A Hybrid Geometric Morphometric Deep Learning Approach for Cut and Trampling Mark Classification
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
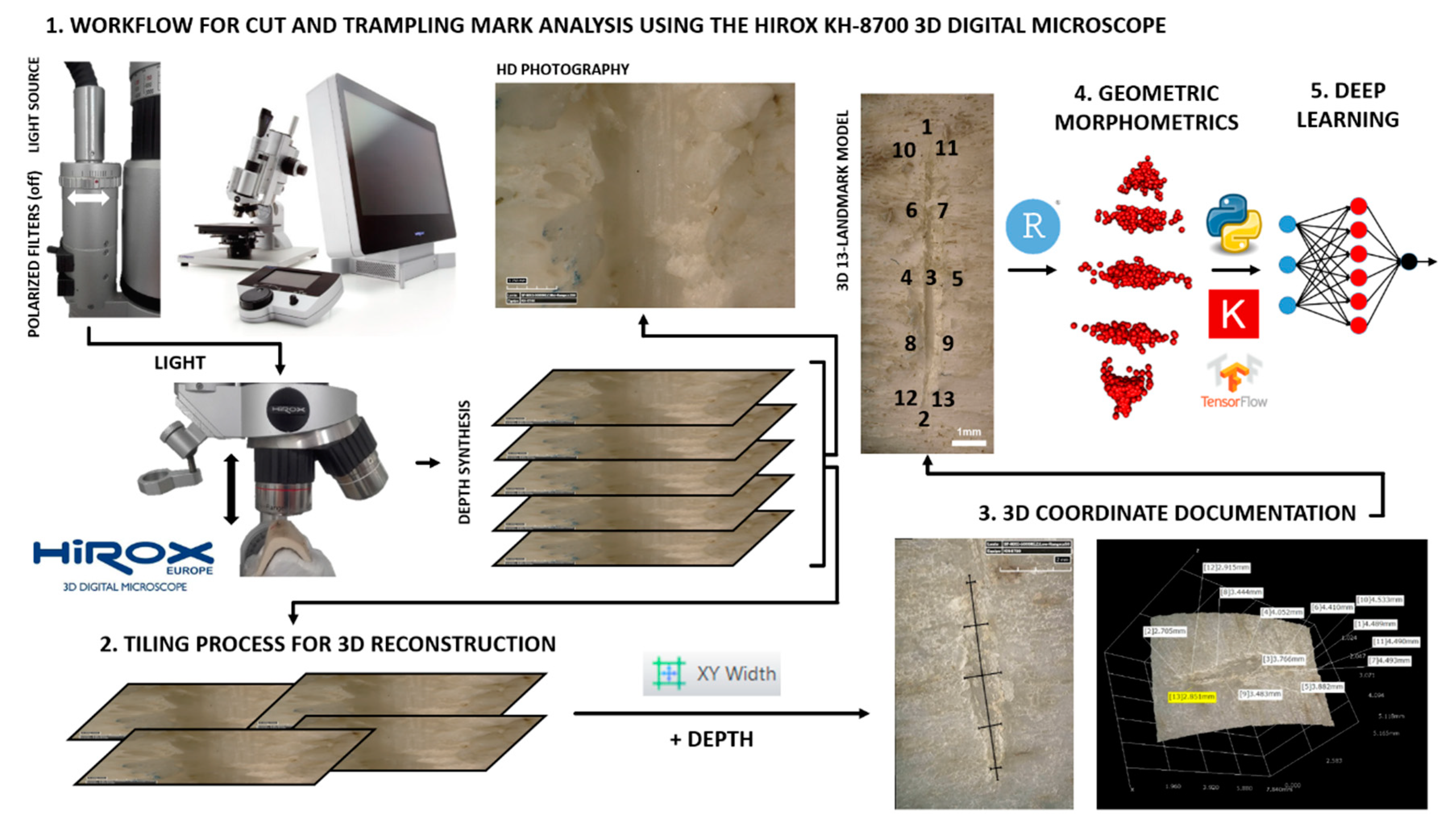
2.1. Digital Reconstruction Technique
2.2. Geometric Morphometrics
2.3. Deep Learning
3. Results
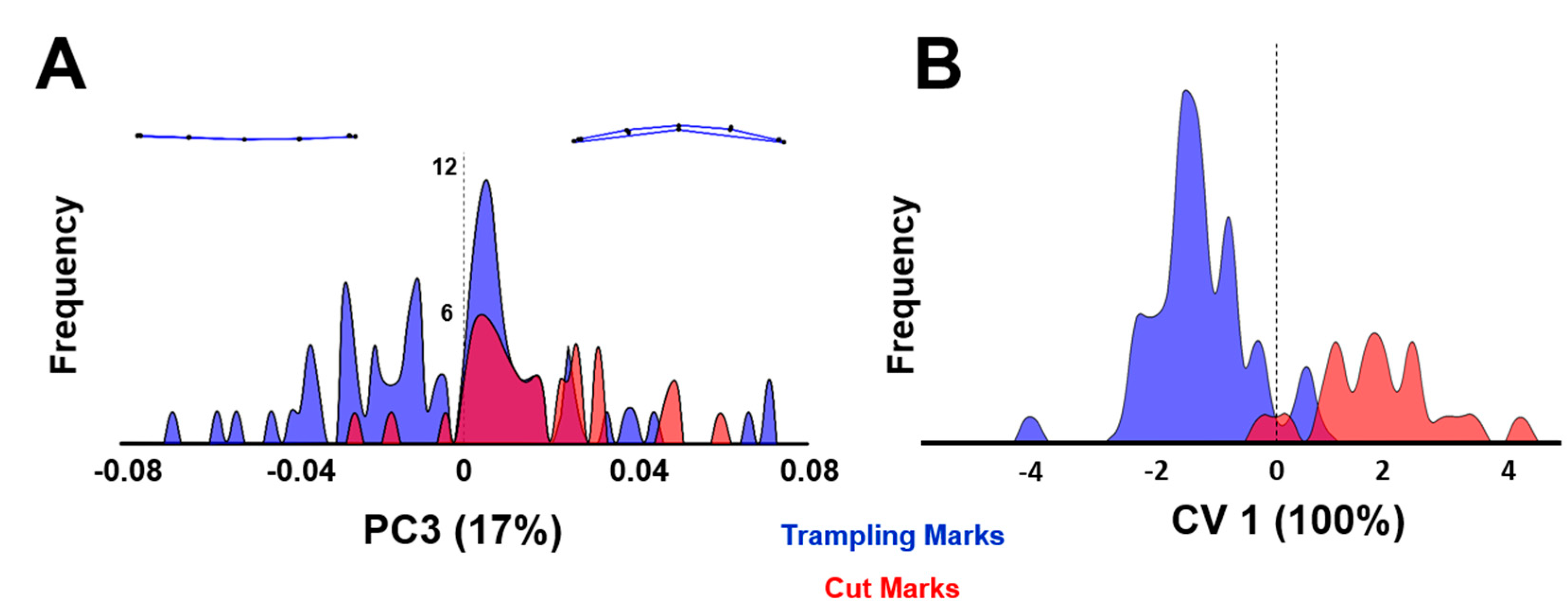
3.1. Geometric Morphometrics
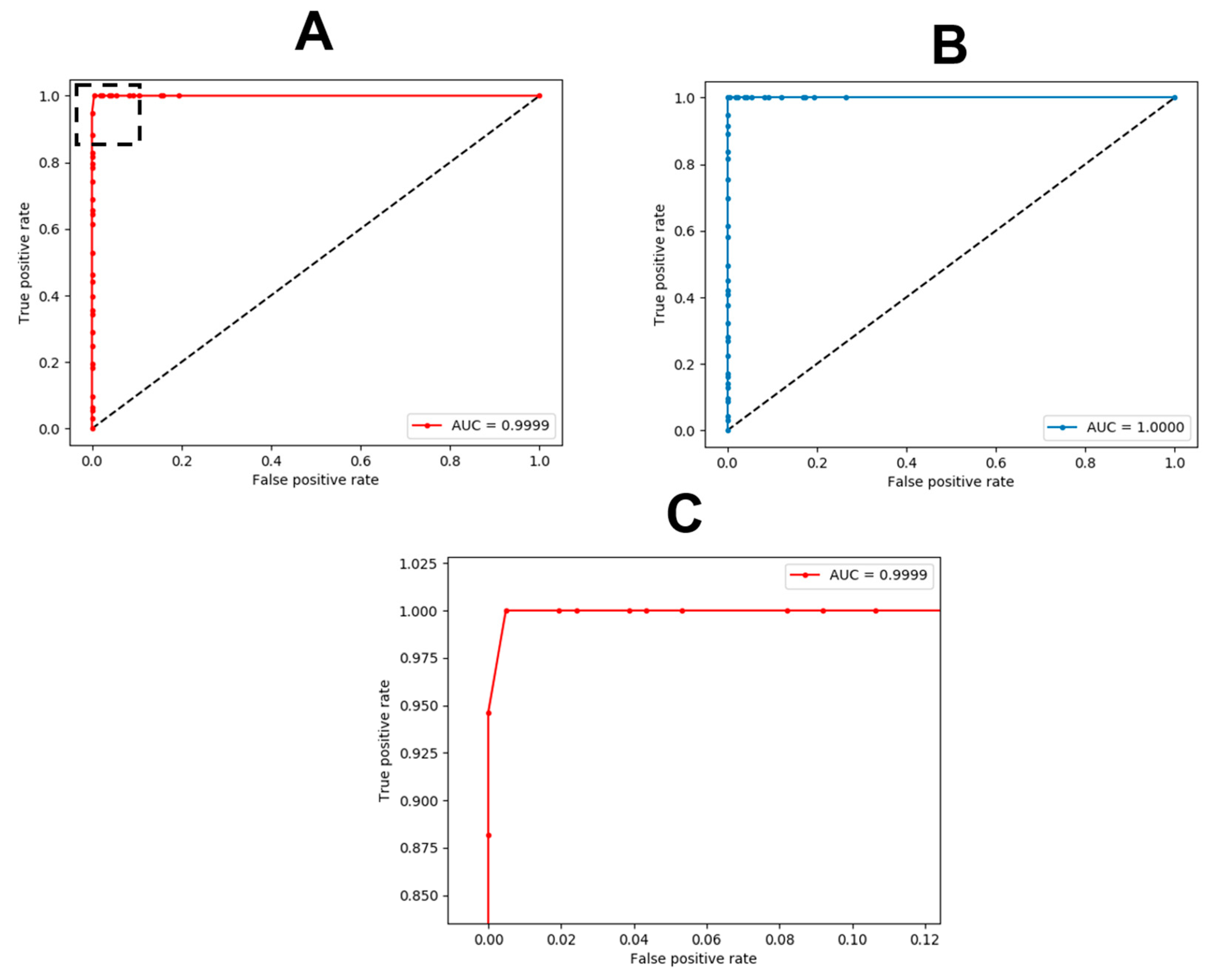
3.2. Deep Learning
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Library | Used For: | Link |
|---|---|---|
| Geomorph | Generalized Procrustes Analysis Principal Components Analysis Thin Plate Splines | https://cran.r-project.org/web/packages/geomorph/geomorph.pdf |
| Shapes | Canonical Variate Analysis | https://cran.r-project.org/web/packages/shapes/shapes.pdf |
| RVAideMemoire | Multivariate Variance Analysis | https://cran.r-project.org/web/packages/RVAideMemoire/RVAideMemoire.pdf |
| Library | Used for: | Link |
|---|---|---|
| TensorFlow 2.0. Keras API | Neural Network Construction Hyperparameter Optimization | https://www.tensorflow.org/ https://keras.io/ |
| Numpy | Numerical applications and operations Slicing, indexing and transformation of data | https://www.numpy.org/ |
| Pandas | Loading data | https://pandas.pydata.org/ |
| Matplotlib | Plotting learning curves and ROC results | https://matplotlib.org/ |
| Scikit-Learn | Model evaluation | https://scikit-learn.org/ |
References
- McPherron, S.P.; Alemseged, Z.; Marean, C.W.; Wynn, J.G.; Reed, D.; Geraads, D.; Bobe, R.; Béarat, H. Evidence for stone-tool-assisted consuption of animal tissues before 3.39 million years ago at Dikika, Ethiopia. Nat. Lett. 2010, 466, 857–860. [Google Scholar] [CrossRef]
- Domínguez-Rodrigo, M.; Pickering, T.R.; Bunn, H.T. Experimental study of cut marks made with rocks unmodified by human flaking and its bearing on claims of ~3.4-Million-Year-Old butchery evidence from Dikika. J. Arch. Sci. 2012, 39, 205–214. [Google Scholar] [CrossRef]
- Domínguez-Rodrigo, M.; Alcalá, L. 3.3 million year old stone tools and butchery traces? More evidence needed. Paleoanthropology. 2016, 2016, 46–53. [Google Scholar]
- Malassé, A.D.; Moigne, A.M.; Singh, M.; Calligaro, T.; Karir, B.; Gaillard, C.; Kaur, A.; Bharwaj, V.; Pal, S.; Abdessadok, S.; et al. Intentional cut marks on bovid from the Quranwala Zone, 2.6 Ma, Siwalik Frontal Range, Northwestern India. Comptes Rendus Palevol 2016, 15, 317–339. [Google Scholar] [CrossRef]
- Holen, S.R.; Deméré, T.A.; Fisher, D.C.; Fullagar, R.; Paces, J.B.; Jefferson, G.T.; Beeton, J.M.; Cerutti, R.A.; Rountret, A.N.; Vescera, L.; et al. A 130,000 year old archaeological site in Southern California, USA. Nature 2017, 544, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Rodrigo, M.; Pickering, T.R.; Semaw, S.; Rogers, M.J. Cutmarked bones from Pliocene archaeological sites at Gona, Afar, Ethiopia: Implications for function of the world’s oldest stone tools. J. Hum. Evol. 2005, 48, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Sahnouni, M.; Parés, J.M.; Duval, M.; Cáceres, I.; Harichane, Z.; van der Made, J.; Pérez-González, A.; Abdessadok, S.; Kandi, N.; Derradji, A.; et al. 1.9-Million-Year and 2.4-Million-Year-Old Artifacts and Stone Tool-Cutmarked bones from Ain Boucherit, Algeria. Science 2019, 362, 1297–1301. [Google Scholar] [CrossRef] [PubMed]
- Bunn, H.T. Meat Eating and Human Evolution: Studies on the Diet and Subsistence Patterns of Plio-Pleistocene Hominids in East Africa. Ph.D. Thesis, University of California, Oakland, CA, USA, 1982. [Google Scholar]
- Milton, K. Primate Diets and Gut Morphology: Implications for Hominid Evolution. In Food and Evolution: Toward a Theory of Human Food Habits; Harris, M., Ross, E.B., Eds.; Temple University: Philadelphia, PA, USA, 1987; pp. 93–115. [Google Scholar]
- Aiello, L.C.; Wheeler, P. The expensive tissue hypothesis. Curr. Anthropol. 1995, 36, 199–221. [Google Scholar] [CrossRef]
- Stanford, C.B.; Bunn, H.T. Meat Eating and Human Evolution; Oxford University: Oxford, UK, 2001. [Google Scholar]
- Binford, L.R. Bones: Ancient Men and Modern Myths; Academic Press: New York, NY, USA, 1981. [Google Scholar]
- Blumenschine, R. Percussion marks, tooth marks and experimental determinations of hominid and carnivore access to long bones at FLK Zinjanthropus, Olduvai Gorge, Tanzania. J. Hum. Evol. 1995, 29, 21–51. [Google Scholar] [CrossRef]
- Domínguez-Rodrigo, M. Meat-eating by early hominids at the FLK-22 Zinjanthropus Site, Olduvai Gorge, Tanzania: An experimental approach using cut mark data. J. Hum. Evol. 1997, 33, 669–690. [Google Scholar] [CrossRef]
- Domínguez-Rodrigo, M.; Barba, R. New estimates of tooth mark and percussion mark frequencies at the FLK-Zinj Site: The carnivore-hominid-carnivore hypothesis falsified. J. Hum. Evol. 2006, 50, 170–194. [Google Scholar] [CrossRef] [PubMed]
- Toth, N.; Schick, K. The Oldowan: Case Studies into the Earliest Stone Age; Stone Age Institute Press: Gosport, England, 2006. [Google Scholar]
- Key, A.J.M.; Dunmore, C.J. The evolution of the Hominin thumb and the influence exerted by non-dominant hand during stone tool production. J. Hum. Evol. 2015, 78, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Toth, N.; Schick, K. An Overview of the Cognitive Implications of the Oldowan Industrial Complex. Azania Arch. Res. Afr. 2018, 53, 3–39. [Google Scholar] [CrossRef]
- Semaw, S.; Roberts, M.J.; Quade, J.; Renne, P.R.; Butler, R.F.; DOmínguez-Rodrigo, M.; Stout, D.; Hart, W.S.; Pickering, T.; Simpson, S.W. 2.6 million year old stone tools and associated bones from OGS-6 and OGS-7, Gona, Afar, Ethiopia. J. Hum. Evol. 2003, 45, 169–177. [Google Scholar] [CrossRef]
- Domalain, M.; Bertin, A.; Daver, G. Was Australopithecus Afarensis able to make the Lomekwian Stone Tools? Towards a realistic biomechanical simulation of hand force capability in fossil hominins and new insights on the role of the fifth digit. Comptes Rendus Palevol 2017, 16, 572–584. [Google Scholar] [CrossRef]
- Courtenay, L.A.; Yravedra, J.; Maté-González, M.Á.; Aramendi, J.; González-Aguilera, D. 3D analysis of cut marks using a new geometric morphometric methodological approach. J. Arch. Anthr. Sci. 2019, 11, 651–665. [Google Scholar] [CrossRef]
- Maté-González, M.Á.; Palomeque-González, J.F.; Yravedra, J.; González-Aguilera, D.; Domínguez-Rodrigo, M. Micro-photogrammetric and morphometric differentiation of cut marks on bones using metal knives, quartzite and flint flakes. J. Arch. Anthr. Sci. 2016, 10, 805–816. [Google Scholar] [CrossRef]
- Courtenay, L.A.; Yravedra, J.; Aramendi, J.; Maté-González, M.Á.; Martín-Perea, D.M.; Uribelarrea, D.; Baquedano, E.; González-Aguilera, D.; Domínguez-Rodrigo, M. Cut marks and raw material exploitation in the Lower Pleistocene Site of Bell’s Korongo (BK, Olduvai Gorge, Tanzania): A geometric morphometric analysis. Quat. Int. 2019, 526, 155–168. [Google Scholar] [CrossRef]
- Aramendi, J.; Maté-González, M.A.; Yravedra, J.; Cruz Ortega, M.; Arriaza, M.C.; González-Aguilera, D.; Baquedano, E.; Domínguez-Rodrigo, M. Discerning carnivore agency through the three-dimensional study of tooth pits: Revisiting crocodile feeding leistoc at FLK-Zinj and FLK NN3 (Olduvai Gorge, Tanzania). Palaeogeog. Palaeoclimat., Palaeoecol. 2017, 488, 93–102. [Google Scholar] [CrossRef]
- Yravedra, J.; García Vargas, E.; Maté González, M.A.; Aramendi, J.; Palomeque-González, J.; Vallés-Iriso, J.; Matasanz-Vicente, J.; González-Aguilera, D.; Domínguez-Rodrigo, M. The use of micro-photogrammetry and geometric morphometrics for identifying carnivore agency in bone assemblage. J. Arch. Sci. Rep. 2017, 14, 106–115. [Google Scholar] [CrossRef]
- Courtenay, L.A.; Yravedra, J.; Huguet, R.; Aramendi, J.; Maté-González, M.Á.; González-Aguilera, D.; Arriaza, M.C. Combining Machine Learning Algorithms and Geometric Morphometrics: A Study of Carnivore Tooth Marks. Palaeogeo Palaeoclim. Palaeoecol. 2019, 522, 28–29. [Google Scholar] [CrossRef]
- Yravedra, J.; Maté-González, M.Á.; Courtenay, L.A.; González-Aguilera, D.; Fernández Fernández, M. The use of canid tooth marks on bone for the identification of livestock predation. Sci. Rep. 2019, 9, 16301. [Google Scholar] [CrossRef] [PubMed]
- Arriaza, M.C.; Domínguez-Rodrigo, M. When Felids and Hominins ruled at Olduvai Gorge: A Machine Learning Analysis of Skeletal Profiles of the Non-Anthropogenic Bed I Sites. Quat. Sci. Rev. 2016, 139, 43–52. [Google Scholar] [CrossRef]
- Domínguez-Rodrigo, M. Successful classification of experimental Bone Surface Modifications (BSM) through Machine Learning algorithms: A solution to the controversial use of BSM in paleoanthropology? J. Arch. Anthro. Sci. 2019, 11, 2711–2725. [Google Scholar] [CrossRef]
- Byeon, W.; Domínguez-Rodrigo, M.; Arampatzis, G.; Baquedano, E.; Yravedra, J.; Maté-González, M.A.; Koumoutsakos, P. Automated identification and deep classification of cut marks on bones and its palaeoanthropological implications. J. Comp. Sci. 2019, 32, 36–43. [Google Scholar] [CrossRef]
- Moclán, A.; Domínguez-Rodrigo, M.; Yravedra, J. Classifying agency in bone breakage: An experimental analysis of fracture planes to differentiate between hominin and carnivore dynamic and static loading using machine learning (ML) algorithms. Archae. Anthro. Sci. 2019, 11, 4463–4680. [Google Scholar] [CrossRef]
- Domínguez-Rodrigo, M.; Saladié, P.; Cáceres, I.; Huguet, R.; Yravedra, J.; Rodríguez-Hidalgo, A.; Martín, R.; Pineda, A.; Marín, J.; Gené, C.; et al. use and abuse of cut mark analyses: The Rorschach Effect. J. Arch. Sci. 2017, 86, 14–23. [Google Scholar] [CrossRef]
- Domínguez-Rodrigo, M.; Juana, S.; Galán, A.B.; Rodríguez, M. A New Protocol to Differentiate Trampling Marks from Butchery Marks. J. Archaeol. Sci. 2009, 36, 2643–2654. [Google Scholar] [CrossRef]
- Cohen, J. Statistical Power Analysis for Behavioural Sciences; Lawrence Erlbaum Assoc.: Mahwah, NJ, USA, 1988. [Google Scholar]
- Courtenay, L.A.; Maté-González, M.Á.; Aramendi, J.; Yravedra, J.; González-Aguilera, D.; Domínguez-Rodrigo, M. Testing Accuracy in 2D and 3D Geometric Morphometric methods for cut mark identification and classification. PeerJ 2018, 6, e5133. [Google Scholar] [CrossRef]
- Yravedra, J.; Diez-Martín, F.; Egeland, C.P.; Maté-González, M.Á.; Palomeque-González, J.F.; Arriaza, M.C.; Aramendi, J.; García Vargas, E.; Estaca-Gómez, V.; Sánchez, P.; et al. FLK-West (Lower Bed II, Olduvai Gorge, Tanzania): A newearly Acheulean site with evidence for human exploitation of fauna. Boreas 2017, 46, 486–502. [Google Scholar] [CrossRef]
- Courtenay, L.A.; Yravedra, J.; Huguet, R.; Ollé, A.; Aramendi, J.; Maté-González, M.Á.; González-Aguilera, D. New taphonomic advances in 3D digital microscopy: A morphological characterisation of trampling marks. Quat Int. 2019, 517, 55–66. [Google Scholar] [CrossRef]
- Dryden, I.L.; Mardia, K.V. Statistical Shape Analysis; John Wiley & Sons: Chichester, UK, 1998. [Google Scholar]
- Bookstein, F. Morphometric Tools for Landmark Data: Geometry and Biology; Cambridge University Press: New York, NY, USA, 1991. [Google Scholar]
- Klingenberg, C.P.; Monteiro, L.R. Distances and Directions in Multidimensional Shape Spaces: Implications for Morphometric Applications. Soc. Syst. Biol. 2005, 54, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Deep Learning with Python; Manning: New York, NY, USA, 2017. [Google Scholar]
- Bishop, C. Neural Networks for Pattern Recognition; Oxford University Press: New York, UK, USA, 1995. [Google Scholar]
- Bishop, C. Pattern Recognition and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Brownlee, J. Better Deep Learning: Train. Faster, Reduce Overfitting and Make Better Predictions; Machine Learning Mastery: Melbourne, Australia, 2019. [Google Scholar]
- Zhang, T. Solving Large Scale Linear Prediction Problems Using Stochastic Gradient Descent Algorithms. In Proceedings of the Twenty-First International Conference on Machine Learning – ICML ’04, Alberta, Canada, 4–8 July 2004. [Google Scholar] [CrossRef]
- Hinton, G. Neural Networks for Machine Learning Online Course. Available online: https://www.coursera.org/learn/neural-netoworks/home/welcome (accessed on 28 November 2019).
- Duchi, J.; Hazan, E.; Singer, Y. Adaptive Subgradient Methods for Online Learning and Stochastic Optimization. J. Mach. Learn. Res. 2011, 12, 2121–2159. [Google Scholar]
- Kingma, D.P.; Ba, J.L. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference for Learning Representations, San Diego, CA, USA, 5–7 May 2015; arXiv: 1412.6980. [Google Scholar]
- Glorot, X.; Bordes, A.; Bengio, Y. Deep Sparse Rectifier Neural Networks. In Proceedings of the 14th International Conference on Artificial Intelligence, Lauderdale, FL, USA, 22–24 June 2011. [Google Scholar]
- Maas, A.L.; Hannun, A.Y.; Ng, A.Y. Rectifier Nonlinearities Improve Neural Network Acoustic Models. In Proceedings of the 30th International Conference on Machine Learning, Atlanta, GA, USA, 16–21 June 2013. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving Deep into Rectifiers: Surpassing Human-Level Perfomanca on ImageNet Classification. arXiv 2015, arXiv:1502.01852v1. [Google Scholar]
- Klambauer, G.; Unterthiner, T.; Mayr, A. Self-Normalizing Neural Networks. Adv. Neural Inf. Process. Syst. 2017, arXiv:1706.02515v5. [Google Scholar]
- Krogh, A.; Hertz, J.A. A simple weight decay can improve generalization. Adv. Neural Inf. Process. Syst. 1991, 4, 950–957. [Google Scholar]
- Srivastava, N. Improving Neural Networks with Dropout. Master’s Thesis, University of Toronto, Toronto, ON, Canada, 2013; pp. 12–13. [Google Scholar]
- Kuhn, M.; Johnson, K. Applied Predictive Modelling; Springer: New York, NY, USA, 2013. [Google Scholar]
- Sing, T.; Sander, O.; Beerenwinkel, N.; Lengauer, T. ROCR: Visualizing classifier performance in R. Bioinform. Apps. Note 2005, 21, 3940–3941. [Google Scholar] [CrossRef]
- Fawcett, T. An introduction to ROC analysis. Pattern Recog. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Blumenschine, R.J.; Marean, C.W.; Capaldo, S.D. Blind tests of inter-analyst correspondence and accuracy in the identification of cut marks, percussion marks, and carnivore tooth marks on bone surfaces. J. Arch. Sci. 1996, 23, 493–507. [Google Scholar] [CrossRef]
- Pante, M.C.; Muttart, M.V.; Keevil, T.L.; Blumenschine, R.J.; Njau, J.K.; Merritt, S.R. A new high resolution 3D quantitative method for identifying Bone Surface Modifications with implications for the Early Stone Age archaeological record. J. Hum. Evol. 2017, 102, 1–11. [Google Scholar] [CrossRef]
- Najafabadi, M.M.; Villanustre, F.; Khoshgoftaar, T.M.; Seliya, N.; Wald, R.; Muharemagic, E. Deep Learning Applications and Challenges in Big Data Analytics. J. Big Data 2015, 2, 1–21. [Google Scholar] [CrossRef]
- Manusco, S. Elliptic Fourier Analysis (EFA) and Artificial Neural Networks (ANNs) for the identification of grapevine (Vitis vinifera L.) genotypes. Vitis 1999, 38, 73–77. [Google Scholar]
- Dobigny, G.; Baylac, M.; Denys, C. Geometric morphometrics, neural networks and diagnosis of sibling Teterillus species (Rodentia, Gerbillinae). Biol. J. Linn. Soc. 2002, 77, 319–327. [Google Scholar] [CrossRef]
- Baylac, M.; Villemant, C.; Simbolotti, G. Combining geometric morphometrics with pattern recognition for the investigation of species complexes. Biol. J. Linn. Soc. 2003, 80, 89–98. [Google Scholar] [CrossRef]
- Bocxlaer, B.V.; Schultheiβ, R. Comparison of morphometric techniques for shapes with few homologous landmarks based on machine learning approaches to biological discrimination. Paleobiology 2010, 36, 497–515. [Google Scholar] [CrossRef]
- Lorenz, C.; Ferraudo, A.S.; Suesdek, L. Artificial Neural Network applied as a methodology of mosquito species identification. Acta Tropica. 2015, 152, 165–169. [Google Scholar] [CrossRef]
- Soda, K.J.; Slice, D.E.; Naylor, G.J.P. Artificial neural networks and geometric morphometric methods as a means of classification: A case study using teeth from Carcharhinus sp. (Carcharinidae). J. Morphol. 2016, 278, 131–141. [Google Scholar] [CrossRef]
- Richter, A.N.; Khohgoftaar, T.M. A review of statistical and machine learning methods for modeling cancer risk using structured clinical data. Artif. Intell. Med. 2018, 90, 1–14. [Google Scholar] [CrossRef]
- Ekins, S. The Next Era: Deep Learning in pharmaceutical research. Pharm. Res. 2016, 33, 259–2603. [Google Scholar] [CrossRef]
- Nolle, T.; Luettgen, S.; Seeliger, A.; Mühlhäuser, M. Analyzing business process anomalies using autoencoders. Mach. Learn. 2018, 107, 1875–1893. [Google Scholar]
- Ibrahim, M.R.; Haworth, J.; Cheng, T. Understanding cities with machine eyes: A review of deep computer vision in urban analytics. Cities 2019, 96, 1–13. [Google Scholar] [CrossRef]
- Pineda, A.; Saladié, P.; Vergés, J.M.; Huguet, R.; Cáceres, I.; Vallverdú, J. Trampling versus Cut Marks on chemically altered surfaces: An experimental approach and archaeological application at the barranc de la Boella Site (la Canonja, Tarragona, Spain). J. Arch. Sci. 2014, 50, 84–93. [Google Scholar] [CrossRef]
- Gümrükçu, M.; Pante, M.C. Assessing the Effects of Fluvial Abrasion on Bone Surface Modifications using High Resolution 3D Scanning. J. Arch. Sci. Rep. 2018, 21, 208–221. [Google Scholar]
- Bharadwak, S.; Bhatt, H.S.; Vatsa, M.; Sing, R. Domain specific learning for newborn face recognition. IEEE Trans. Info. Forens. Sec. 2016, 11, 1630–1641. [Google Scholar] [CrossRef]
- Keshari, R.; Vatsa, M.; Singh, R. Learning structure and strength of CNN filters for small sample size training. arXiv 2018, arXiv:1803.11405v1. [Google Scholar]
- D’Souza, R.N.; Huang, P.Y.; Yeh, F.C. Small data challenge: Structural analysis and optimization of Convolutional Neural Networks with small sample size. bioRxiv 2018. [Google Scholar] [CrossRef]
- Feng, S.; Zhou, H.; Dong, H. Using deep neural network with small dataset to predict material defects. Mater. Des. 2019, 162, 300–310. [Google Scholar] [CrossRef]



| Hyperparameter | Tested Settings | References |
|---|---|---|
| Number of Layers | Between 1 and 7 | [41,42,44] |
| Node Density * | Between 3 and 20 | [41,42,44] |
| Activation Function * | ReLU, Leaky ReLU, Tanh | [49,50,51,52] |
| Kernel Initializer | None, Uniform | [51,53] |
| Dropout ** | None, Present with a threshold of 0.5 between 0.5 and 0.9 | [54] |
| Weight Regularizer | None, l2 with a threshold between 0.01 and 0.00001 | [53] |
| Weight Constraint | UnitNorm, MaxNorm, MinMaxNorm | [54] |
| Training Epochs | Between 150 and 2000 | [41,42,44] |
| Batch Size | 4, 8, 16, 32, 64, 128, 256 | [41,42,44] |
| Optimizer | Stochastic Gradient Descent, RMSProp, AdaGrad, Adam | [45,46,47,48] |
| Learning Rate | Between 0.1 and 0.00001 | |
| Decay | Between 0.9 and 0.0001 | |
| Momentum | Between 0.99 and 0.1 |
| Training | Validation | Testing | ||
|---|---|---|---|---|
| Accuracy | Max | 100.00 | 100.00 | 100.00 |
| Mean | 99.56 | 99.50 | 99.59 | |
| Upper CI | 100.00 | 100.00 | 100.00 | |
| Lower CI | 98.97 | 98.74 | 98.95 | |
| Min | 98.43 | 97.63 | 98.00 | |
| Loss | Max | 0.13 | 0.09 | 0.02 |
| Mean | 0.05 | 0.02 | 0.01 | |
| Upper CI | 0.08 | 0.05 | 0.01 | |
| Lower CI | 0.02 | 0.00 | 0.00 | |
| Min | 0.00 | 0.00 | 0.00 |
| Mean | 2 SD | |
|---|---|---|
| Sensitivity | 1.000 | 0.000 |
| Specificity | 0.999 | 0.004 |
| Kappa | 0.997 | 0.008 |
| AUC | 1.000 | 0.000 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Courtenay, L.A.; Huguet, R.; González-Aguilera, D.; Yravedra, J. A Hybrid Geometric Morphometric Deep Learning Approach for Cut and Trampling Mark Classification. Appl. Sci. 2020, 10, 150. https://doi.org/10.3390/app10010150
Courtenay LA, Huguet R, González-Aguilera D, Yravedra J. A Hybrid Geometric Morphometric Deep Learning Approach for Cut and Trampling Mark Classification. Applied Sciences. 2020; 10(1):150. https://doi.org/10.3390/app10010150
Chicago/Turabian StyleCourtenay, Lloyd A., Rosa Huguet, Diego González-Aguilera, and José Yravedra. 2020. "A Hybrid Geometric Morphometric Deep Learning Approach for Cut and Trampling Mark Classification" Applied Sciences 10, no. 1: 150. https://doi.org/10.3390/app10010150
APA StyleCourtenay, L. A., Huguet, R., González-Aguilera, D., & Yravedra, J. (2020). A Hybrid Geometric Morphometric Deep Learning Approach for Cut and Trampling Mark Classification. Applied Sciences, 10(1), 150. https://doi.org/10.3390/app10010150








