.K.; Sharma, S.; Shukla, K.; Pradhan, S.; Aggarwal, L.M. Segmentation and classification of medical images using texture-primitive features: Application of BAM-type artificial neural network. Med. Phys. 2008, 33, 119. [Google Scholar] [CrossRef]
- Han, C.; Kitamura, Y.; Kudo, A.; Ichinose, A.; Rundo, L.; Furukawa, Y.; Umemoto, K.; Li, Y.; Nakayama, H. Synthesizing diverse lung nodules wherever massively: 3D multi-conditional GAN-based CT image augmentation for object detection. In Proceedings of the 2019 International Conference on 3D Vision (3DV), Quebec City, QC, Canada, 16–19 September 2019; pp. 729–737. [Google Scholar]
- Candemir, S.; Jaeger, S.; Palaniappan, K.; Musco, J.P.; Singh, R.K.; Xue, Z.; Karargyris, A.; Antani, S.; Thoma, G.; McDonald, C.J. Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans. Med. Imaging 2013, 33, 577–590. [Google Scholar] [CrossRef]
- Narayanan, B.N.; Hardie, R.C. A Computationally Efficient U-Net Architecture for Lung Segmentation in Chest Radiographs. In Proceedings of the 2019 IEEE National Aerospace and Electronics Conference (NAECON), Dayton, OH, USA, 15–19 July 2019; pp. 279–284. [Google Scholar]
- Sultana, F.; Sufian, A.; Dutta, P. Evolution of Image Segmentation using Deep Convolutional Neural Network: A Survey. arXiv 2020, arXiv:2001.04074. [Google Scholar] [CrossRef]
- Aviles-Rivero, A.I.; Papadakis, N.; Li, R.; Sellars, P.; Fan, Q.; Tan, R.T.; Schönlieb, C.B. GraphXNet: Chest X-Ray Classification Under Extreme Minimal Supervision. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; Springer: New York, NY, USA, 2009; pp. 504–512. [Google Scholar]
- Li, J.; Yuan, G.; Fan, H. Multifocus image fusion using wavelet-domain-based deep CNN. Comput. Intell. Neurosci. 2019, 2019, 4179397. [Google Scholar] [CrossRef] [Green Version]
- Aymaz, S.; Köse, C.; Aymaz, Ş. Multi-focus image fusion for different datasets with super-resolution using gradient-based new fusion rule. Multimed. Tools Appl. 2020, 79, 13311–13350. [Google Scholar] [CrossRef]
- Georgescu, M.I.; Ionescu, R.T.; Verga, N. Convolutional Neural Networks with Intermediate Loss for 3D Super-Resolution of CT and MRI Scans. arXiv 2020, arXiv:2001.01330. [Google Scholar] [CrossRef]

| Scale | Model | Proposed Loss | PSNR | SSIM | MS-SSIM |
|---|---|---|---|---|---|
| 2 | Proposed | 0.0072 | 34.822 | 0.9366 | 0.9911 |
| VDSR | 0.0091 | 32.564 | 0.9355 | 0.9801 | |
| 3 | Proposed | 0.0161 | 32.274 | 0.9000 | 0.9861 |
| VDSR | 0.0157 | 31.972 | 0.8858 | 0.9815 | |
| 4 | Proposed | 0.0201 | 30.764 | 0.8706 | 0.9745 |
| VDSR | 0.0250 | 30.691 | 0.8652 | 0.9722 | |
| 8 | Proposed | 0.0454 | 27.306 | 0.7839 | 0.9233 |
| VDSR | 0.0478 | 25.087 | 0.7644 | 0.9088 |
| Model | Accuracy | AUC | Sensitivity | Specificity | Precision | F-Score |
|---|---|---|---|---|---|---|
| Baseline (HR) | 0.8283 | 0.8502 | 0.7800 | 0.8776 | 0.8667 | 0.8211 |
| Baseline (scale 2) | 0.8182 | 0.8586 | 0.7600 | 0.8561 | 0.8636 | 0.8085 |
| Hydra Head (scale 2) | 0.8512 | 0.8905 | 0.7700 | 0.9082 | 0.8953 | 0.8280 |
| Baseline (scale 3) | 0.7980 | 0.8492 | 0.6800 | 0.8475 | 0.8947 | 0.7727 |
| Hydra Head (scale 3) | 0.8384 | 0.8748 | 0.7500 | 0.9184 | 0.9000 | 0.8000 |
| Baseline (scale 4) | 0.7929 | 0.8632 | 0.6900 | 0.8980 | 0.8734 | 0.7709 |
| Hydra Head (scale 4) | 0.8219 | 0.8416 | 0.7100 | 0.9388 | 0.9189 | 0.7816 |
| Baseline (scale 8) | 0.7727 | 0.8537 | 0.7000 | 0.8469 | 0.8041 | 0.7568 |
| Hydra Head (scale 8) | 0.8081 | 0.8608 | 0.7800 | 0.8761 | 0.8235 | 0.7991 |
| Model | Accuracy | Loss | IoU |
|---|---|---|---|
| Baseline (HR) | 0.9863 | 0.0355 | 0.9464 |
| Baseline (scale 2) | 0.9700 | 0.0545 | 0.9263 |
| Hydra Head (scale 2) | 0.9858 | 0.0316 | 0.9516 |
| Baseline (scale 3) | 0.9613 | 0.0496 | 0.9285 |
| Hydra Head (scale 3) | 0.9897 | 0.0248 | 0.9599 |
| Baseline (scale 4) | 0.9390 | 0.0557 | 0.9201 |
| Hydra’s Head (scale 4) | 0.9801 | 0.0325 | 0.9229 |
| Baseline (scale 8) | 0.9516 | 0.0816 | 0.8839 |
| Hydra Head (scale 8) | 0.9830 | 0.0411 | 0.9391 |
| Hydra Scale | Train | Test | Accuracy | Loss | IoU |
|---|---|---|---|---|---|
| 2 | Shenzhen | Shenzhen | 0.9762 | 0.0125 | 0.9521 |
| Shenzhen | JSRT | 0.9832 | 0.0351 | 0.9265 | |
| Montgomery & JSRT | Shenzhen | 0.9768 | 0.0561 | 0.9294 | |
| 3 | Shenzhen | Shenzhen | 0.9647 | 0.0297 | 0.8986 |
| Shenzhen | JSRT | 0.9824 | 0.0132 | 0.9461 | |
| Montgomery & JSRT | Shenzhen | 0.9727 | 0.0444 | 0.9178 | |
| 4 | Shenzhen | Shenzhen | 0.9719 | 0.0454 | 0.9163 |
| Shenzhen | JSRT | 0.9663 | 0.0536 | 0.9175 | |
| Montgomery & JSRT | Shenzhen | 0.9742 | 0.0711 | 0.9116 | |
| 8 | Shenzhen | Shenzhen | 0.9612 | 0.0663 | 0.9132 |
| Shenzhen | JSRT | 0.9364 | 0.0653 | 0.8947 | |
| Montgomery & JSRT | Shenzhen | 0.9641 | 0.0753 | 0.8816 |
| Method | IoU | Computation Time (per image) |
|---|---|---|
| Anatomical Atlas [40] | 0.9500 | 20–25 s |
| Efficient U-Net [41] | 0.9200 | 0.08 s |
| Hydra Head (Scale 2) | 0.9742 | 0.03 s |
| Approach | Task | Train Parameters | Train Time (Seconds) | Test Time (Seconds) |
|---|---|---|---|---|
| VDSR [21] | SISR | 668,227 | 72,490.26 | 0.23 |
| Hydra | SISR (Trunk) | 395,717 | 21,917.64 | 0.09 |
| Classification Head | 295,715 | 180.75 | 0.01 | |
| Segmentation Head | 786,497 | 387.87 | 0.03 | |
| Traditional | Baseline Classification | 1,573,506 | 650.72 | 0.05 |
| Baseline Segmentation | 786,497 | 8238.98 | 0.02 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zamzmi, G.; Rajaraman, S.; Antani, S. Accelerating Super-Resolution and Visual Task Analysis in Medical Images. Appl. Sci. 2020, 10, 4282. https://doi.org/10.3390/app10124282
Zamzmi G, Rajaraman S, Antani S. Accelerating Super-Resolution and Visual Task Analysis in Medical Images. Applied Sciences. 2020; 10(12):4282. https://doi.org/10.3390/app10124282
Chicago/Turabian StyleZamzmi, Ghada, Sivaramakrishnan Rajaraman, and Sameer Antani. 2020. "Accelerating Super-Resolution and Visual Task Analysis in Medical Images" Applied Sciences 10, no. 12: 4282. https://doi.org/10.3390/app10124282
APA StyleZamzmi, G., Rajaraman, S., & Antani, S. (2020). Accelerating Super-Resolution and Visual Task Analysis in Medical Images. Applied Sciences, 10(12), 4282. https://doi.org/10.3390/app10124282