Raman Spectral Analysis for Quality Determination of Grignard Reagent
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. Raman Spectral Measurement
2.3. Data Preprocessing and Multivariate Analysis
2.4. Variable Selection
3. Results and Discussion
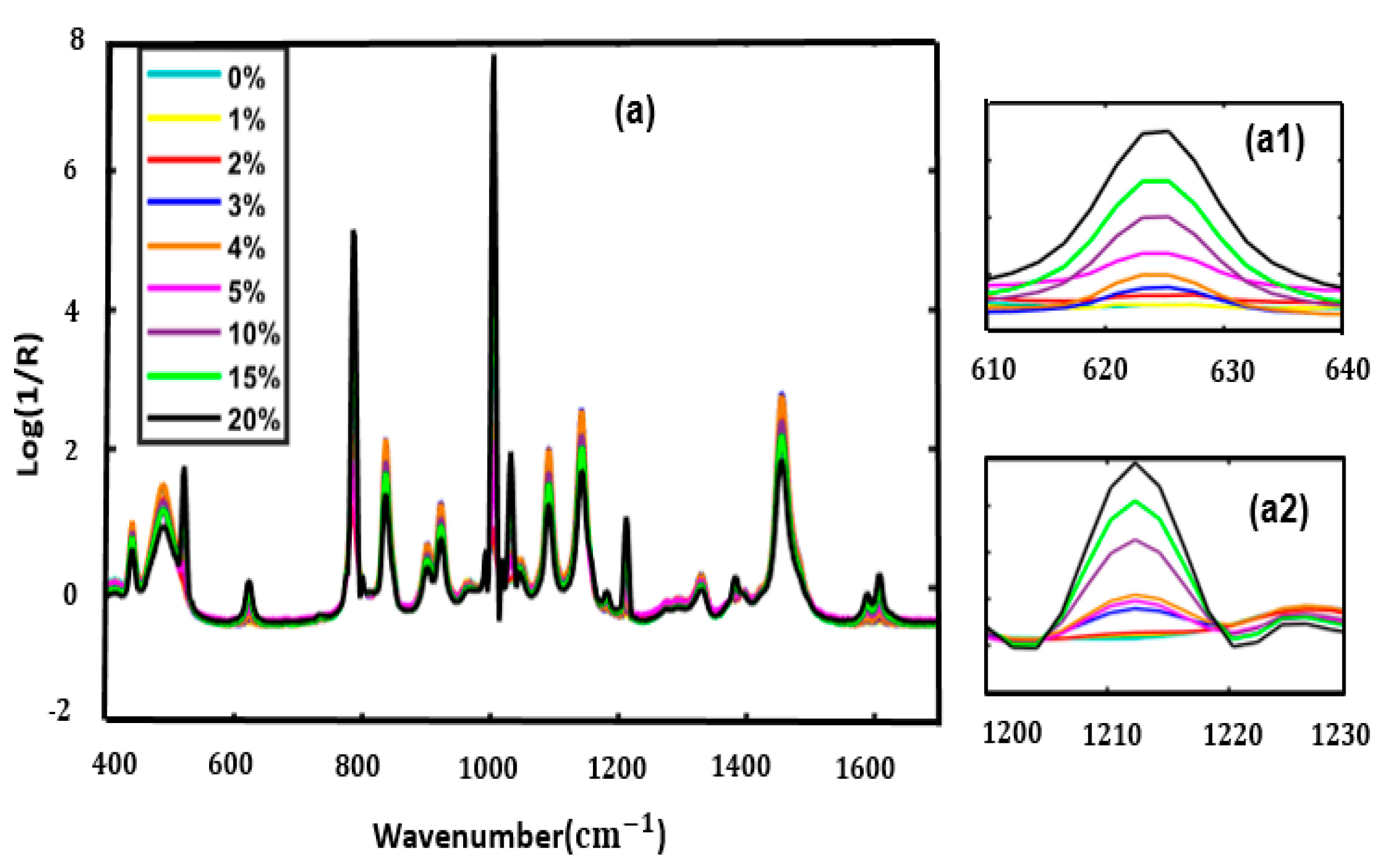
3.1. Spectral Interpretation
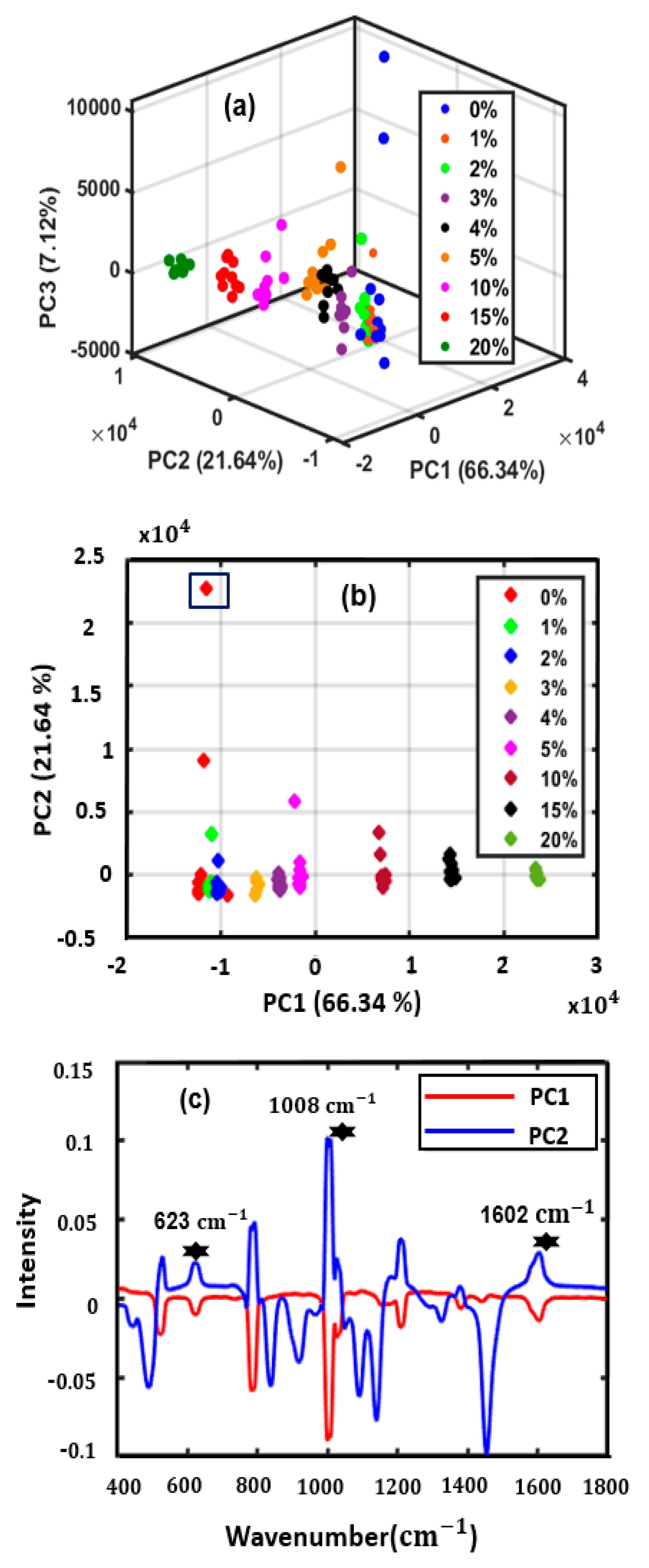
3.2. Principal Component Analysis (PCA) Model Data Visualization
3.3. PLSR Models to Predict Toluene-Adulterated Grignard Reagent
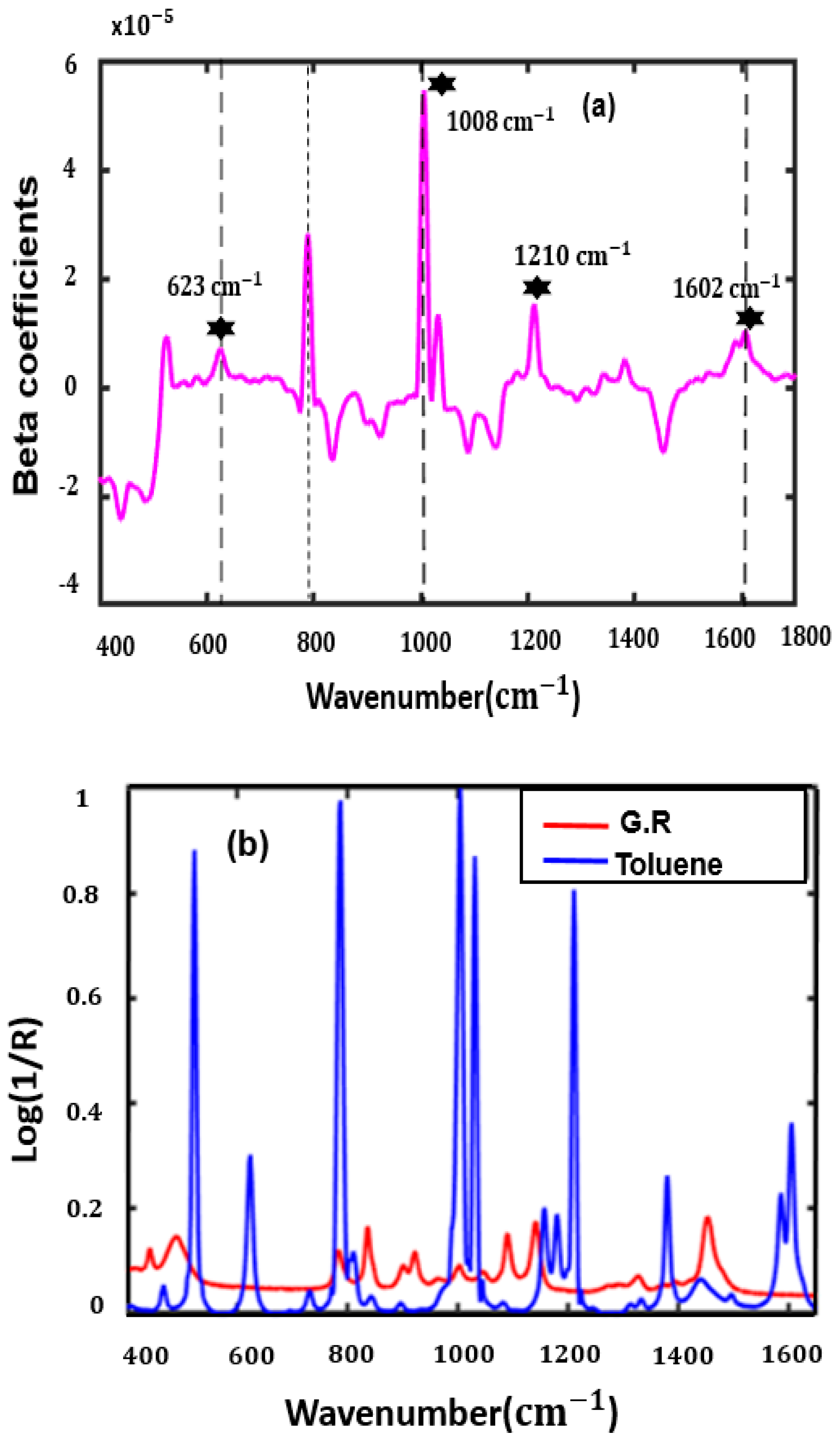
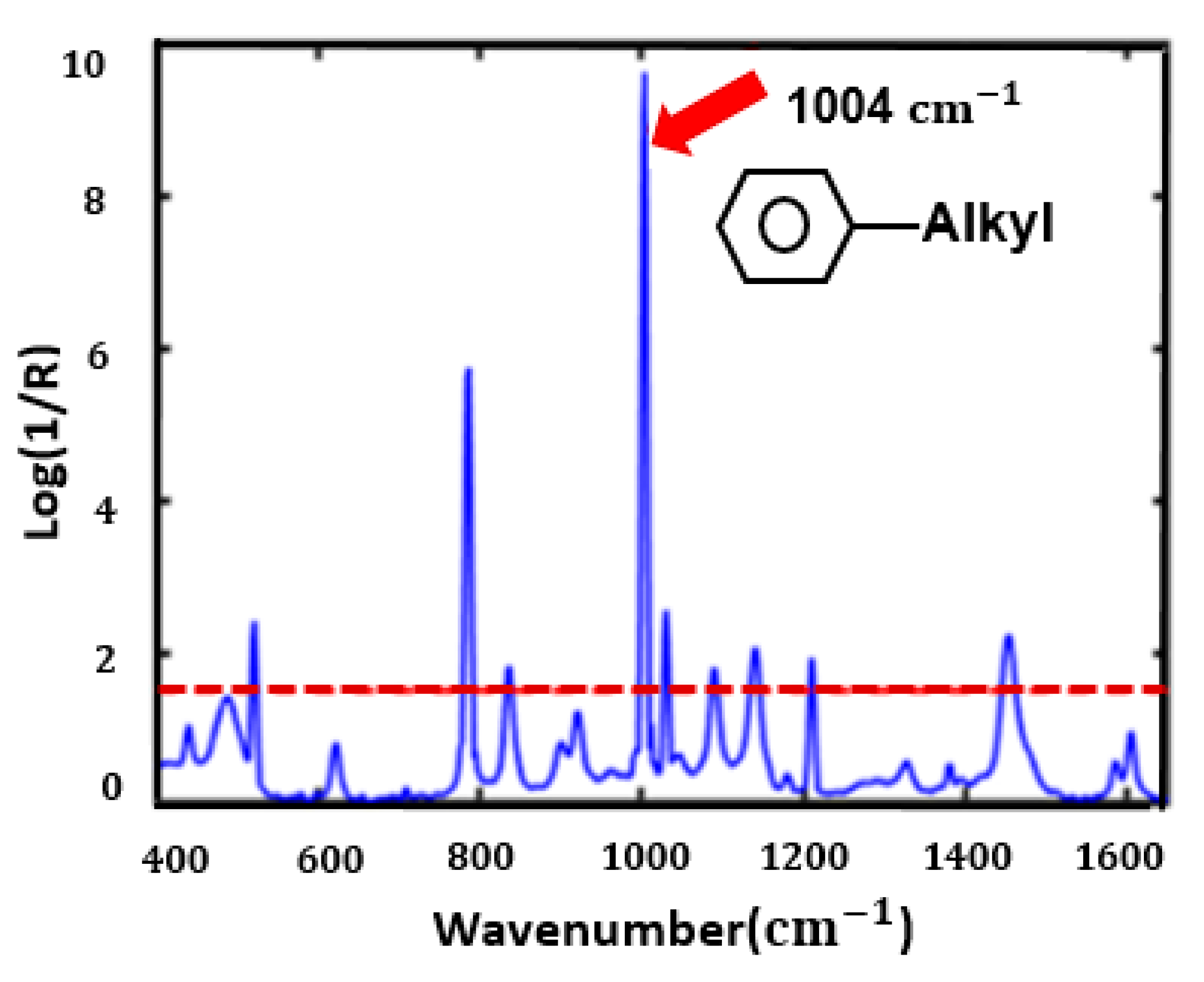
3.4. VIP-PLSR Analysis to Predict Toluene-Adulterated Grignard Reagent
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cervera-Padrell, A.E.; Nielsen, J.P.; Jønch Pedersen, M.; Müller Christensen, K.; Mortensen, A.R.; Skovby, T.; Dam-Johansen, K.; Kiil, S.; Gernaey, K.V. Monitoring and control of a continuous Grignard reaction for the synthesis of an active pharmaceutical ingredient intermediate using inline NIR spectroscopy. Org. Process Res. Dev. 2012, 16, 901–914. [Google Scholar] [CrossRef] [Green Version]
- Richey, H. Grignard Reagents: New Developments; Wiley: Chichester, UK, 2000. [Google Scholar]
- Jordan, V.C. A current view of tamoxifen for the treatment and prevention of breast cancer. Br. J. Pharmacol. 1993, 110, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Kantevari, S.; Nair, C.K.S.; Pardhasaradhi, M. A practical synthesis of 5-(4’-methylbiphenyl-2-yl)-1H-tetrazole. J. Heterocycl. Chem. 2006, 43, 1353–1356. [Google Scholar] [CrossRef]
- Hong, S.J.; Lee, A.Y.; Han, Y.H.; Park, J.; So, J.D.; Kim, G. Rancidity Prediction of Soybean Oil by Using Near-Infrared Spectroscopy Techniques. J. Biosys. Eng. 2018, 43, 219–228. [Google Scholar]
- Zhang, X.F.; Zou, M.Q.; Qi, X.H.; Liu, F.; Zhu, X.H.; Zhao, B.H. Detection of melamine in liquid milk using surface-enhanced Raman scattering spectroscopy. J. Raman Spectrosc. 2010, 41, 1655–1660. [Google Scholar] [CrossRef]
- Nedeljkovic, A.; Tomasevic, I.; Miocinovic, J.; Pudja, P. Feasibility of discrimination of dairy creams and cream-like analogues using Raman spectroscopy and chemometric analysis. Food Chem. 2017, 232, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Lohumi, S.; Joshi, R.; Kim, M.S.; Qin, J.; Baek, I.; Cho, B.K. Raman Spectral Analysis for Non-invasive Detection of External and Internal Parameters of Fake Eggs. Sens. Actuators B Chem. 2020, 303, 127243. [Google Scholar] [CrossRef]
- Joshi, R.; Cho, B.K.; Joshi, R.; Lohumi, S.; Faqeerzada, M.A.; Amanah, H.Z.; Lee, J.; Mo, C.; Lee, H. Raman spectroscopic analysis to detect olive oil mixtures in argan oil. Korean J. Agric. Sci. 2019, 46, 183–194. [Google Scholar] [CrossRef]
- Paudel, A.; Raijada, D.; Rantanen, J. Raman spectroscopy in pharmaceutical product design. Adv. Drug Deliv. Rev. 2015, 89, 3–20. [Google Scholar] [CrossRef] [Green Version]
- Yonzon, C.R.; Donovan, B.A. Raman spectroscopic method for content Uniformity of a dry powder inhaler. Am. Pharm. Rev. 2008, 11, 70–76. [Google Scholar]
- Schulze, G.; Jirasek, A.; Yu, M.M.L.; Lim, A.; Turner, R.F.B.; Blades, M.W. Investigation of selected baseline removal techniques as candidates for automated implementation. Appl. Spectrosc. 2005, 59, 545–574. [Google Scholar] [CrossRef] [PubMed]
- Mo, C.; Kang, S.; Lee, K.; Kim, G.; Cho, B.-K.; Lim, J.G.; Lee, H.S.; Park, J. Germination prediction of cucumber (cucumis sativus) seed using raman spectroscopy. J. Biosys. Eng. 2012, 37, 404–410. [Google Scholar] [CrossRef]
- Lee, H.; Cho, B.K.; Kim, M.S.; Lee, W.H.; Tewari, J.; Bae, H.; Sohn, S.I.; Chi, H.Y. Prediction of crude protein and oil content of soybeans using Raman spectroscopy. Sens. Actuators. B Chem. 2013, 185, 694–700. [Google Scholar] [CrossRef] [Green Version]
- Handbook of Near-Infrared Analysis—Google Books. Available online: https://books.google.co.kr/books?hl=en&lr=&id=6EEd1a0uka0C&oi=fnd&pg=PP1&dq=D.A.+Burns,+E.W.+Ciurczak,+Handbook+of+Near-Infrared+Analysis,+3rd+ed.%3B+Taylor+%26+Francis+Group:+New+York,+2007.&ots=L3b8O0HPrv&sig=BMt4VSyfi0eVfRJ6pawZtcP6CvA#v=onepage&q&f=false (accessed on 6 November 2019).
- Joshi, R.; Cho, B.K.; Lohumi, S.; Joshi, R.; Lee, J.; Lee, H.; Mo, C. Evaluation of benzene residue in edible oils using Fourier transform infrared (FTIR) spectroscopy. Korean J. Agric. Sci. 2019, 46, 257–271. [Google Scholar]
- Gowena, A.A.; Downey, G.; Esquerre, C.; O’Donnell, C.P. Preventing over-fitting in PLS calibration models of near-infrared (NIR) spectroscopy data using regression coefficients. J. Chemom. 2011, 25, 375–381. [Google Scholar] [CrossRef]
- Lohumi, S.; Lee, S.; Cho, B.K. Optimal variable selection for Fourier transform infrared spectroscopic analysis of starch-adulterated garlic powder. Sens. Actuators B Chem. 2015, 216, 622–628. [Google Scholar] [CrossRef]
- Cho, J.H.; Lee, D.; Jin, H.P.; Kim, K.; Lee, I.B. Optimal approach for classification of acute leukemia subtypes based on gene expression data. Biotechnol. Prog. 2002, 18, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y. Statistical Approaches for Detection of Relevant Genes and Pathway in Analysis of Gene Expression Data. Ph.D. Thesis, University of California, Davis, CA, USA, 2008. [Google Scholar]
- Pérez-Enciso, M.; Tenenhaus, M. Prediction of clinical outcome with microarray data: A partial least squares discriminant analysis (PLS-DA) approach. Hum. Genet. 2003, 112, 581–592. [Google Scholar]
- Kwok, C.S. Separations with Raman Spectroscopy for Identification of Oxidatively Modified Components in Parkinson’s Disease. Ph.D. Thesis, McGill University, Montreal, QC, Canada, 2008. [Google Scholar]
- Okparanma, R.N.; Mouazen, A.M. Visible and near-infrared spectroscopy analysis of a polycyclic aromatic hydrocarbon in soils. Sci. World J. 2013, 2013, 1–9. [Google Scholar] [CrossRef] [PubMed]
- William, P.; Norris, K. Near-Infrared Technology in the Agricultural and Food Industrie, 2nd ed.; American Association of Cereal Chemists: St. Paul, MN, USA, 2001; pp. 145–169. [Google Scholar]
- LIU, C.; BERG, R.W. Nonlinearity in Intensity versus Concentration Dependence for the Deep UV Resonance Raman Spectra of Toluene and Heptane. Appl. Spectrosc. Rev. 2013, 48, 425–437. [Google Scholar] [CrossRef]

| Toluene (%) | Toluene (mL) | Grignard Reagent (mL) |
|---|---|---|
| 0 | 0.0 | 30.0 |
| 1 | 0.3 | 29.7 |
| 2 | 0.6 | 29.4 |
| 3 | 0.9 | 29.1 |
| 4 | 1.2 | 28.8 |
| 5 | 1.5 | 28.5 |
| 10 | 3.0 | 27.0 |
| 15 | 4.5 | 25.5 |
| 20 | 6.0 | 24.0 |
| Total amount used = 18 mL | Total amount used = 252 mL |
| Methods | SEC (%) | SECV (%) | SEP (%) | LVs | Bias (%) | RPD | |||
| Whole spectra | 0.94 | 0.65 | 0.94 | 0.68 | 0.95 | 0.79 | 4 | −0.61 | 5.24 |
| VIP-PLSR | 0.98 | 0.60 | 0.96 | 0.66 | 0.97 | 0.71 | 4 | −2.21 | 7.15 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joshi, R.; Joshi, R.; Mo, C.; Faqeerzada, M.A.; Amanah, H.Z.; Masithoh, R.E.; Kim, M.S.; Cho, B.-K. Raman Spectral Analysis for Quality Determination of Grignard Reagent. Appl. Sci. 2020, 10, 3545. https://doi.org/10.3390/app10103545
Joshi R, Joshi R, Mo C, Faqeerzada MA, Amanah HZ, Masithoh RE, Kim MS, Cho B-K. Raman Spectral Analysis for Quality Determination of Grignard Reagent. Applied Sciences. 2020; 10(10):3545. https://doi.org/10.3390/app10103545
Chicago/Turabian StyleJoshi, Rahul, Ritu Joshi, Changyeun Mo, Mohammad Akbar Faqeerzada, Hanim Z. Amanah, Rudiati Evi Masithoh, Moon S. Kim, and Byoung-Kwan Cho. 2020. "Raman Spectral Analysis for Quality Determination of Grignard Reagent" Applied Sciences 10, no. 10: 3545. https://doi.org/10.3390/app10103545







