Effects of Pollutants in Urban Wastewater on Rhizoplane Microbial Communities in Constructed Wetlands: Resistance and Resilience of Macrophyte-Associated Microbiomes
Abstract
1. Introduction
2. Materials and Methods
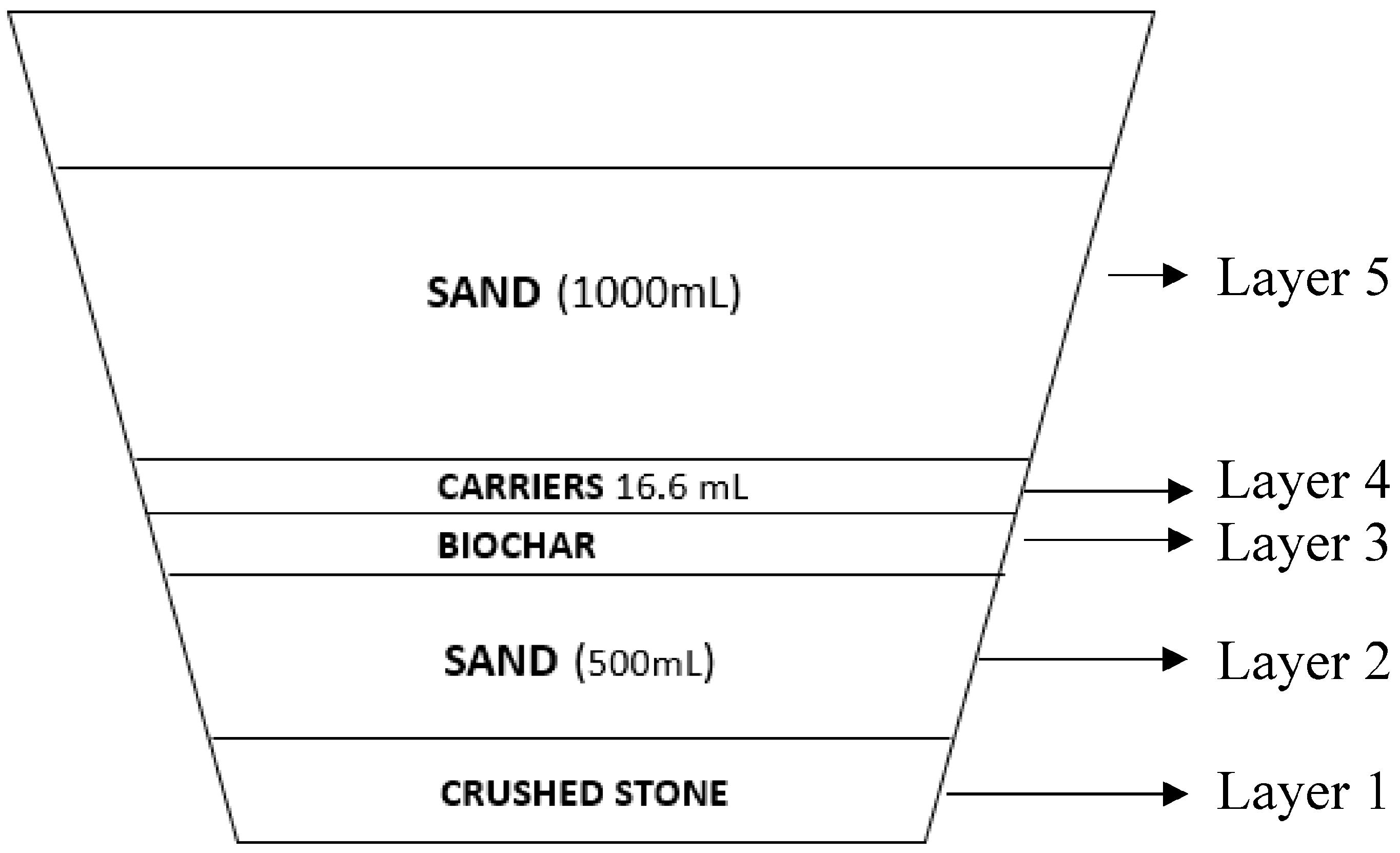
2.1. Vertical Submerged Flow System (VSFS-CW) Set Up
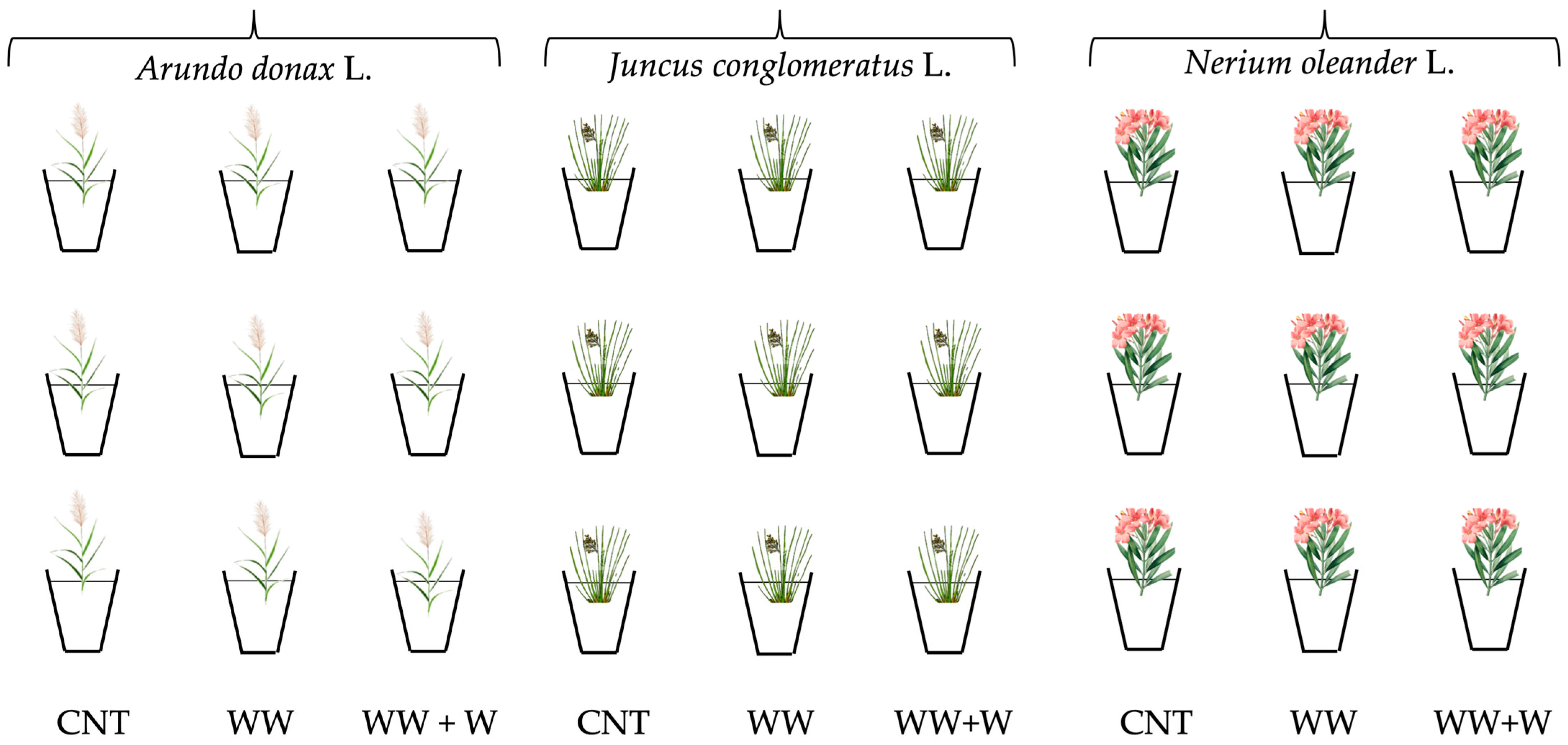
2.2. Set Up of the Experimental Plan
2.3. Wastewater Physical–Chemical Characterization
2.3.1. Chemical Oxygen Demand (COD)
2.3.2. Nitrate (NO3−), Nitrite (NO2−), and Ammonia (NH4+)
2.3.3. Chloride (Cl−)
2.4. Microbial Community Composition
2.4.1. Wastewater DNA Purification
2.4.2. Rhizo-Microbial Collection and DNA Purification
2.4.3. Amplicon Library Preparation and Next-Generation Sequencing (NGS)
2.5. Sequences Analysis
2.6. Microbiome Diversity Indices
- c = Pre-treatment (control);
- d = Post-treatment (after disturbance);
- ResI = 1 → no change = high resistance;
- ResI = 0 → complete change = no resistance.
- D_: dissimilarity between pre- and post-treatment;
- : dissimilarity between pre- and recovered;
- C = pre-treatment (CNT);
- D (disturbed) = immediately post-treatment (WW);
- R (recovered) = after recovery period (WW + W);
- RI = 1 indicates full recovery;
- RI < 1 indicates partial recovery;
- RI > 1 indicates overcompensation (a new stable state).
3. Results
3.1. Wastewater Treatment Efficiency
3.2. Microbiome Analysis
3.2.1. Wastewater Microbiome Characterization
3.2.2. Rhizoplane Microbial Community Characterizing Different Macrophyte Plant Species
Rhizoplane Communities of Arundo donax L.
Rhizoplane Communities of Juncus conglomeratus L.
Rhizoplane Communities of Nerium oleander L.
3.3. Effects of Wastewater on Rhizospheric Biodiversity of the Microbial Community
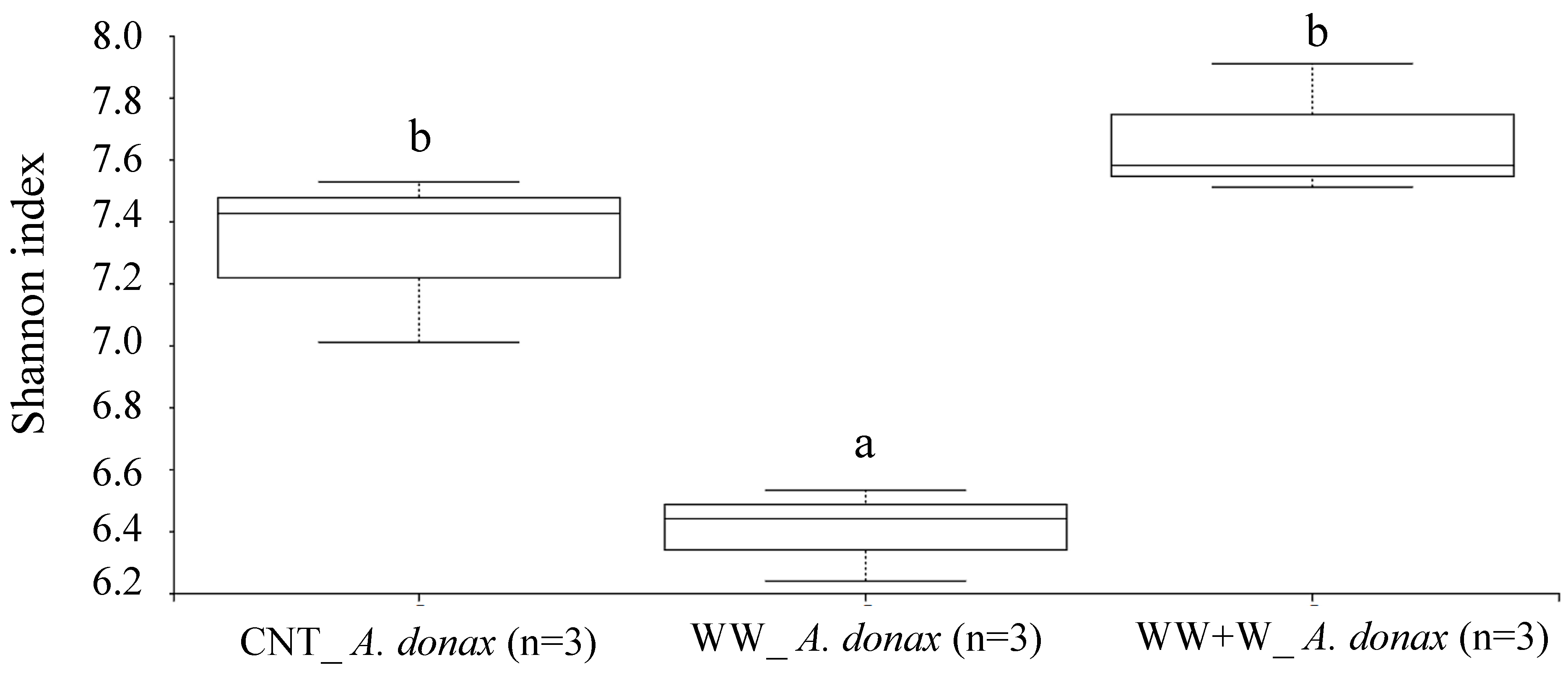
3.3.1. Rhizoplane Biodiversity Index of Arundo donax L.
3.3.2. Rhizoplane Biodiversity Index in Juncus conglomeratus L.
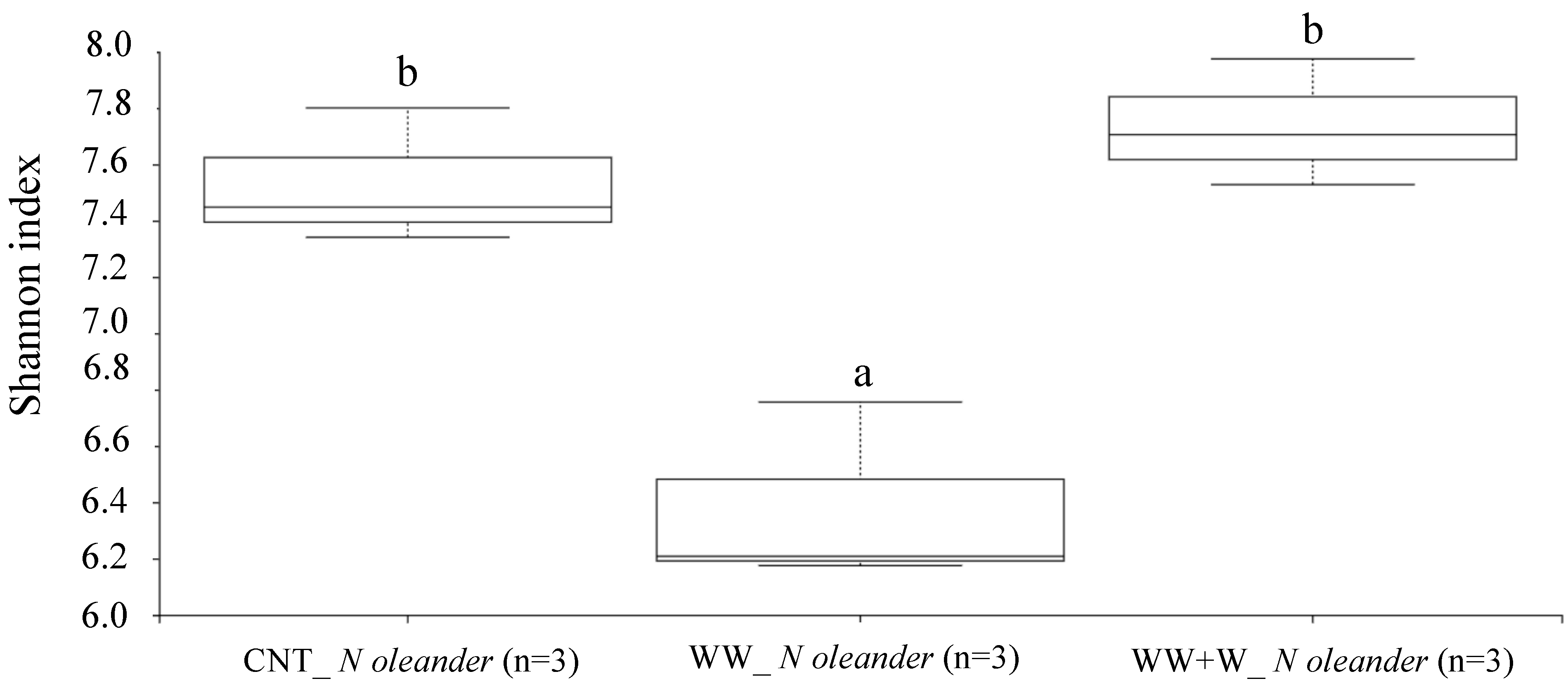
3.3.3. Rhizoplane Biodiversity Index in Nerium oleander L.
3.4. Normalization Index Definition
3.4.1. Normalization Index for Bacteria
3.4.2. Normalization Index for Fungi
4. Discussions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hernández-Canseco, J.; Bautista-Cruz, A.; Sánchez-Mendoza, S.; Aquino-Bolaños, T.; Sánchez-Medina, P.S. Plant Growth-Promoting Halobacteria and Their Ability to Protect Crops from Abiotic Stress: An Eco-Friendly Alternative for Saline Soils. Agronomy 2022, 12, 804. [Google Scholar] [CrossRef]
- Wang, J.W.; Long, Y.N.; Yu, G.L.; Wang, G.L.; Zhou, Z.Y.; Li, P.Y.; Zhang, Y.M.; Yang, K.; Wang, S.T. A Review on Microorganisms in Constructed Wetlands for Typical Pollutant Removal: Species, Function, and Diversity. Front. Microbiol. 2022, 13, 845725. [Google Scholar] [CrossRef] [PubMed]
- Gebru, S.B.; Werkneh, A.A. Werkneh, Applications of constructed wetlands in removing emerging micropollutants from wastewater: Occurrence, public health concerns, and removal performances-a review. S. Afr. J. Chem. Eng. 2024, 48, 395–416. [Google Scholar]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. Raaijmakers, The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Koskey, G.; Mburu, S.W.; Awino, R.; Njeru, E.M.; Maingi, J.M. Potential Use of Beneficial Microorganisms for Soil Amelioration, Phytopathogen Biocontrol, and Sustainable Crop Production in Smallholder Agroecosystems. Front. Sustain. Food Syst. 2021, 5, 606308. [Google Scholar] [CrossRef]
- Gentile, A.; Piccolo, P.; Iannece, P.; Cicatelli, A.; Castiglione, S.; Guarino, F. Reduction of antimicrobial resistance: Advancements in nature-based wastewater treatment. J. Hazard. Mater. 2024, 471, 134330. [Google Scholar] [CrossRef]
- Zheng, X.H.; Zhang, J.; Li, M.T.; Zhuang, L.L. Optimization of the pollutant removal in partially unsaturated constructed wetland by adding microfiber and solid carbon source based on oxygen and carbon regulation. Sci. Total Environ. 2021, 752, 141919. [Google Scholar] [CrossRef]
- Di Lenola, M.; Caracciolo, A.B.; Grenni, P.; Ancona, V.; Rauseo, J.; Laudicina, V.A.; Uricchio, V.F.; Massacci, A. Effects of Apirolio Addition and Alfalfa and Compost Treatments on the Natural Microbial Community of a Historically PCB-Contaminated Soil. Water Air Soil Pollut. 2018, 229, 143. [Google Scholar] [CrossRef]
- Urakawa, H.; Bernhard, A.E. Wetland management using microbial indicators. Ecol. Eng. 2017, 108, 456–476. [Google Scholar] [CrossRef]
- Alexandrino, D.A.M.; Mucha, A.P.; Almeida, C.M.R.; Gao, W.; Jia, Z.J.; Carvalho, M.F. Biodegradation of the veterinary antibiotics enrofloxacin and ceftiofur and associated microbial community dynamics. Sci. Total Environ. 2017, 581, 359–368. [Google Scholar] [CrossRef]
- Zhao, J.; Ni, T.; Li, J.; Lu, Q.; Fang, Z.Y.; Huang, Q.W.; Zhang, R.F.; Li, R.; Shen, B.; Shen, Q.R. Effects of organic-inorganic compound fertilizer with reduced chemical fertilizer application on crop yields, soil biological activity and bacterial community structure in a rice-wheat cropping system. Appl. Soil Ecol. 2016, 99, 1–12. [Google Scholar] [CrossRef]
- Brix, H. Functions of macrophytes in constructed wetlands. Water Sci. Technol. 1994, 29, 71–78. [Google Scholar] [CrossRef]
- Kausar, S.; Mahmood, Q.; Raja, I.A.; Khan, A.; Sultan, S.; Gilani, M.A.; Shujaat, S. Potential of Arundo donax to treat chromium contamination. Ecol. Eng. 2012, 42, 256–259. [Google Scholar] [CrossRef]
- Papazoglou, E.G.; Karantounias, G.A.; Vemmos, S.N.; Bouranis, D.L. Photosynthesis and growth responses of giant reed (Arundo donax L.) to the heavy metals Cd and Ni. Environ. Int. 2005, 31, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Borin, M.; Barbera, A.C.; Milani, M.; Molari, G.; Zimbone, S.M.; Toscano, A. Biomass production and N balance of giant reed (Arundo donax L.) under high water and N input in Mediterranean environments. Eur. J. Agron. 2013, 51, 117–119. [Google Scholar] [CrossRef]
- Corno, L.; Pilu, R.; Adani, F. Arundo donax L.: A non-food crop for bioenergy and bio-compound production. Biotechnol. Adv. 2014, 32, 1535–1549. [Google Scholar] [CrossRef]
- Mingorance, M.D.; Valdés, B.; Rossini Oliva, S. Strategies of heavy metal uptake by plants growing under industrial emissions. Environ. Int. 2007, 33, 514–520. [Google Scholar] [CrossRef]
- Tammone, C.; Tangredi, D.N.; Cicatelli, A.; Giuliano, F.; Spiniello, I.; Guarino, F.; Castiglione, S. Efficient phytoremediation of first-flush storm water using an innovative constructed wetland: A pilot-scale study towards sustainable water reuse. Plant Biosyst. Int. J. Deal. All Asp. Plant Biol. 2025, 159, 66–78. [Google Scholar] [CrossRef]
- Paolo, P. Valutazione Dell’impatto dei Reflui Urbani sul Microbioma Rizosferico. Master’s Thesis, Department Chemistry and Biology, University of Salerno, Fisciano, Italy, 2023. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High resolution sample inference from amplicon data. bioRxiv 2015, 024034. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Mandal, S.; Van Treuren, W.; White, R.A.; Eggesbø, M.; Knight, R.; Peddada, S.D. Analysis of composition of microbiomes: A novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015, 26, 27663. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation Evaluation and Phylogenetic Diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Rani, R.; Jain, S.; Garg, H. A review of nature-inspired algorithms on single-objective optimization problems from 2019 to 2023. Artif. Intell. Rev. 2024, 57, 126. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Colmer, Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef]
- Syranidou, E.; Christofilopoulos, S.; Kalogerakis, N. Juncus spp.-The helophyte for all (phyto)remediation purposes? New Biotechnol. 2017, 38, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, O.; Nguyen, T.T.H.; Al-Mamun, A.; Chang, I.S.; Ng, H.Y. T-RFLP reveals high β-Proteobacteria diversity in microbial fuel cells enriched with domestic wastewater. J. Appl. Microbiol. 2010, 109, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.M.; Hu, J.D.; Teng, Y.G.; Wang, J.S.; Chen, H.Y.; Guo, X.R.; Zhai, Y.Z. Response of microbial community to different media in start-up period of Annan constructed wetland in Beijing of China. Environ. Pollut. 2023, 337, 122529. [Google Scholar] [CrossRef] [PubMed]
- Ogwugwa, V.H.; Oyetibo, G.O.; Amund, O.O. Taxonomic profiling of bacteria and fungi in freshwater sewer receiving hospital wastewater. Environ. Res. 2021, 192, 110319. [Google Scholar] [CrossRef]
- Romanis, C.S.; Timms, V.J.; Crosbie, N.D.; Neilan, B.A. 16S rRNA gene amplicon sequencing data from an Australian wastewater treatment plant. Microbiol. Resour. Announc. 2024, 13, e01237-23. [Google Scholar] [CrossRef]
- Mawang, C.-I.; Azman, A.-S.; Fuad, A.-S.M.; Ahamad, M. Actinobacteria: An eco-friendly and promising technology for the bioaugmentation of contaminants. Biotechnol. Rep. 2021, 32, e00679. [Google Scholar] [CrossRef]
- Allison, S.D.; Martiny, J.B.H. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Yin, M.; He, T.; Xie, S.G. Influence of substrate type on microbial community structure in vertical-flow constructed wetland treating polluted river water. Environ. Sci. Pollut. Res. 2015, 22, 16202–16209. [Google Scholar] [CrossRef] [PubMed]
- Mesacasa, L.; Cabral, F.S.; Fochi, D.A.T.; Oliveira, W.D.; Oliveira, F.; Kersting, M.; Colares, G.S.; Rodriguez, A.L.; Lutterbeck, C.A.; Konrad, O.; et al. Constructed Wetlands and the role of the fungal community for wastewater treatment: A review. Ecohydrol. Hydrobiol. 2025, 25, 493–501. [Google Scholar] [CrossRef]
- He, Y.T.; Huang, Z.L.; Li, H.; Huang, J.; Qin, X.L.; Wu, Z.J. Sediment Fungal Communities of Constructed Wetlands Dominated by Zizania latifolia and Phragmites communis and Their Effect on Organic Pollutant Removal. Water 2023, 15, 2291. [Google Scholar] [CrossRef]
- Verma, P.; Yadav, A.N.; Kumar, V.; Singh, D.P.; Saxena, A.K. Beneficial plant-microbes interactions: Biodiversity of microbes from diverse extreme environments and its impact for crop improvement. In Plant-Microbe Interactions in Agro-Ecological Perspectives: Volume 2: Microbial Interactions and Agro-Ecological Impacts; Springer: Singapore, 2017; pp. 543–580. [Google Scholar]
- Ban, Y.H.; Xiao, Z.; Wu, C.; Lv, Y.C.; Meng, F.K.; Wang, J.Y.; Xu, Z.Y. The positive effects of inoculation using arbuscular mycorrhizal fungi and/or dark septate endophytes on the purification efficiency of CuO-nanoparticles-polluted wastewater in constructed wetland. J. Hazard. Mater. 2021, 416, 126095. [Google Scholar] [CrossRef]
- Hu, B.; Hu, S.S.; Chen, Z.B.; Vymazal, J. Employ of arbuscular mycorrhizal fungi for pharmaceuticals ibuprofen and diclofenac removal in mesocosm-scale constructed wetlands. J. Hazard. Mater. 2021, 409, 124524. [Google Scholar] [CrossRef]
- Treseder, K.K.; Lennon, J.T. Fungal traits that drive ecosystem dynamics on land. Microbiol. Mol. Biol. Rev. 2015, 79, 243–262. [Google Scholar] [CrossRef]
- Griffiths, B.S.; Philippot, L. Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol. Rev. 2013, 37, 112–129. [Google Scholar] [CrossRef]
- Shade, A.; Peter, H.; Allison, S.D.; Baho, D.; Berga, M.; Buergmann, H.; Huber, D.H.; Langenheder, S.; Lennon, J.T.; Martiny, J.B.; et al. Fundamentals of Microbial Community Resistance and Resilience. Front. Microbiol. 2012, 3, 2012. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Bay, G.; Lee, C.; Chen, C.L.; Mahal, N.K.; Castellano, M.J.; Hofmockel, K.S.; Halverson, L.J. Agricultural Management Affects the Active Rhizosphere Bacterial Community Composition and Nitrification. mSystems 2021, 6. [Google Scholar] [CrossRef]
- Shearer, C.A.; Descals, E.; Kohlmeyer, B.; Kohlmeyer, J.; Marvanová, L.; Padgett, D.; Porter, D.; Raja, H.A.; Schmit, J.P.; Thorton, H.A.; et al. Fungal biodiversity in aquatic habitats. Biodivers. Conserv. 2007, 16, 49–67. [Google Scholar] [CrossRef]
- Challacombe, J.F.; Hesse, C.N.; Bramer, L.M.; McCue, L.A.; Lipton, M.; Purvine, S.; Nicora, C.; Gallegos-Graves, L.; Porras-Alfaro, A.; Kuske, C.R. Genomes and secretomes of Ascomycota fungi reveal diverse functions in plant biomass decomposition and pathogenesis. BMC Genom. 2019, 20, 976. [Google Scholar] [CrossRef] [PubMed]
- Maza-Márquez, P.; Vilchez-Vargas, R.; Kerckhof, P.M.; Aranda, E.; González-López, J.; Rodelas, B. Community structure, population dynamics and diversity of fungi in a full-scale membrane bioreactor (MBR) for urban wastewater treatment. Water Res. 2016, 105, 507–519. [Google Scholar] [CrossRef]
- Kovacs, M.H.; Nghia, N.K.; Kovacs, E.D. Urban Forest Fragmentation Reshapes Soil Microbiome–Carbon Dynamics. Diversity 2025, 17, 545. [Google Scholar] [CrossRef]
- Wang, J.; Appidi, M.R.; Burdick, L.H.; Abraham, P.E.; Hettich, R.L.; Pelletier, D.A.; Doktycz, M.J. Formation of a constructed microbial community in a nutrient-rich environment indicates bacterial interspecific competition. Msystems 2024, 9, e0000624. [Google Scholar] [CrossRef]
- Oren, A. Thermodynamic limits to microbial life at high salt concentrations. Environ. Microbiol. 2011, 13, 1908–1923. [Google Scholar] [CrossRef]
- Ansola, G.; Arroyo, P.; de Miera, L.E.S. Characterisation of the soil bacterial community structure and composition of natural and constructed wetlands. Sci. Total Environ. 2014, 473, 63–71. [Google Scholar] [CrossRef]

| Pollutants (mg L−1) | T0 | After 48 h | ||
|---|---|---|---|---|
| Influent | A. donax (n = 3) | N. oleander (n = 3) | J. conglomeratus (n = 3) | |
| COD | 1353 ± 112 | 186 * ± 7 | 190 * ± 3 | 177 * ± 3 |
| N-NH4 | 10.5 ± 0.8 | 1.3 * ± 0.2 | ND | ND |
| N-NO3 | 3.5 ± 1.6 | 18.5 *± 0.2 | 16.0 *± 0.1 | 15.3 * ± 0.1 |
| N-NO2 | 1.2 ± 0.1 | ND | ND | ND |
| Cl− | 283 ± 24 | 211 ± 22 | 193 ± 15 | 123 * ± 9 |
| INDEX | Frequencies % |
|---|---|
| Archaea; Crenarchaeota | 0.02 |
| Archaea; Euryarchaeota | 0.29 |
| Bacteria; Acidobacteria | 2.86 |
| Bacteria; Actinobacteria | 6.76 |
| Bacteria; Bacteroidetes | 1.47 |
| Bacteria; Cyanobacteria | 0.04 |
| Bacteria; Firmicutes | 18.20 |
| Bacteria; Patescibacteria | 1.99 |
| Bacteria; Proteobacteria | 58.28 |
| Bacteria; Spirochaetes | 0.06 |
| Bacteria | 9.94 |
| Unassigned | 0.04 |
| INDEX | Frequencies % |
|---|---|
| k_Fungi;_ | 26.43 |
| K_fungi; p_Basidiomycota | 5.88 |
| K_fungi; p_Chytridiomycota | 3.29 |
| K_fungi; p_Glomeromycota | 3.30 |
| K_fungi; p_Mortirellomycota | 2.34 |
| K_fungi; p_Rozellomycota | 3.54 |
| K_fungi; p_Ascomycota | 44.27 |
| K_fungi; p_Blastocladiomycota | 2.29 |
| K_fungi; p_Fungi_phy_Incertae_sedis | 2.64 |
| K_fungi; p_Monoblepharmomycota | 2.29 |
| K_fungi; p_Olpidiomycoita | 3.69 |
| INDEX | WW/CNT TNI J. conglomeratus | WW/CNT TNI N. oleander | WW/CNT TNI A. donax |
|---|---|---|---|
| Thermoprotei | 0 | 0 | 0 |
| Halobacteria | 0 | 0.05 | 0.73 |
| Subgroup 6 | 1.57 | 1.83 | 2.32 |
| Actinobacteria | 3.32 | 4.04 | 0.62 |
| Bacteroidia | 0.77 | 0.99 | 1.72 |
| Oxyphotobacteria | 0 | 0 | 0 |
| Bacilli | 1.9 | 1.78 | 0.49 |
| Clostridia | 0.39 | 0.28 | 1.13 |
| Firmicutes | 0.65 | 0.3 | 1.12 |
| Patescibacteria-ABY1 | 1.84 | 1.93 | 1.72 |
| Planctomycetacia | 1.72 | 0 | 0 |
| Alphaproteobacteria | 0.64 | 0.7 | 0.87 |
| Deltaproteobacteria | 1.53 | 1.14 | 3.62 |
| Gammaproteobacteria | 0.93 | 1.01 | 0.97 |
| Proteobacteria | 1.2 | 0.75 | 1.63 |
| Spirochaetia | 0 | 0 | 0 |
| Bacteria | 0.67 | 0.4 | 1.02 |
| INDEX | TNI WW + W/CNT J. Conglomeratus | TNI WW + W/CNT N. oleander | TNI WW + W/CNT A. donax |
|---|---|---|---|
| Thermoprotei | 0 | 0 | 0 |
| Halobacteria | 1.06 | 1.27 | 0.73 |
| Euryarchaeota | 0 | 0 | 0 |
| Subgroup 6 | 2.35 | 2.22 | 2.32 |
| Actinobacteria | 0.62 | 1.92 | 0.62 |
| Bacteroidia | 1.99 | 1.41 | 1.72 |
| Oxyphotobacteria | 0 | 0 | 0 |
| Bacilli | 0.40 | 0.53 | 0.49 |
| Clostridia | 1.15 | 0.95 | 1.13 |
| Firmicutes | 1.07 | 0.79 | 1.12 |
| Patescibacteria-ABY1 | 2.05 | 3.25 | 1.72 |
| Patescibacteria | 0 | 0 | 0 |
| Planctomycetacia | 0 | 0 | 0 |
| Alphaproteobacteria | 0.76 | 0.75 | 0.87 |
| Deltaproteobacteria | 3.86 | 4.45 | 3.62 |
| Gammaproteobacteria | 0.91 | 0.96 | 0.97 |
| Proteobacteria | 2.05 | 1.56 | 1.63 |
| Spirochaetia | 0 | 0.02 | 0 |
| Bacteria | 1.34 | 0.83 | 1.02 |
| INDEX | TNI WW/CNT N. oleander | TNI WW/CNT A. donax | TNI WW/CNT J. conglomeratus |
|---|---|---|---|
| k_Fungi | 1.18 | 0.94 | 0.35 |
| Ascomycota | 0 | 0 | 0.03 |
| Ascomycota_cls_Incertae_sedis | 0 | 0 | 6.01 |
| Dothideomycetes | 0 | 0 | 2.06 |
| Eurotiomycetes | 0 | 1.02 | 0.72 |
| Lecanoromycetes | 0 | 0.68 | 0 |
| Leotiomycetes | 0 | 0.49 | 2.53 |
| Saccharomycetes | 0 | 1.30 | 4.31 |
| Sordariomycetes | 12.47 | 0 | 1.64 |
| Taphrinomycetes | 0.22 | 0 | 0 |
| Basidiomycota | 14.51 | 0 | 3.04 |
| Agaricomycetes | 0.92 | 0 | 1.71 |
| Cystobasidiomycetes | 0 | 0 | 4.25 |
| Exobasidiomycetes | 0 | 0 | 0 |
| Malasseziomycetes | 0 | 0 | 0.91 |
| Microbotryomycetes | 0 | 0 | 0 |
| Tremellomycetes | 0 | 0 | 0.09 |
| Ustilaginomycetes | 0 | ND | 0 |
| Blastocladiomycetes | 0 | ND | 0 |
| Chytridiomycetes | 0 | ND | 0 |
| Lobulomycetes | 0 | 0 | 19.80 |
| Fungi_cls_Incertae_sedis | 0 | ND | 0 |
| Glomeromycota | 0 | ND | 0 |
| Glomeromycetes | 0 | 0 | 0.19 |
| Monoblepharidomycetes | 0 | ND | 0 |
| Olpidiomycetes | 0 | 0 | 1.62 |
| Rozellomycota_cls_Incertae_sedis | 0 | 0 | 0.41 |
| INDEX | TNI WW + W/CNT N. oleander | TNI WW + W/CNT A. donax | TNI WW + W/CNT J. conglomeratus |
|---|---|---|---|
| k_Fungi | 1.04 | 1.14 | 0.85 |
| Cladosporiaceae | 0 | 0 | 0.22 |
| Pleosporaceae | 0 | 0 | 0.01 |
| Orbiliaceae | 0 | 0 | 1.33 |
| Plectosphaerellaceae | 0 | 1.68 | 0.62 |
| Hypocreaceae | 0 | 0.37 | 0 |
| Nectriaceae | 0 | 0.12 | 0 |
| Stachybotryaceae | 0 | 0.76 | 6.23 |
| Basidiomycota | 7.00 | 0 | 1.10 |
| Agaricomycetes | 0.16 | ND | 0 |
| Psathyrellaceae | 1.39 | 0 | 1.47 |
| Lobulomycetales_fam_Incertae_sedis | 0 | 0 | 0 |
| Fungi_fam_Incertae_sedis | 0 | 0 | 0 |
| Malasseziomycetes | 0 | 0 | 2.09 |
| Malasseziomycetes | 0 | 0 | 0 |
| Microbotryomycetes | 0 | 0 | 0.34 |
| Tremellomycetes | 0 | 0 | 0 |
| Ustilaginomycetes | 0 | ND | 0 |
| Blastocladiomycetes | 0 | ND | 0 |
| Chytridiomycetes | 0 | ND | 0 |
| Lobulomycetes | 0 | 0 | 0.52 |
| Fungi_cls_Incertae_sedis | 0 | ND | 61.95 |
| Glomeromycota | 0 | ND | 13.13 |
| Glomeromycetes | 0 | 0 | 0.59 |
| Monoblepharidomycetes | 0 | ND | 1.45 |
| Olpidiomycetes | 0 | 0 | 0.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piccolo, P.; Gentile, A.; Cicatelli, A.; Guarino, F.; Castiglione, S. Effects of Pollutants in Urban Wastewater on Rhizoplane Microbial Communities in Constructed Wetlands: Resistance and Resilience of Macrophyte-Associated Microbiomes. Environments 2025, 12, 414. https://doi.org/10.3390/environments12110414
Piccolo P, Gentile A, Cicatelli A, Guarino F, Castiglione S. Effects of Pollutants in Urban Wastewater on Rhizoplane Microbial Communities in Constructed Wetlands: Resistance and Resilience of Macrophyte-Associated Microbiomes. Environments. 2025; 12(11):414. https://doi.org/10.3390/environments12110414
Chicago/Turabian StylePiccolo, Paolo, Annamaria Gentile, Angela Cicatelli, Francesco Guarino, and Stefano Castiglione. 2025. "Effects of Pollutants in Urban Wastewater on Rhizoplane Microbial Communities in Constructed Wetlands: Resistance and Resilience of Macrophyte-Associated Microbiomes" Environments 12, no. 11: 414. https://doi.org/10.3390/environments12110414
APA StylePiccolo, P., Gentile, A., Cicatelli, A., Guarino, F., & Castiglione, S. (2025). Effects of Pollutants in Urban Wastewater on Rhizoplane Microbial Communities in Constructed Wetlands: Resistance and Resilience of Macrophyte-Associated Microbiomes. Environments, 12(11), 414. https://doi.org/10.3390/environments12110414












