Abstract
Electrochemical biosensors that combine high selectivity of biochemical affinity with precise electrochemical detection are one of the most necessary and powerful tools for assessing environmental pollution. This review addresses electrochemical biosensors that assess environmental pollutant toxicity. Electrochemical biosensors using enzyme activity inhibition, DNA, whole cells, and cytochrome P450 will be introduced, their advantages and applications will be discussed, and trends and challenges for designing reliable sensors for practical use will be addressed.
1. Introduction
Environmental pollution has existed at least since the time that people started using fire, and has been an increasing problem worldwide since the rapid development of industry during the 19th century. In the modern world, a wide variety of toxicants, including heavy metals, pesticides, detergents, endocrine disruptors, antibiotics, and carcinogens, are on the list of these pollutants. According to the Royal Commission on Environmental Pollution in their 24th report, there are some 30,000 chemicals that are commonly being used nowadays, and each of them may pollute the environment during processing, use, and discard [1]. Environmental pollution reduces the number of wild animals, degrades the functioning of ecosystems, and poses a threat to human health. Therefore, for environmental safety, the prompt and precise evaluation of pollutant toxicity has become an urgent task. However, various chemical substances are mixed in the environment. The measurement of the toxicity of an individual chemical substance is complicated. To evaluate toxicity under these circumstances, evaluating the overall toxicity to organisms may be more efficient than determining the toxicity of individual chemicals. In this context, biosensors may be a good alternative or a complementary analytical tool.
A biosensor comprises an integral device involving a biological recognition element and an element that converts a recognition event into an output signal. Biosensors are classified into basic groups according to their signal transmission methods and biological recognition principles. Biosensors can be categorized by their transducing element as electrochemical, optical, piezoelectric, and thermal sensors; and based on the biorecognition principle, they are immunochemical, enzymatic, non-enzymatic receptor, and DNA and whole-cell biosensors. In recent times, biosensors have extensively been used for toxicity determination. Several research works for the toxicity determination of water [2,3], phenol, and nitrophenols [4], copper (II) ions [5], and other metal ions are available. The biosensors measured the toxic response of certain chemicals on the enzymatic activity, the respiratory rate, the assessment of various growth parameters, etc. The microtox method based on the bioluminescence attenuation in the presence of toxins is a popular method used for toxicity determination [6]. It is an acute toxicity test biosensor based on the bioluminescence of the marine bacterium Vibrio fischeri. MICREDOX, a rapid biosensor developed by Lincoln Technology was developed for the direct assessment of toxic chemicals on environmental biological materials [7]. This biosensor is superior for rapid toxicity assessment in terms of sensitivity, detection time, and reproducibility. Another well-known biosensor for toxicity determination is ToxTell, a mediated amperometric microbial biosensor [8]. Multiple bacterial species can be used as biological components to determine precise values of toxicants in real samples. This bacterial biosensor generates an electric current by passive electron extraction from the electron transport system, the central metabolic pathway of the cell. The currents produced with/without the toxin are proportional to the cell metabolic activity and indicate the inhibition. Several bacterial strains have been produced that can comprehensively detect the effects of contaminants in samples that may cause genotoxicity or other toxicity to the human body [9]. By using these toxicity biosensors, the monitoring of environmental pollutants becomes very easy and convenient. The advantages of these biosensors are their simplicity, high sensitivity, direct transduction, miniaturization, facilitation of continuous monitoring, ease of use, and low cost compared to conventional methods. Their potential to complement laboratory-based and field analytical methods for environmental monitoring are demonstrated in [10,11,12,13].
Electrochemical biosensors combine a biological recognition element immobilized on the electrode surface and a physicochemical detector. It allows the application of electrochemical methods to biological processes to generate electrical signals corresponding to the concentration of biological analytes [14]. These biosensors involve biocomponents and electrodes in different combinations where enzymes, antibodies, nucleic acids, hormone receptors, microorganisms or whole cells, and tissues are immobilized onto the electrode surfaces [15]. A general schematic for operating the electrochemical biosensor for evaluating the toxicity of the pollutants is shown in Figure 1. In recent years, materials, technologies, and applications of biosensors have been studied by various researchers. Some studies addressed enzyme-based biosensors and the electrocatalytic detection of small molecules [16,17], while others have provided an overview on the application of the nanostructured film in biosensors [18]. The development of glucose sensors based on carbon nanotubes (CNTs) and electrostatic assembly was reviewed by Harper and Anderson [19]. Additionally, some reviews address specific technologies, such as the development of DNA biosensors using peptide nucleic acids [20] and advances in technology using encapsulated enzymes [21]. Su et al. reported the application of microorganisms to measure analytes [22]. Among all electrochemical biosensors, some of them focused on enzyme-based, DNA-based, whole cell-based, and cytochrome P450-based biosensors. High specificity, sensitivity, fast response, and ease of operation are the advantages of electrochemical biosensors, which meet the requirements for environmental monitoring in the field [23]. The current technological progress makes it possible to miniaturize electrochemical biosensors with nanoscale instrumentation; thus, it becomes very useful for some sophisticated applications with a small volume of samples. The automation of electrochemical biosensors and the ability to analyze the opaque medium makes it advantageous over other biosensors. The appeal of electrochemical biosensors is noticed in many practical applications.
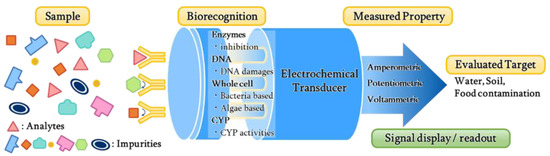
Figure 1.
Type of electrochemical biosensors to evaluate the toxicity of environmental pollutants.
In this review, the article presents an overview of the research and development of electrochemical biosensors for the toxicity assessment of environmental pollutants and describes important progress, as well as trends and challenges for designing reliable sensors for practical applications. We will review the electrochemical biosensors using inhibition of enzyme activity, DNA, whole-cell, cytochrome P450, and their uses, advantages, and limitations.
2. Biosensors Based on Enzyme Inhibition Activity
Enzyme-based biosensors measure the physical response, which yields a quantity related to the enzyme-catalyzed reaction rate. Enzyme activity is usually evaluated by direct measuring the electroactive products and co-substrates involved in the enzymatic reaction. To develop an enzyme-based biosensor, an indispensable step is the enzyme conjugation on the transducers for analyte quantification. The substrates are catalyzed by the enzyme and yield products. There are different methods for immobilization, the choice of which depends on many factors, including the nature and properties of the biological element, the transducer type, the analyte physicochemical properties, and the biosensor operation conditions. These considerations are essential for the maximum activity of the biological elements in the immobilized microenvironment [24,25,26]. The simplest and fastest method for immobilization is the adsorption of enzymes on the electrode surface. Many substances adsorb enzymes on their surfaces, including glass, silica gel, alumina, kaolin, charcoal, cellulose, and collagen, either by physical or by chemical adsorption. There are also methods to physically encapsulate the enzyme within a synthetic gel layer (a mixture of biomaterial and monomer solution that is polymerized to form a gel), or to covalently bond the functional groups in the biomaterial to the support matrix. Reactions are carried out under mild conditions at low temperature and ionic strength, with pH in the physiological range. Cross-linking is another method for enzyme immobilization. With this method, Biomaterials are usually bonded with a cross-linking agent to enhance adhesion. Glutaraldehyde is a widely used bifunctional agent. Different immobilization issues have also been discussed in the literature [27,28,29,30,31] and are shown in Figure 2. The recent developments in the field of enzyme-based biosensors are largely focused on the improvement of the immobilization and stability of enzymes. Among the various immobilization methods, covalent bonding offers the highest stability, trailed by cross-linking and encapsulation [32]. The least stable method is the adsorption where the enzymes and transducers are linked by the weak van der Waals forces, which are not very stable. However, this immobilization method is very easy to carry out because no reagents are necessary and cleanup is also inessential [32].
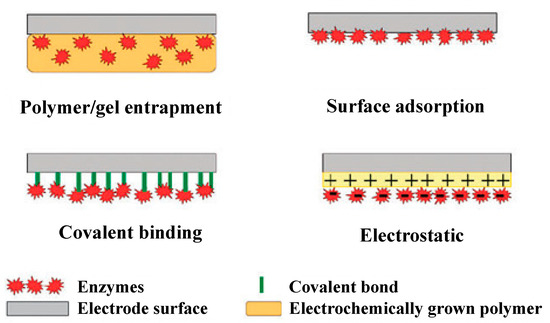
Figure 2.
Commonly used methods for enzyme immobilization onto an electrode surface. Adapted with permission from Ref. [32]. 2010, Royal Society of Chemistry.
Enzyme inhibition-based biosensors have been applied for various kinds of analytes, including organophosphorus and organochlorine pesticides, derivatives of insecticides, and heavy metals [24]. These biosensor systems rely on quantitative analysis of the difference in enzyme activity before/after exposure to a given target analyte. The working principle of enzyme inhibition-based biosensors is shown in Figure 3 [25]. The inhibited enzyme activity percentage after exposure is the quantification of the inhibitor toxicity and the incubation time [33,34]. The percentage inhibition is calculated as follows:
where I (%) is the inhibition percentage; A0 is the enzyme activity before inhibitor exposure; and Ai is the enzyme activity after the inhibitor exposure. There are various types of enzyme inhibition-based biosensors, including glucose oxidase, urease, cholinesterase, alkaline phosphates, etc. They are extensively used for the registration of the environmental toxicity of different types of pollutants.
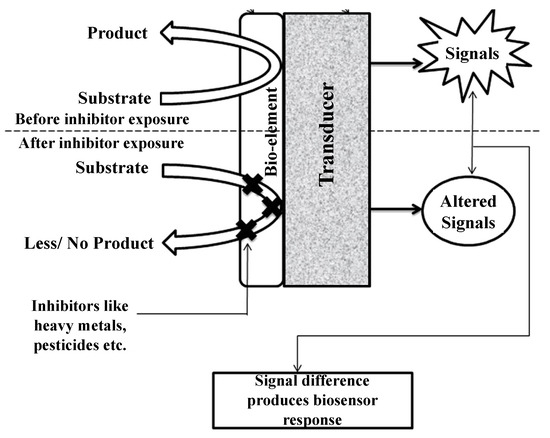
Figure 3.
Working principle of enzyme inhibition-based biosensors. Adapted with permission from Ref. [25]. 2013, Taylor & Francis.
2.1. Glucose Oxidase Inhibition
Glucose oxidase (GOx) is an oxidoreductase enzyme that primarily catalyzes the oxidation of glucose to hydrogen peroxide and gluconolactone. GOx-coated electrodes have been used widely since the pioneering works by Clark and Lyons in the 1950–1960s, known as first-generation biosensors (Figure 4). In these biosensors, the oxidase is immobilized behind a semipermeable membrane on the surface of the platinum electrode. GOx is a widely available, cheap, and stable enzyme that is mainly used in biosensors for industrial processes; it has high specificity for β-D-glucose and can be registered by the following reactions [32]:
β-D-glucose + GOx–FAD → GOx–FADH2 + δ-D-gluconolactone
GOx–FADH2 + O2 → GOx–FAD + H2O2
H2O2 → 2e− + O2 + 2H+
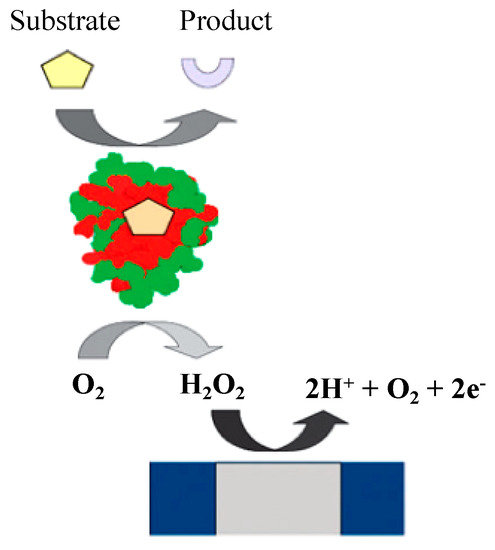
Figure 4.
Oxygen-dependent first-generation amperometric biosensor. Adapted with permission from Ref. [32]. 2010, Royal Society of Chemistry.
Recently, there have been some research works based on glucose oxidase inhibition, and most of them address heavy metals toxicity [35,36,37,38]. Ghica and Brett developed a biosensor using glucose oxidase immobilized on the carbon film electrode to determine copper, cadmium, lead, and zinc using polymerized Neutral Red as a mediator [35]. The biosensor was found to be sensitive to heavy metals with the detection limits of µg/mL. Guascito et al. presented an amperometric glucose oxidase biosensor for the determination of a wide group of heavy metals [36]. The enzyme glucose oxidase was immobilized on electro-synthesized poly-o-phenylenediamine. The inhibition results revealed that sensibility, detection limit, linear range, and standard deviation of the electrode make it suitable for the quantitative environmental analysis of metal ions. An electrochemical biosensor for mercury detection in compost extract was developed by enzyme immobilization on an aniline membrane supported on a platinum electrode by glutaraldehyde, with ferrocene as an electron transfer mediator [37]. A lower detection limit was achieved compared with a glucose oxidase-based biosensor immobilized on a carbon paste electrode modified with manganese dioxide [38].
2.2. Urease Inhibition
Urease was the first enzyme crystallized out from jack bean [39]. Biosensors based on urease inhibition are widely used for measuring heavy metals such as copper, cadmium, chromium, lead, and mercury [25]. A potentiometric biosensor with urease enzyme immobilized on self-assembled gold nanoparticles was developed and found to possess a low detection limit (0.05 µmol/L), fast response, and regeneration [40]. Do et al. developed another amperometric biosensor for the determination of mercury by immobilizing the enzyme on a polyaniline-Nafion film/Au/Al2O3-sensing electrode and achieved the detection level of (0.01 ppm) [41]. An optical fiber-based biosensor has been developed for detecting heavy metals in an aqueous sample considering the sol-gel approach [42]. A conductometric biosensor has been developed to determine the metal ions and it was found that the sensitivity for different metals was Cd2 + > Cu2 + > Pb2+ [43].
2.3. Cholinesterase Inhibition
Cholinesterase is an enzyme that hydrolyzes the neurotransmitter acetylcholine into choline and acetic acid, necessary reactions for cholinergic neurons to return to a quiescent state after activation. This enzyme has been widely used in direct electrochemical biosensors for pesticide detection. Analyzers based on cholinesterase inhibition are popular for detecting organophosphorus compounds [24]. Recently, this type of biosensor was designed based on the amperometric detection [39]. The toxic pollutants detected by this transduction mode include parathion, methyl parathion, paraoxon, methyl paraoxon, carbaryl, aldicarb, carbofuran, ppirimicarb, chloropyrifosoxon-methyl, nerve agents, fenthion, trichlorofon, and triazophosetc [44,45,46,47]. Based on their respective preferences for substrates, cholinesterase enzymes are of two types: acetylcholinesterase (AChE) hydrolyzes acetylcholine and butyrylcholinesterase (BChE) hydrolyzes butyrylcholine. Liu and Lin have developed an amperometric biosensor using AChE immobilized on CNT-modified glassy carbon (GC) electrode for detecting organophosphate pesticides and nerve agents [45]. The biosensor integrated with a flow injection system for measuring paraxon reached the detection limit of 0.4 pM with a 6-min inhibition time. In another study, an assay for organophosphate paraoxon was developed on the base of AChE immobilized on gelatin capable of detecting 200 pg for one minute [46]. Sinha et al. also developed a biosensor for paraxon determination using zinc oxide matrix for the AChE immobilization and paraoxon detection in the range of 0.035–1.38 ppm [47]. The biosensor has excellent storage ability of up to 3 months in phosphate-buffered saline (pH 7.4) at 4 °C.
2.4. Alkaline Phosphatase Inhibition
Alkaline phosphatase (ALP) is the most common binding enzyme in immunoassays due to its high turnover rate, broad substrate specificity, and potential for applications. Its activity has been determined electrochemically using phenyl phosphate, naphthyl phosphate, ascorbic acid 2-phosphate, and p-nitrophenyl phosphate stable substrates. Among them, p-nitrophenyl phosphate is probably one of the most popular substrates for ALP because it can be used to electrochemically detect enzymatically generated p-nitrophenol. Numerous studies using alkaline phosphatase enzyme inhibition were carried out by several researchers [48,49,50]. Alvarado-Gámez et al. developed an electrochemical chronoamperometric biosensor for the determination of vanadium ions based on ALP enzyme inhibition [48]. ALP has been immobilized by cross-linking to the screen-printed carbon electrodes (SPCEs) modified by gold nanoparticles (ALP-AuNPs-SPCEs). p-Nitrophenyl phosphate sodium salt (PAPP) was used as substrate and the detection limit of 0.39 ± 0.06 μM was found for vanadium ions with repeatability of 7.7% (n = 4) and reproducibility of 8% (n = 3). For detecting carbofuran using ALP enzyme inhibition, another indirect amperometric biosensor was designed where ALP was immobilized on a carbon nano-powder paste electrode (CNPPE) and chronoamperometric monitoring of the enzyme activity inhibition was performed [49]. The decrease of the response signal is monitored after the addition of the inhibitor keeping the substrate (disodium phenyl phosphate) concentration constant. The biosensor principle is given in Figure 5. The biosensor is satisfactory in terms of detection limit, dynamic range, stability, and precision; facilitating qualitative and quantitative determination of carbofuran. The limit of detection was found to be 10 µg/L for carbofuran, with a repeatability of 2.3% (n = 3), and reproducibility was 3.4% (n = 3). Tekaya et al. measured the ALP inhibition from the cyanobacterium, Arthrospira platensis using physical adsorption of the cells on the ceramic part of gold-interdigitated transducers [50]. ALP inhibition with ф detection limit of 10−20 M was registered with cadmium and mercury. The half-maximal inhibitory concentration (IC50) was evaluated as 10−19 M and 10−17 M for Cd2+ and Hg2+, respectively. The biosensor is not metal-specific and provides an integral response to the traces of heavy metals in natural waters and effluents.
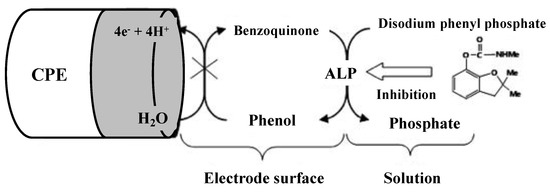
Figure 5.
Inhibition of the ALP/CPE biosensor with carbofuran. Adapted with permission from [49].
2.5. Other Enzymes Inhibition
The previously described enzymes are commonly used for biosensor construction. In addition to these enzymes, choline oxidase, polyphenol oxidase, nitrate reductase, invertase, horseradish peroxidase/catalase, and protein phosphatase have been reported in the literature. These enzymes have not been the subject of extensive research for the development of inhibitory biosensors. The electrochemical assay for MC-LR that we have developed uses the enzymatic inhibition of protein phosphatase 2A (PP2A). The rapid and simple measurement of PP2A activity was performed by the p-aminophenol (PAP) electrochemical oxidation upon enzymatic conversion using hydrodynamic chronoamperometry. The IC50 value calculated from the change in PP2A activity under the influence of MC-LR is well below the WHO provisional standard value of 0.08 μg/L [51].
2.6. Comparison of Different Types of Enzyme Inhibition-Based Biosensor
Table 1 summarizes the comparison of various enzyme inhibition-based biosensor.

Table 1.
Enzyme inhibition-based biosensors with comparative analysis of their inhibitors and detection limit.
3. DNA-Based Biosensor
To develop the environmental monitoring strategies for specific toxicants, the use of DNA as a recognition tool is promising due to its simplicity, speed, and low cost of gene analysis and testing compared to the traditional techniques [52]. Various aptamer sensors have also been reported, but are not presented in this review. An electrochemical DNA biosensor is a convenient tool to determine the DNA damages caused by different toxic events like chemical, drug, and radioactive agents [53,54]. The DNA biosensors are based on the molecular interaction between surface-bound DNA and the target analyte. The change occurring due to the binding molecules at the recognition layer being measured by the transducer is converted to electronic signals. The difference between the signals before and after the interaction is used for the quantification of these analytes [55,56,57]. It is also possible to detect toxic molecules interacting with DNA by monitoring changes in the guanine oxidation signal in relation to electrochemical DNA biosensors.
DNA immobilization onto the transducer surface strongly affects the performance of the electrochemical DNA-based biosensors. The main goal of this method is to ensure proper molecular orientation of single-stranded DNA (ssDNA) probes and high accessibility to target DNA fragments [58]. Various immobilization methods have been developed, including physical and electrochemical adsorption, film encapsulation, affinity binding (e.g., avidin-biotin complex), chemisorption, and covalent bonding [8,58,59,60,61] (Figure 6). The self-assembled monolayers (SAM) formed from alkanethiols on gold substrates are very appropriate linkers for covalent immobilization of ssDNA probes on the electrode surface [62,63,64,65]. To develop dynamic surfaces that can tune their biochemical functionality is the new challenge in the development of DNA biosensors [65].
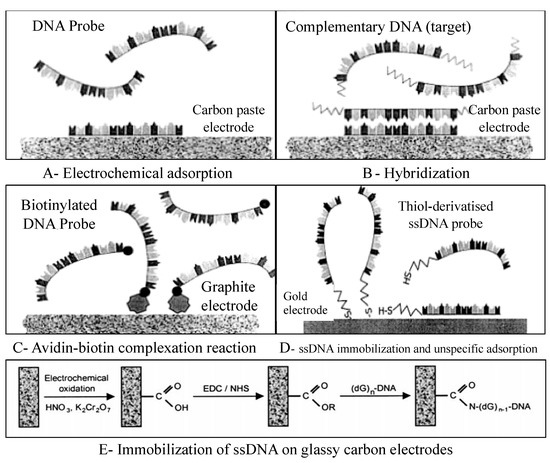
Figure 6.
DNA immobilization onto the electrode surface. (A) Electrochemical adsorption of DNA probe applying a positive potential to an electrochemical transducer. (B) Hybridization between the probe and the target with the same positive potential. (C) DNA immobilization involving avidin-biotin complexation between avidin and biotinylated DNA probe. (D) DNA immobilization by covalent attachment of thiol-derivatized probe on the surface of the gold electrode. (E) ssDNA immobilization on glassy carbon electrodes through deoxyguanosine group (dG)n -DNA) by carbodiimide method (EDC: 1-3(-dimethylaminopropyl)-3-ethyl-carbodiimide; NHS: N-hydroxysulfosuccinimide). Adapted with permission from Ref. [59]. 2017, Elsevier.
To determine the suitability of the DNA biosensor, the performances of a disposable electrochemical DNA biosensor and Toxalert®100 commercial toxicity test based on the bioluminescence inhibition of the bacterium Vibrio fischeri were compared for detecting toxicants in water samples [66]. Electrochemical measurements were performed using an electrochemical cell comprising three screen-printed electrodes, which can be “dropped and sensed” with as little as 10 µL of sample solution. The study found that DNA biosensors are efficient due to their rapid and easily registered response to the presence of small compounds compared to the commercial toxicity test. This single use of a disposable biosensor prevents contamination among the samples and can be used in situ because of its portability. The main advantages include the easy immobilization of the DNA layer and fast measurement process. Another study investigated the voltammetric and spectrophotometric study of complexing heavy metals (Fe3+, Cu2+, Pb2+, and Cd2+) with DNA, especially with denatured ssDNA [67]. A biosensor was constructed using a fixed mercury membrane electrode (SMFE) with and without silver support as the working electrode. The detection limit was found to be 4.0 × 10−11, 1.0 × 10−10, 1.0 × 10−9, and 5.0 × 10−9 M for Cu2+, Pb2+, Cd2+, and Fe3+.
Electrochemical DNA biosensors have been under development in recent years based on nanosized labels and amplification platforms. These nanosized materials lead to rapid current response for target molecules through the acceleration of electron transfer [52]. There are two types of nanomaterials: polymeric nanoparticles and metal nanoparticles. Polyaniline, polypyrrole, polythiophene, polyacetylene, and polyindole are widely studied nanoparticles, and numerous studies have been reported that explore the properties of Au, Ag, Pt, and Pd nanoparticles for the design of amphoteric bioelectronic devices [68,69,70,71,72]. Nanomaterials provide ultrasensitive biosensors with their favorable electronic properties, large surface area, and electrolytic activity [73]. Recently, magnetic microbeads [74] and carbon nanotubes [75,76] also gained popularity as nanomaterials. Galandova et al. developed a disposable electrochemical biosensor for detecting deep DNA damage [77] based on the adsorption of a multiwall carbon nanotubes–chitosan composite followed by depositing double stranded DNA. The presence of immobilized DNA was evaluated using conventional differential pulse voltammetry (DPV) using the [Co(phen)3]3+ DNA marker, and by cyclic voltammetry and electrochemical impedance spectroscopy (EIS) with [Fe(CN)6]3− as a redox probe in solution. The CV and EIS methods have been introduced instead of a complicated procedure with the [Co(phen)3]3+ marker. They found the DNA-based biosensors with bionanocomposite interfaces formed by multiwalled carbon nanotubes and chitosan are effective, inexpensive, and non-toxic compared to mercury-based elements and the highly stable disposable sensors of chemical toxicity that are easy to prepare.
4. Whole-Cell Biosensor
Whole-cell biosensors (WCBs) are a very useful alternative to classical biosensors and have recently been introduced by several authors [78,79]. Entire prokaryotic or eukaryotic cells are used in WCBs as a single element integrating both bioreporter and transducer (Figure 7). They provide the qualitative or quantitative information regarding the substance present in the experimental medium. They also allows the measurement of the total bioavailability of given toxicants rather than its free form [7]. In a WCB, a sufficiently quantifiable signal is obtained using microorganisms with a high growth rate and short generation time [80]. There are several advantages of using microorganisms in WCBs over the purified enzymes as the biocomponent of biosensors. In the natural environment, the existence of different kinds of microorganisms allows the choice of suitable strains for specific purposes, and it is also possible to grow them in the culture medium [81]. The ability to analyze samples with multiple enzyme involvement is another important advantage of WCBs. Different types of microorganisms are used in WCBs, including bacteria [82], algae [83], yeast [84], fungi [85], and plant cells [86]. Among them, bacteria and algae are the most-used microorganisms.
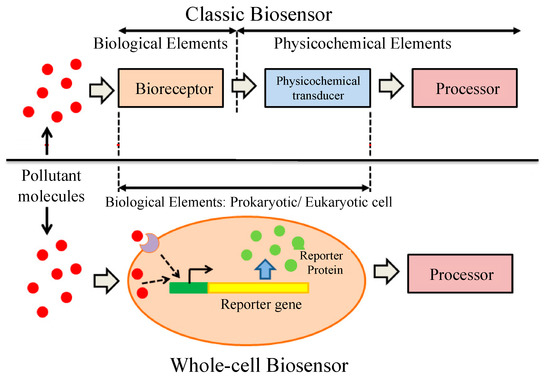
Figure 7.
Classic and whole-cell biosensor elements [80]. In a classic biosensor, bioreceptor and transducer are two different things whereas in a whole-cell biosensor both bioreceptor and transducer are incorporated within the same cell. Adapted with permission from [80].
4.1. Bacteria Based Whole-Cell Biosensor
Bacteria have high adaptability to different extreme conditions such as high temperatures, excessive salinity, pH, and environments polluted with a high concentration of heavy metals, and are highly favored by the researcher in WCB development [87]. Recently, researchers have been working with Escherichia coli (E. coli) for the toxicity assessment of toxic chemicals, metals, pesticides, organic pollutants, etc. [88,89,90,91,92,93,94]. Liu et al. performed a direct toxicity assessment based on an electrochemical method using E. coli and the chemicals 3,5-dichlorophenol (DCP), KCN, and As2O3, and some heavy metal ions Cu2+, Pb2+, Ni2+, and Hg2+ were chosen as toxicants [88]. Chronoamperometry incorporating ferricyanide as a redox probe was employed to understand the overall effect of chemical toxicity on E. coli respiration, rather than chemical identification. Yong et al. developed an electrochemical biosensor using E. coli as a model organism for the rapid determination of pesticide toxicity by adopting ferricyanide instead of natural electron acceptor O2 [91]. Various chemicals, such as 3,5-dichlorophenol (DCP), Ametryn, Fenamiphos, and Endosulfan, were chosen as toxicants. In both of the studies, it was found that the electrochemical method using E. coli is an inexpensive, rapid, reliable, and sensitive method for toxicity assays and is able to be used in a range of applications. Another study used recombinant E. coli to produce a heavy metal biosensor for Zn and Cu [95]. Verma and Singh constructed a biosensor based on Bacillus sphaericus for the monitoring of nickel ions in industrial effluents and foods [96]. The urease enzyme (synthesized within the cell) inhibition was used to quantify the concentration of Ni. Sochor et al. used the growth and metabolic rate of Staphylococcus aureus for cadmium (Cd) detection [97], and Oh et al. used the sulfur-oxidizing ability of sulfur-oxidizing bacteria for chromium (Cr) detection [2].
4.2. Algae-Based Whole-Cell Biosensor
Algae are highly sensitive to environmental pollutants and provide an excellent option for scientists to develop biosensors for toxicity determination [98,99]. Generally, they constitute the bioreceptor element as an integrated whole-cell sensor. In biosensor applications, microalgae are commonly used due to their microscopic size, which make them easy to culture and immobilize, and have a high reproductive rate [87]. Among the algal species used in biosensors development, Chlorella vulgaris is the most common and widely used by several researchers. Around the extracellular membrane, several enzymes act as reporters in the presence of heavy metals, making them a very functional species. One of these enzymes is alkaline phosphatase [100].
Some research works address alkaline phosphatase activity (APA) of the microalgae C. vulgaris [83,100,101,102,103]. Singh and Mittal [83] designed a biosensor based on inhibitory action of mercury toward alkaline phosphatase enzyme present in the cell wall of C. vulgaris. The algal species was immobilized on the glassy carbon surface using bovine serum albumin and glutaraldehyde crosslinker. The quantification was carried out amperometrically by measuring the current generated by oxidation of enzymatically formed electroactive p-nitrophenol. The sensitivity of 10−14 M to 10−6 M for mercury was not interfered by the presence of alkali, alkaline earth and other transition metal ions. Chouteau et al. [100,102] and Guedri and Durrieu [101] developed conductometric biosensor using immobilized C. vulgaris microalgae as bioreceptors and tested for APA analysis to detect the changes in conductivity induced by the catalytic reaction of the enzyme after the exposure to heavy metals, including Cd, Pb, and Zn. A very low ppb detection level was obtained for heavy metals that was more sensitive than bioassays. Berezhetskyy et al. also used a conductometric microtransducer to detect Cd, Cu, Ni, Pb, and Zn in wastewater and found detection limits of about 1 ppb for Cd2+, Co2+, Ni2+, Pb2+ and 10 ppb for Zn2+ [103]. Amperometric algal biosensors were developed by Shitanda et al. to evaluate water toxicity using C. vulgaris [104,105]. Oxygen generated by photosynthetic activity of the algae was registered amperometrically. The biosensor responses to toxic chemicals including 6-chloro-N-ethyl-N-isopropyl-1,3,5-triazine-2,4-diamine (atrazine) and 3-(3,4-dichlorophenyl)-1,1-diethylurea (DCMU), toluene, and benzene were evaluated by inhibition ratios of the reduction current. They found the biosensor could be fabricated faster and at a lower cost than the conventional Clarke oxygen electrode-based algae biosensor.
Another algal species, Chlamydomonas reinhardtii, was used for whole-cell biosensors by other researchers. The species was used by Shitanda et al. to develop electrochemical biosensors for detecting toxic chemicals such as toluene, Cu2+, and Ni2+ based on motility and negative gravity [106]. The changes in the collective flagellar motility of the algae induced by the coexisting toxicants were registered as redox currents variations. The gravitational axis-based flagellate biosensor was more strongly affected by the coexistence of toluene than the conventional algal biosensor employing photosynthetic activity. We have also successfully measured the toxicity of heavy metals (Hg, Cd, Pb, Zn, Cu) from inhibition of alkaline phosphatase (ALP) enzyme in C. reinhardtii [107]. ALP activity was induced in C. reinhardtii by phosphate starvation, and electrochemical oxidation of PAP associated with enzymatic conversion, which was altered by the inhibitory effect of heavy metals, was measured. Rapid measurement of enzyme activity was provided by hydrodynamic voltammetry using a rotating disk electrode (RDE) mounted on a 50-µL micro-droplet (Figure 8). With this method, a detection limit of the enzyme activity was 5.4 × 10−7 U, with a reaction time of only 60 s.
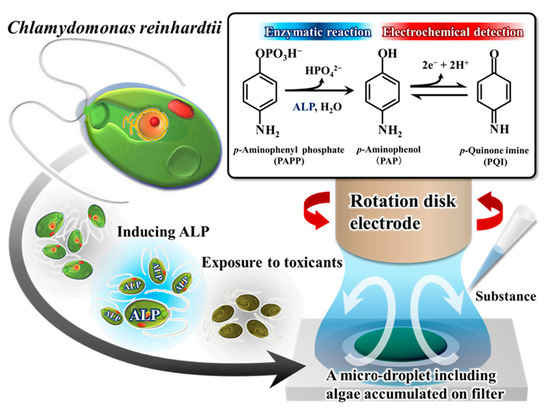
Figure 8.
Microalgal bioassay based on ALP with hydrodynamic electrochemical detection using RDE in a micro-droplet. Adapted with permission from Ref. [107]. 2017, Elsevier.
4.3. Other Types of Whole-Cell Biosensors
Besides bacteria and algae, protozoa and plant cells are also used for biosensor development. The protozoa with no cell walls increase the sensitivity of the biosensors and have more similar metabolic characteristics with human cells [87]. The research work performed by Amaro et al. primarily reported a ciliated protozoan Tetrahymena thermophila as a whole-cell biosensor of the bioavailable heavy metals in polluted soil or water [108]. They created a transformed T. thermophila containing a metallothionein promoter that activated in the presence of heavy metals and successfully expressed the linked luciferase genes. Biosensors based on the T. thermophila metallothionein promoter are the most sensitive eukaryotic metal biosensors and are more sensitive than some prokaryotic biosensors.
The application of widely available natural cell-bound dyes, such as carotenoids, in biosensors is less familiar. In plants, carotenoids absorb energy from chlorophyll and dissipate it as heat through internal conversion, thereby reducing the formation of reactive oxygen species. Their production increases in the presence of heavy metals and at the same time increase oxidative stress in plants. Thus, the effect of natural cell-bound carotenoids on heavy metal exposure is well anticipated. A rapid method for prompt detection of heavy metals (Cu, Pb, Zn) based on the response of natural cell-bound carotenoids in Daucus carota was reported [86]. D. carota cell suspensions were exposed to heavy metals and the response of intracellular carotenoids was detected.
5. Biosensors Based on Cytochrome P450
Cytochromes P450s (CYPs) are members of a family involving over 3000 hemeproteins that catalyze the NADPH-related monooxygenation and other biotransformation reactions such as epoxidation, hydroxylation, and heteroatom oxidation. They are involved in the metabolism of over 1,000,000 different xenobiotic compounds [109]. CYP enzymes have been identified in almost all domains of life, including animals, plants, fungi, protists, bacteria, archaea, and even viruses [110,111,112]. More than 7700 individual CYPs are known, among which, 57 CYP species were found in the human liver. Only fifteen human CYP members can metabolize drugs and other chemicals; and five of them (CYP1A2, 2C9, 2C19, 2D6, and 3A4) are responsible for approximately 95% of CYP-mediated drug metabolism in the body, while each one is specific for certain types of reactions involving different substrates [113]. Due to the involvement of distinct classes of biotransformation reactions, CYPs are very promising for the pharmaceutical industry.
The CYP family has attracted the attention of enzyme engineers since the early 1970s due to their specific catalytic activities. However, the significant advancement in this area was achieved with the development of appropriate CYP immobilization methods [114]. By immobilizing CYP, it can function as a biosensor, receiving electrons directly from the electrode without the need for an electron donor or an enzyme as an electron transfer partner. The change of current flow upon the electric potential application correlates with the catalytic activity of the enzyme against certain substrates. Immobilization of biosensors for CYPs are fabricated mainly by adsorption on bare electrodes [115,116], and the electrode environment has been found to have a significant effect on the electrical activity of CYPs [109]. To design a better CYP biosensor, electrodes were modified with protein-friendly materials that adequately prevent the denaturation on the electrode surface. The electrodes were modified by various strategies such as layer-by-layer (LbL) adsorption, encapsulation in polymers and gels, and covalent attachment, such as self-assembled monolayers (SAMs) on gold electrodes. The electrode modifications used in CYP biosensors are presented in Table 2. These studies use the combination of analytical and electrochemical techniques to confirm that it is possible to detect different xenobiotic compounds with a CYP biosensor using its electroactivity.

Table 2.
The outline of different types of electrodes in CYP biosensors.
There have been several review articles focused on CYP enzymes and biosensors. A review by Schneider and Clark focused on the immobilization strategies and different electrode materials for CYP biosensors [130]. Moreover, they placed special emphasis on the CYP electrode surface characterization and electrochemically-mediated catalysis. They mainly focused on a biosensor based on human-drug-metabolizing CYPs. The CYP biosensors based on mediated electron transfer, protein electrochemistry, and direct electron transfer of CYP were overviewed by Bistolas et al. [114].
Among the CYP members present in the human body, cytochrome P450 3A4 (CYP3A4) is considered the most important drug-metabolizing enzyme in the P450 family, which is involved in the oxidation of almost half of the drugs metabolized by CYP members [113,130]. Therefore, the development of a biosensor for drug detection based on the changes in CYP3A4 activity using both metabolism and inhibition becomes an obvious goal. The CYP3A4 activity changes are usually determined from the metabolites formation rate by electrons donated from NADPH to CYP3A4. Later, an electrode was used as an alternative electron source to NADPH for electrochemically driven catalysis of P450 enzymes. Electrochemical biosensors based on the direct electron transfer (DET) between CYP3A4 and an electrode were recently studied for drug metabolites detection. An amperometric biosensor based on the redox properties of human CYP3A4 was developed by Joseph et al. [117], which addressed the biocatalysis of CYP3A4 for four substrates (verapamil, midazolam, quinidine, and progesterone) at a poly(dimethyldiallylammoniumchloride) (PDDA) and 3-mercapto-1-propenesulfonic acid (MPS)-modified gold electrode. Dodhia et al. demonstrated that coupling efficiency and catalytic activity at electrode surfaces could be significantly enhanced by tuning the electron flow to CYP3A4 [120]. They obtained DET at thiol reactive cystamine–maleimide-modified gold and polyion-modified GC electrodes. Artificial redox chains were applied to regulate the electron flow. Some other researchers additionally found DET at a gold electrode modified with aromatic compounds and CYP3A4/CPR microsomes [131,132]. All of the above-mentioned studies used modified layers for immobilizing proteins and accelerating DET, because electrochemical processing of CYP directly on an electrode without a modifying layer is complicated. Niwa and co-workers developed an electrochemically driven CYP3A4 biosensor based on DET for detecting drug metabolism and its inhibition using a carbon nanofiber (CNF)-modified film electrode [133]. The electrocatalytic activity of CYP3A4 co-immobilized with different carbon nanomaterials such as CNTs and carbon black (CB) was also investigated. CYP3A4 and CNF (CYP3A4/CNF)-modified film electrodes exhibit higher reduction current than that in CNTs and CB-modified film electrodes. CNFs possessing a high conductivity and large surface area provide a suitable microenvironment, providing excellent DET and biocatalytic properties.
In another study, Niwa and co-workers successfully detected the DET from a human CYP layer adsorbed on a bare indium tin oxide (ITO) film (polycrystalline and amorphous) electrode without any modifying layers [134]. They applied this technique to the evaluation of drug metabolism and inhibitors, and found that polycrystalline ITO films were superior to amorphous ITO films and other conventional electrodes as biocatalytic electrodes for more efficient DET. With these techniques, it is possible to construct simple CYP drug-metabolizing biosensors with high throughput. There are different types of electrode modification strategies mentioned in the Table 2 and presented in Figure 9.
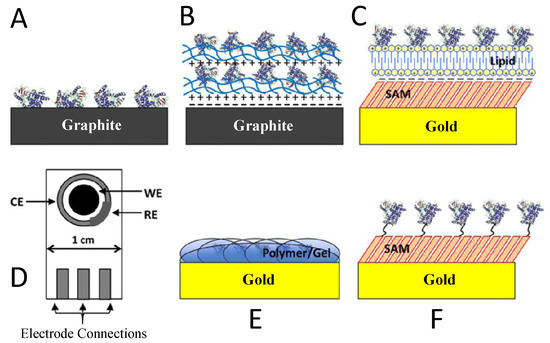
Figure 9.
Strategies of electrode immobilization for fabrication of CYP biosensors [130]. (A) Adsorption to a bare electrode. (B) Layer-by-layer (LbL) adsorption. (C) Adsorption to a thin film. (D) Screen-printed electrode. (E) Encapsulation in polymers or gels. (F) Covalent attachment to SAMs on a gold electrode. Adapted with permission from Ref. [130]. 2013, Elsevier.
5.1. LbL Adsorption
The LbL adsorption provides a more protein friendly environment for electrochemically driven CYP catalysis. Using this process, it is possible to build numerous layers of oppositely charged films and CYPs on the electrode surface. Joseph et al. [117] developed a biosensor by the CYP3A4 immobilization on a gold electrode sequentially modified with MPS and PDDA. Similar modification with PEI and PSS films on pyrolytic graphite (PG) was made by Sultana et al. to detect electrochemically driven epoxidation of styrene [118]. Krishnan et al. studied LbL films in the form of recombinant CYP1A2 or 2E1 and CPR/cytochrome b5-containing microsomes on basal-plane pyrolytic graphite (BPG) electrodes [119]. They found that the electron transfer at the CYP electrode follows a natural catalytic pathway, via CPR to P450.
5.2. Adsorption to Thin Films
Tween 80 nonionic detergent. The aminopyrine and benzphetamine drug substrates were detected by amperometry [135]. The highest amount of electroactive 2B4 was obtained when using the mixture of sodium montmorillonite clay, Tween, and 2B4 deposited on the GC electrode. Furthermore, a composite membrane of nanosodium montmorillonite clay and detergent dihexadecyl phosphate was used to immobilize rat CYP1A1 on EPG electrodes for detection of benzo[a]pyrene [136]. The first study detected reversible reduction and oxidation peaks at a formal potential of −295 mV for Ag/AgCl at the GC/Clay + Tween/2B4 electrode, whereas the second study revealed a pair of redox peaks at a potential of −0.360 mV. These studies evidence the efficiency of thin films made of clay nanoparticles for the fabrication of efficient CYP biosensors.
5.3. Screen-Printed Electrodes
Screen-printed electrodes remarkably simplify the fabrication of CYP biosensors due to their low cost and ease of conductive additives mixing into the electrode paste. It is possible to construct many different sizes and configurations of electrodes using screen-printed electrodes [130]. Antonini et al. studied the direct electrochemical deposition of CYPs on riboflavin–graphite screen-printed electrodes to fabricate a CYP1A2 biosensor [137]. The immobilization was performed by encapsulation of 1A2 in a glycerol and agarose gel matrix. They successfully detected the drug clozapine electrochemically. To construct CYP biosensors, screen-printed electrodes were combined with lipid DDAB. Shumyantseva et al. [138] developed a biosensor using screen-printed graphite electrodes modified with gold nanoparticles and DDAB and compared with cytochromes P450 2B4 (CYP2B4), P450 1A2 (CYP1A2), sterol14a-demethylase (CYP51b1) DET. Gold nanoparticles were found to increase the reduction current observed in the aerobic CV scan. They also studied the electrochemically driven benzphetamine catalysis with DDAB/AuNP/2B4 electrodes [123].
5.4. Encapsulation in Polymers or Gels
To enhance the electroactivity of electrodes, numerous works addressed the encapsulation of biosensors in polymers or gels for immobilizing P450s on the electrode surface. Some commonly used conductive polymers are polypyrrole and polyacetylene, etc. The first study of a CYP biosensor based on this technique was published by Sugihara et al. by immobilizing camphor-induced cytochrome P-450 (P450CAM) in polypyrrole polymerized on an ITO electrode [139]. A recent study by Alonso-Lomillo et al. [125] used the encapsulation in polypyrrole technique for the 2B4 immobilization on a gold working electrode in a CYP2B4 biosensor. The chronoamperometric analysis revealed a current signal for the substrate Phenobarbital with a detection limit of 0.289 µmol/dm3. In another study, a CYP3A4 biosensor was fabricated by immobilizing 3A4 in a matrix composed of epoxy copolymers P (GMA-co-MPC) and acetylene black on a glassy carbon electrode [140]. The composite significantly accelerated the electron transfer between 3A4 and the electrode by providing a biocompatible microenvironment. The biosensor responded amperometrically to diethylstilbestrol and displayed a detection limit of 5.9 × 10−8 mol/L. CYPs were encapsulated on electrodes in another hydrophilic gel comprising sol-gel, chitosan, and agarose to facilitate the electron transfer [130].
5.5. Covalent Attachment to Self-Assembled Monolayers on Gold
The covalent attachment of CYPs to SAMs on a gold electrode provides a simple and strong technique for CYP biosensor construction. Various finishing reagents, such as carboxyls, amines, and maleimides, are usually used to modify gold electrodes. However, it is unclear from the literature on CYP biosensors whether short- or long-chain SAMs accelerate the electron transfer. Some biosensors used long-chain alkanethiols, for example, 11-mercaptoundecanoic acid (MUA) or hexanoicacid, and some used shorter chain thiols such as dithio-bismaleimidoethane (DTME) for CYP immobilization [129]. Different immobilizations strategies are used for covalent attachment of CYPs to SAMs formed on gold electrodes; among which, amine coupling is the most common strategy. Fantuzzi et al. successfully immobilized a CYP3A4 fusion protein and flavodoxin (FLD) on a gold electrode using amine coupling of 6-hexanethiol (6HT) and 7-mercaptoheptanoic acid (7MHA) to a mixed SAM [128]. The formation of inactive CYP species was observed in bacterial and mammalian CYP immobilized on SAMs [130].
5.6. Recent Advancements in CYP Biosensing
Recently, micro-fabricated electrodes and nanostructured materials such as multiwall carbon nanotubes (MWCNTs) have been widely used for CYP biosensor construction. Cararra et al. developed a CYP biosensor using MWCNT-modified screen-printed electrodes from 3A4, 2B4, and 2C9 CYPs for detecting the Benzphetamine, Cyclophosphamide, Dextromethorphan, Naproxen and Flurbiprofen drugs [141]. Comparing 3A4 biosensors prepared with bare or MWCNT electrodes to the drug cyclophosphamide, it was found that MWCNTs incorporation greatly enhances the sensitivity. Nanostructured gold and quantum dots are also being used to construct CYP biosensors. It was found that the electroactivity of CYP3A4 adsorbed on the surface could be enhanced through the naphthalenethiolate SAM built on the surface of the sputtered gold electrode [142]. In another study, the direct electrochemistry of 3A4 was examined by immobilizing 3A4 on gold nanodome arrays made by micro-fabrication [143]. The effects of a nanostructured surface on the direct electrochemical reaction of CYPs were demonstrated using ZnSe quantum dots [144].
6. Conclusions
Biosensors combining a selective biochemical recognition and a high sensitivity of electrochemical detection are widely used in practice. They are currently applied in medical, pharmaceutical, and environmental monitoring. Recent progress regarding electrochemical analysis has been published in various review articles [145,146,147,148,149,150]. Currently, electrochemical biosensors have become reliable techniques for the determination of environmental toxicity. These biosensors have shown progress in several areas, such as the use of genetic modification of enzymes and microorganisms, creative ways to immobilize recognition elements, and improved sensor interfaces. The technological progress of these biosensors have made them rapid, reliable, able to be used in real time, cost effective, and portable. They also require a minimal volume of sample solution. However, there are still barriers to a broader range of applications.
There have been significant research activities regarding the development of enzyme inhibition-based biosensors. However, analytical applications of these biosensors are still limited due to their inability to distinguish different toxic compounds in the same sample, for example, pesticides and toxic metal ions. These are the barriers to their use for regulatory purposes. Despite their limitations, the enzyme inhibition-based biosensors show high sensitivity, and provide simple and cost-effective techniques, especially when used for pesticides. The development of a screen-printed electrode to avoid electrode fouling is also possible with this technique. Currently, some researchers are also working to use genetically modified enzymes for biosensor design.
The development of DNA biosensors is of interest to many researchers due to their ability to quickly obtain sequence-specific information, their simplicity, and their cost-effectiveness. Electrochemical DNA biosensors are particularly interesting due to their sensitivity and miniaturization of technology. Nanomaterials are important for the development of these biosensors, and they provide high reactivity and the ultimate miniaturization level. Future research on DNA biosensors could include the development of reliable devices that can be used by non-specialized personnel or the development of compact and portable devices.
Whole-cell biosensors can respond to changes that occur in their environment and are suitable for eco-toxicity testing and environmental monitoring use. These biosensors have remarkable sensitivity and accuracy in environmental applications. These biosensors are continuously developing in terms of their size, detection time, and field applicability. However, they have some limitations in their selectivity, stability, and storability, and need to be improved for proper use. Future research should address these issues.
The uses of whole-cell-based biosensors have some advantages and disadvantages over enzyme-based biosensors. Some enzymes are deactivated when their molecular structure is altered by separation or immobilization; therefore, the use of whole cells avoids enzyme extraction. The enzymes occurring within the cells in nature remain with coenzymes and activators of biochemical pathways, which eliminates the need to add them to the medium. Whole cells must be used to get functional information, that is, to determine how some factors affect the organism. The reduced signal generation rate and the low selectivity are the most common drawbacks of whole-cell biosensors.
Cytochrome P450 cytochrome-based biosensors are of great interest to researchers due to their ability to measure the toxicity of drugs and other foreign compounds, since the other available methods are expensive and time consuming. These biosensors provide a cheap, simple, selective, and rapid alternative for the quick measurement of drugs and metabolites. Direct electron transfer from electrode to CYP is the most suitable approach for developing CYP biosensors. Proper CYP immobilization onto the electrode surface is essential to optimize the DET to the CYP. There are different types of electrode modification techniques that can be used to optimize the DET. Future research should deal with electrode miniaturization using nanostructured materials to test the real feasibility of this biosensor to detect drugs and metabolites.
Author Contributions
Conceptualization, M.S.I. and H.K.; methodology, M.S.I., K.S. (Kazuto Sazawa), K.S. (Kazuharu Sugawara) and H.K.; writing—original draft preparation, M.S.I. and H.K.; writing—review and editing, M.S.I., K.S. (Kazuto Sazawa), K.S. (Kazuharu Sugawara), and H.K.; supervision, H.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Not applicable.
Acknowledgments
This research work was financially supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Royal Commission on Environmental Pollution. Chemicals in Products: Safeguarding the Environment and Human Health. 24th Report; The Stationery Office: Norwich, UK, 2003. [Google Scholar]
- Oh, S.E.; Hassan, S.H.A.; Van Ginkel, S.W. A novel biosensor for detecting toxicity in water using sulfur-oxidizing bacteria. Sens. Actuators B Chem. 2011, 154, 17–21. [Google Scholar] [CrossRef]
- Eltzov, E.; Yehuda, A.; Marks, R.S. Creation of a new portable biosensor for water toxicity determination. Sens. Actuators B Chem. 2015, 221, 1044–1054. [Google Scholar] [CrossRef]
- Liu, C.; Yong, D.; Yu, D.; Dong, S. Cell-based biosensor for measurement of phenol and nitrophenols toxicity. Talanta 2011, 84, 766–770. [Google Scholar] [CrossRef]
- Wang, X.; Wang, X.; Zhang, J.; Bu, Y.; Yan, X.; Chen, J.; Huang, J.; Zhao, J. Direct toxicity assessment of copper (II) ions to activated sludge process using a p-benzoquinone-mediated amperometric biosensor. Sens. Actuators B Chem. 2015, 208, 554–558. [Google Scholar] [CrossRef]
- Volpi Ghirardini, A.V.; Girardini, M.; Marchetto, D.; Pantani, C. Microtox® solid phase test: Effect of diluent used in toxicity test. Ecotoxicol. Environ. Saf. 2009, 72, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Tizzard, A.; Webber, J.; Gooneratne, R.; John, R.; Hay, J.; Pasco, N. MICREDOX: Application for rapid biotoxicity assessment. Anal. Chim. Acta 2004, 522, 197–205. [Google Scholar] [CrossRef]
- Wang, X.; Liu, M.; Wang, X.; Wu, Z.; Yang, L.; Xia, S.; Chen, L.; Zhao, J. p-benzoquinone-mediated amperometric biosensor developed with Psychrobacter sp. for toxicity testing of heavy metals. Biosens. Bioelectron. 2013, 41, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Davidov, Y.; Rozen, R.; Smulski, D.R.; Van Dyk, T.K.; Vollmer, A.C.; Elsemore, D.A.; LaRossa, R.A.; Belkin, S. Improved bacterial SOS promoter∷ lux fusions for genotoxicity detection. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2000, 466, 97–107. [Google Scholar] [CrossRef]
- Rodriguez-Mozaz, S.; de Alda, M.J.L.; Marco, M.P.; Barceló, D. Biosensors for environmental monitoring A global perspective. Talanta 2005, 65, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Mattias, K.J.M.; Turner, A.P.F. Biosensors in air monitoring. J. Environ. Monit. 1999, 1, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Soelberg, S.D.; Chinowsky, T.; Geiss, G.; Spinelli, C.B.; Stevens, R.; Near, S.; Kauffman, P.; Yee, S.; Furlong, C.E. A portable surface plasmon resonance sensor system for real-time monitoring of small to large analytes. J. Ind. Microbiol. Biotechnol. 2005, 32, 669–674. [Google Scholar] [CrossRef]
- Badihi-Mossberg, M.; Buchner, V.; Rishpon, J. Electrochemical Biosensors for Pollutants in the Environment. Electroanalysis 2007, 19, 2015–2028. [Google Scholar] [CrossRef]
- Power, A.C.; Morrin, A. Electroanalytical sensor technology. In Electrochemistry; Khalid, M.A.A., Ed.; IntechOpen: London, UK, 2013; pp. 141–177. [Google Scholar]
- Wang, J. Analytical Electrochemistry, 3rd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006. [Google Scholar]
- Shao, Y.; Wang, J.; Wu, H.; Liu, J.; Aksay, I.; Lin, Y. Graphene based electrochemical sensors and biosensors: A review. Electroanalysis 2010, 22, 1027–1036. [Google Scholar] [CrossRef]
- Zhao, Z.; Lei, W.; Zhang, X.; Wang, B.; Jiang, H. ZnO-based amperometric enzyme biosensors. Sensors 2010, 10, 1216–1231. [Google Scholar] [CrossRef]
- Siqueira, J.R.; Caseli, L.; Crespilho, F.N.; Zucolotto, V.; Oliveira, O.N. Immobilization of biomolecules on nanostructured films for biosensing. Biosens. Bioelectron. 2010, 25, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Harper, A.; Anderson, M.R. Electrochemical glucose sensors—Developments using electrostatic assembly and carbon nanotubes for biosensor construction. Sensors 2010, 10, 8248–8274. [Google Scholar] [CrossRef]
- Singh, R.P.; Oh, B.K.; Choi, J.W. Application of peptide nucleic acid towards development of nanobiosensor arrays. Bioelectrochemistry 2010, 79, 153–161. [Google Scholar] [CrossRef]
- Park, B.W.; Yoon, D.Y.; Kim, D.S. Recent progress in bio-sensing techniques with encapsulated enzymes. Biosens. Bioelectron. 2010, 26, 1–10. [Google Scholar] [CrossRef]
- Su, L.; Jia, W.; Hou, C.; Lei, Y. Microbial biosensors: A review. Biosens. Bioelectron. 2011, 26, 1788–1799. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical biosensors–Sensor principles and architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Amine, A.; Mohammadi, H.; Bourais, I.; Palleschi, G. Enzyme inhibition-based biosensors for food safety and environmental monitoring. Biosens. Bioelectron. 2006, 21, 1405–1423. [Google Scholar] [CrossRef]
- Bachan Upadhyay, L.S.; Verma, N. Enzyme inhibition based biosensors: A review. Anal. Lett. 2013, 46, 225–241. [Google Scholar] [CrossRef]
- Singh, M.; Verma, N.; Garg, A.K.; Redhu, N. Urea biosensors. Sens. Actuators B Chem. 2008, 134, 345–351. [Google Scholar] [CrossRef]
- Zejli, H.; Hidalgo-Hidalgo de Cisneros, J.L.; Naranjo-Rodriguez, I.; Liu, B.; Temsamani, K.R.; Marty, J.L. Alumina sol-gel/sonogel-carbon electrode based on acetylcholinesterase for detection of organophosphorus pesticides. Talanta 2008, 77, 217–221. [Google Scholar] [CrossRef]
- Du, D.; Ding, J.; Cai, J.; Zhang, J.; Liu, L. In situ electrodeposited nanoparticles for facilitating electron transfer across self-assembled monolayers in biosensor design. Talanta 2008, 74, 1337–1343. [Google Scholar] [CrossRef]
- Anitha, K.; Mohan, S.V.; Reddy, S. Development of acetylcholinesterase silica sol–gel immobilized biosensor—An application towards oxydemeton methyl detection. Biosens. Bioelectron. 2004, 20, 848–856. [Google Scholar] [CrossRef] [PubMed]
- Andreescu, S.; Bucur, B.; Marty, J.L. Affinity immobilization of tagged enzymes. In Immobilization of Enzymes and Cells, 2nd ed.; Guisan, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2006; pp. 97–106. [Google Scholar]
- Bucur, B.; Danet, A.F.; Marty, J.L. Cholinesterase immobilisation on the surface of screen-printed electrodes based on concanavalin A affinity. Anal. Chim. Acta 2005, 530, 1–6. [Google Scholar] [CrossRef]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef]
- Guerrieri, A.; Monaci, L.; Quinto, M.; Palmisano, F. A disposable amperometric biosensor for rapid screening of anticholinesterase activity in soil extracts. Analyst 2002, 127, 5–7. [Google Scholar] [CrossRef]
- Ivanov, A.; Evtugyn, G.; Budnikov, H.; Ricci, F.; Moscone, D.; Palleschi, G. Cholinesterase sensors based on screen-printed electrodes for detection of organophosphorus and carbamic pesticides. Anal. Bioanal. Chem. 2003, 377, 624–631. [Google Scholar] [CrossRef]
- Ghica, M.E.; Brett, C.M.A. Glucose oxidase inhibition in poly(neutral red) mediated enzyme biosensors for heavy metal determination. Microchim. Acta 2008, 163, 185–193. [Google Scholar] [CrossRef]
- Guascito, M.R.; Malitesta, C.; Mazzotta, E.; Turco, A. Inhibitive determination of metal ions by an amperometric glucose oxidase biosensor. Sens. Actuators B Chem. 2008, 131, 394–402. [Google Scholar] [CrossRef]
- Liu, J.X.; Xu, X.; Tang, L.; Zeng, G. Determination of trace mercury in compost extract by inhibition based glucose oxidase biosensor. Tran. Nonferrous Metal Soc. 2009, 19, 235–240. [Google Scholar] [CrossRef]
- Samphao, A.; Rerkchai, H.; Jitcharoen, J.; Nacapricha, D.; Kalcher, K. Indirect determination of mercury by inhibition of glucose oxidase immobilized on a carbon paste electrode. Int. J. Electrochem. Sci. 2012, 7, 1001–1010. [Google Scholar]
- Balasubramanian, A.; Ponnuraj, K. Crystal structure of the first plant urease from jack bean: 83 years of journey from its first crystal to molecular structure. J. Mol. Biol. 2010, 400, 274–283. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, Z.; Yang, M.; Guo, M.; Wu, Z.; Shen, G.; Yu, R. Inhibitive determination of mercury ion using a renewable urea biosensor based on self-assembled gold nanoparticles. Sens. Actuators B Chem. 2006, 114, 1–8. [Google Scholar] [CrossRef]
- Do, J.S.; Lin, K.H.; Ohara, R. Preparation of urease/nano-structured polyaniline-Nafion®/Au/Al2O3 electrode for inhibitive detection of mercury ion. J. Taiwan Inst. Chem. Eng. 2011, 4, 662–668. [Google Scholar] [CrossRef]
- Gani, A.A.; Ashari, M.R.; Kuswandi, B. An optical fiber biosensor for heavy metal ions based on a modified single sol–gel film of urease and chlorophenol red in flow system. Sens. Lett. 2010, 8, 320–327. [Google Scholar] [CrossRef]
- Ilangovan, R.; Daniel, D.; Krastanov, A.; Zachariah, C.; Elizabeth, R. Enzyme based biosensor for heavy metal ions determination. Biotechnol. Biotechnol. Equip. 2006, 20, 184–189. [Google Scholar] [CrossRef]
- Arduini, F.; Ricci, F.; Tuta, C.S.; Moscone, D.; Amine, A.; Palleschi, G. Detection of carbamic and organophosphorous pesticides in water samples using a cholinesterase biosensor based on Prussian Blue-modified screen-printed electrode. Anal. Chim. Acta 2006, 580, 155–162. [Google Scholar] [CrossRef]
- Liu, G.; Lin, Y. Biosensor based on self-assembling acetylcholinesterase on carbon nanotubes for flow injection/amperometric detection of organophosphate pesticides and nerve agents. Anal. Chem. 2006, 78, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Pohanka, M.; Jun, D.; Kuca, K. Amperometric biosensors for real time assays of organophosphates. Sensors 2008, 8, 5303–5312. [Google Scholar] [CrossRef]
- Sinha, R.; Ganesana, M.; Andreescu, S.; Stanciu, L. AChE biosensor based on zinc oxide sol-gel for the detection of pesticides. Anal. Chim. Acta 2010, 661, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Alvarado-Gámez, A.L.; Alonso-Lomillo, M.A.; Domínguez-Renedo, O.; Arcos-Martínez, M.J. A disposable alkaline phosphatase-based biosensor for vanadium chronoamperometric determination. Sensors 2014, 14, 3756–3767. [Google Scholar] [CrossRef] [PubMed]
- Samphao, A.; Suebsanoh, P.; Wongsa, Y.; Pekec, B.; Jitchareon, J.; Kalcher, K. Alkaline phosphatase inhibition based amperometric biosensor for the detection of carbofuran. Int. J. Electrochem. Sci. 2013, 8, 3254–3264. [Google Scholar]
- Tekaya, N.; Saiapina, O.; Ben Ouada, H.; Lagarde, F.; Ben Ouada, H.; Jaffrezic-Renault, N. Ultrasensitive conductometric detection of heavy metals based on inhibition of alkaline phosphatase activity from Arthrospira platensis. Bioelectrochemistry 2013, 90, 24–29. [Google Scholar] [CrossRef]
- Islam, M.S.; Sazawa, K.; Sugawara, K.; Kuramitz, H. Micro-droplet hydrodynamic voltammetry for the determination of microcystin-LR based on protein phosphatase. J. Water Environ. Technol. 2019, 17, 18–26. [Google Scholar] [CrossRef]
- Abu-Salah, K.M.; Alrokyan, S.A.; Khan, M.N.; Ansari, A.A. Nanomaterials as analytical tools for genosensors. Sensors 2010, 10, 963–993. [Google Scholar] [CrossRef]
- Wang, J. Real time electrochemical monitoring toward green analytical chemistry. Acc. Chem. Res. 2002, 35, 811–816. [Google Scholar] [CrossRef]
- Mehrvar, M.; Abdi, M. Recent developments, characteristics, and potential applications of electrochemical biosensors. Anal. Sci. 2004, 20, 1113–1126. [Google Scholar] [CrossRef]
- Chiti, G.; Marrazza, G.; Mascini, M. Electrochemical DNA biosensor for environmental monitoring. Anal. Chim. Acta 2001, 427, 155–164. [Google Scholar] [CrossRef]
- Lucarelli, F.; Kicela, A.; Palchetti, I.; Marrazza, G.; Mascini, M. Electrochemical DNA biosensor for analysis of wastewater samples. Bioelectrochemistry 2002, 58, 113–118. [Google Scholar] [CrossRef]
- Lucarelli, F.; Palchetti, I.; Marrazza, G.; Mascini, M. Electrochemical DNA biosensor as screening tool for the detection of toxicant in water and wastewater samples. Talanta 2002, 56, 949–957. [Google Scholar] [CrossRef]
- Lucarelli, F.; Marrazza, G.; Turner, A.P.; Mascini, M. Carbon and gold electrodes as electrochemical transducers for DNA hybridization sensors. Biosens. Bioelectron. 2004, 19, 515–530. [Google Scholar] [CrossRef] [PubMed]
- Pividori, M.I.; Merkoçi, A.; Alegret, S. Electrochemical genosensor design: Immobilisation of oligonucleotides onto transducer surfaces and detection methods. Biosens. Bioelectron. 2000, 15, 291–303. [Google Scholar] [CrossRef]
- de-los-Santos-Alvarez, P.; Lobo-Castañón, M.J.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. Current strategies for electrochemical detection of DNA with solid electrodes. Anal. Bioanal. Chem. 2004, 378, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; He, P.; Liu, S.; Ye, J.; Fang, Y. Immobilization of single-stranded deoxyribonucleic acid on gold electrode with self-assembled aminoethanethiol monolayer for DNA electrochemical sensor applications. Talanta 1998, 47, 487–495. [Google Scholar] [CrossRef]
- Chaki, N.K.; Vijayamohanan, K. Self-assembled monolayers as a tunable platform for biosensor applications. Biosens. Bioelectron. 2002, 17, 1–12. [Google Scholar] [CrossRef]
- Kerman, K.; Ozkan, D.; Kara, P.; Meric, B.; Gooding, J.J.; Ozsoz, M. Voltammetric determination of DNA hybridization using methylene blue and self-assembled alkanethiol monolayer on gold electrodes. Anal. Chim. Acta 2002, 462, 39–47. [Google Scholar] [CrossRef]
- Smith, R.K.; Lewis, P.A.; Weiss, P.S. Patterning self-assembled monolayers. Prog. Surf. Sci. 2004, 75, 1–68. [Google Scholar] [CrossRef]
- Cagnin, S.; Caraballo, M.; Guiducci, C.; Martini, P.; Ross, M.; Santaana, M.; Danley, D.; West, T.; Lanfranchi, G. Overview of electrochemical DNA biosensors: New approaches to detect the expression of life. Sensors 2009, 9, 3122–3148. [Google Scholar] [CrossRef] [PubMed]
- Tencaliec, A.M.; Laschi, S.; Magearu, V.; Mascini, M. A comparison study between a disposable electrochemical DNA biosensor and a Vibrio fischeri-based luminescent sensor for the detection of toxicants in water samples. Talanta 2006, 69, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Babkina, S.S.; Ulakhovich, N.A. Complexing of heavy metals with DNA and new bioaffinity method of their determination based on amperometric DNA based biosensor. Anal. Chem. 2005, 77, 5678–5685. [Google Scholar] [CrossRef]
- Malhotra, B.D.; Chaubey, A.; Singh, S.P. Prospects of conducting polymers in biosensors. Anal. Chim. Acta 2006, 578, 59–74. [Google Scholar] [CrossRef]
- Im, Y.; Vasquez, R.P.; Lee, C.; Myung, N.; Penner, R.; Yun, M. Single metal and conducting polymer nanowire sensors for chemical and DNA detections. J. Phys. Conf. Ser. 2006, 38, 61–64. [Google Scholar] [CrossRef]
- Park, S.J.; Taton, T.A.; Mirkin, C.A. Array-based electrical detection of DNA using nanoparticle probes. Science 2002, 295, 1503–1506. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Polsky, R.; Xu, D. Silver-enhanced colloidal gold electrochemical stripping detection of DNA hybridization. Langmuir 2001, 17, 5739–5741. [Google Scholar] [CrossRef]
- Wang, J.; Xu, D.; Kawde, A.N.; Polsky, R. Metal nanoparticle-based electrochemical stripping potentiometric detection of DNA hybridization. Anal. Chem. 2001, 73, 5576–5581. [Google Scholar] [CrossRef]
- Wang, J. Nanomaterial-based electrochemical biosensors. Analyst 2005, 130, 421–426. [Google Scholar] [CrossRef]
- Lermo, A.; Campoy, S.; Barbé, J.; Hernández, S.; Alegret, S.; Pividori, M.I. In situ DNA amplification with magnetic primers for the electrochemical detection of food pathogens. Biosens. Bioelectron. 2007, 22, 2010–2017. [Google Scholar] [CrossRef]
- Rivas, G.A.; Rubianes, M.D.; Rodríguez, M.C.; Ferreyra, N.F.; Luque, G.L.; Pedano, M.L.; Miscoria, S.A.; Parrado, C. Carbon nanotubes for electrochemical biosensing. Talanta 2007, 74, 291–307. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, K.; Ma, H. Electrochemical DNA biosensor based on silver nanoparticles/poly(3-(3-pyridyl) acrylic acid)/carbon nanotubes modified electrode. Anal. Biochem. 2009, 387, 13–19. [Google Scholar] [CrossRef]
- Galandova, J.; Ziyatdinova, G.; Labuda, J. Disposable electrochemical biosensor with multiwalled carbon nanotubes-chitosan composite layer for the detection of deep DNA damage. Anal. Sci. 2008, 24, 711–716. [Google Scholar] [CrossRef]
- Belkin, S. Microbial whole-cell sensing systems of environmental pollutants. Curr. Opin. Microbiol. 2003, 6, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Van der Meer, J.R.; Belkin, S. Where microbiology meets microengineering: Design and applications of reporter bacteria. Nature Reviews. Microbiology 2010, 8, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, J.C.; Amaro, F.; Martín-González, A. Heavy metal whole-cell biosensors using eukaryotic microorganisms: An updated critical review. Front. Microbiol. 2015, 6, 48. [Google Scholar] [CrossRef]
- Yagi, K. Applications of whole-cell bacterial sensors in biotechnology and environmental science. Appl. Microbiol. Biotechnol. 2007, 73, 1251–1258. [Google Scholar] [CrossRef]
- Wong, L.S.; Lee, Y.H.; Surif, S. Whole-cell biosensor using Anabaena torulosa with optical transduction for environmental toxicity evaluation. J. Sens. 2013, 2013, 567272. [Google Scholar] [CrossRef]
- Singh, J.; Mittal, S.K. Chlorella sp. based biosensor for selective determination of mercury in presence of silver ions. Sens. Actuators B Chem. 2012, 165, 48–52. [Google Scholar] [CrossRef]
- Matsuura, H.; Yamamoto, Y.; Muraoka, M.; Akaishi, K.; Hori, Y.; Uemura, K.; Tsuji, N.; Harada, K.; Hirata, K.; Bamba, T.; et al. Development of surface engineered yeast cells displaying phytochelatin synthase and their application to cadmium biosensors by the combined use of pyrene excimer fluorescence. Biotechnol. Prog. 2013, 29, 1197–1202. [Google Scholar] [CrossRef]
- Choe, S.I.; Gravelat, F.N.; Al Abdallah, Q.; Lee, M.J.; Gibbs, B.F.; Sheppard, D.C. Role of Aspergillus niger acrA in arsenic resistance and its use as the basis for an arsenicbiosensor. Appl. Environ. Microbiol. 2012, 78, 3855–3863. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.S.; Choong, C.W. Rapid detection of heavy metals with the response of carotenoids in Daucus carota. Int. J. Environ. Sci. Dev. 2014, 5, 270–273. [Google Scholar] [CrossRef]
- Teo, S.C.; Wong, L.S. Whole-cell-based biosensors for environmental heavy metals detection. Annu. Res. Rev. Biol. 2014, 4, 2663–2674. [Google Scholar] [CrossRef]
- Liu, C.; Sun, T.; Xu, X.; Dong, S. Direct toxicity assessment of toxic chemicals with electrochemical method. Anal. Chim. Acta 2009, 641, 59–63. [Google Scholar] [CrossRef]
- Mcquillan, J.S.; Shaw, A.M. Whole-cell Escherichia coli-based bio-sensor assay for dual zinc oxide nanoparticle toxicity mechanisms. Biosens. Bioelectron. 2014, 51, 274–279. [Google Scholar] [CrossRef]
- Catterall, K.; Robertson, D.; Hudson, S.; Teasdale, P.R.; Welsh, D.T.; John, R. A sensitive, rapid ferricyanide-mediated toxicity bioassay developed using Escherichia coli. Talanta 2010, 82, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Yong, D.; Liu, C.; Yu, D.; Dong, S. A sensitive, rapid and inexpensive way to assay pesticide toxicity based on electrochemical biosensor. Talanta 2011, 84, 7–12. [Google Scholar] [CrossRef]
- Pokhrel, L.R.; Silva, T.; Dubey, B.; El Badawy, A.M.; Tolaymat, T.M.; Scheuerman, P.R. Rapid screening of aquatic toxicity of several metal-based nanoparticles using the MetPLATE™ bioassay. Sci. Total Environ. 2012, 426, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Yong, D.; Liu, L.; Yu, D.; Dong, S. Development of a simple method for biotoxicity measurement using ultramicroelectrode array under non-deaerated condition. Anal. Chim. Acta 2011, 701, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, X.J.; Zhao, J.F.; Chen, L. Toxicity assessment of heavy metals and organic compounds using CellSense biosensor with E. coli. Chin. Chem. Lett. 2008, 19, 211–214. [Google Scholar] [CrossRef]
- Ravikumar, S.; Ganesh, I.; Yoo, I.-k.; Hong, S.H. Construction of a bacterial biosensor for zinc and copper and its application to the development of multifunctional heavy metal adsorption bacteria. Process Biochem. 2012, 47, 758–765. [Google Scholar] [CrossRef]
- Verma, N.; Singh, M. A Bacillus sphaericus based biosensor for monitoring nickel ions in industrial effluents and foods. J. Autom. Methods Manag. Chem. 2006, 2006, 83427. [Google Scholar] [CrossRef]
- Sochor, J.; Zitka, O.; Hynek, D.; Jilkova, E.; Krejcova, L.; Trnkova, L.; Adam, V.; Hubalek, J.; Kynicky, J.; Vrba, R.; et al. Bio-sensing of cadmium (II) ions using Staphylococcus aureus. Sensors 2011, 11, 10638–10663. [Google Scholar] [CrossRef] [PubMed]
- Ferro, Y.; Perullini, M.; Jobbagy, M.; Bilmes, S.A.; Durrieu, C. Development of a biosensor for environmental monitoring based on microalgae immobilized in silica hydrogels. Sensors 2012, 12, 16879–16891. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.P.T.; Cho, C.-W.; Yun, Y.-S. Algal biosensor-based measurement system for rapid toxicity detection. In Advances in Measurement; Sharma, M.K., Ed.; IntechOpen: London, UK, 2010; pp. 273–288. [Google Scholar]
- Chouteau, C.; Dzyadevych, S.; Durrieu, C.; Chovelon, J.M. A bi-enzymatic whole-cell conductometric biosensor for heavy metal ions and pesticides detection in water samples. Biosens. Bioelectron. 2005, 21, 273–281. [Google Scholar] [CrossRef]
- Guedri, H.; Durrieu, C. A self-assembled monolayers based conductometric algal whole-cell biosensor for water monitoring. Microchim. Acta 2008, 163, 179–184. [Google Scholar] [CrossRef]
- Chouteau, C.; Dzyadevych, S.; Chovelon, J.M.; Durrieu, C. Development of novel conductometric biosensors based on immobilized whole-cell Chlorella vulgaris microalgae. Biosens. Bioelectron. 2004, 19, 1089–1096. [Google Scholar] [CrossRef]
- Berezhetskyy, A.L.; Durrieu, C.; Nguyen-Ngoc, H.; Chovelon, J.-M.; Dzyadevych, S.V.; Tran-Minh, C. Conductometric biosensor based on whole-cell microalgae for assessment of heavy metals in wastewater. Biopolym. Cell 2007, 23, 511–518. [Google Scholar] [CrossRef]
- Shitanda, I.; Takada, K.; Sakai, Y.; Tatsuma, T. Compact amperometric algal biosensors for the evaluation of water toxicity. Anal. Chim. Acta 2005, 530, 191–197. [Google Scholar] [CrossRef]
- Shitanda, I.; Takamatsu, S.; Watanabe, K.; Itagaki, M. Amperometric screen-printed algal biosensor with flow injection analysis system for detection of environmental toxic compounds. Electrochim. Acta 2009, 54, 4933–4936. [Google Scholar] [CrossRef]
- Shitanda, I.; Takada, K.; Sakai, Y.; Tatsuma, T. Amperometric biosensing systems based on motility and gravitaxis of flagellate algae for aquatic risk assessment. Anal. Chem. 2005, 77, 6715–6718. [Google Scholar] [CrossRef]
- Islam, M.S.; Sazawa, K.; Hata, N.; Sugawara, K.; Kuramitz, H. Determination of heavy metal toxicity by using a micro-droplet hydrodynamic voltammetry for microalgal bioassay based on alkaline phosphatase. Chemosphere 2017, 188, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Amaro, F.; Turkewitz, A.P.; Martín-González, A.; Gutiérrez, J.C. Whole-cell biosensors for detection of heavy metal ions in environmental samples based on metallothionein promoters from Tetrahymena thermophila. Microb. Biotechnol. 2011, 4, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Shumyantseva, V.V.; Bulko, T.V.; Archakov, A.I. Electrochemical reduction of cytochrome P450 as an approach to the construction of biosensors and bioreactors. J. Inorg. Biochem. 2005, 99, 1051–1063. [Google Scholar] [CrossRef] [PubMed]
- Lamb, D.C.; Lei, L.; Warrilow, A.G.; Lepesheva, G.I.; Mullins, J.G.; Waterman, M.R.; Kelly, S.L. The first virally encoded cytochrome P450. J. Virol. 2009, 83, 8266–8269. [Google Scholar] [CrossRef]
- Snyder, M.J. Aquatic P450 Species. In The Ubiquitous Roles of Cytochrome P450 Proteins, Volume 3: Metal Ions in Life Sciences; Sigel, A., Sigel, H., Sigel, R.K.O., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2007; pp. 97–126. [Google Scholar]
- Danielson, P.B. The cytochrome P450 superfamily: Biochemistry, evolution and drug metabolism in humans. Curr. Drug Metab. 2002, 3, 561–597. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. Cytochrome P450 and chemical toxicology. Chem. Res. Toxicol. 2008, 21, 70–83. [Google Scholar] [CrossRef]
- Bistolas, N.; Wollenberger, U.; Jung, C.; Scheller, F.W. Cytochrome P450 biosensors—A review. Biosens. Bioelectron. 2005, 20, 2408–2423. [Google Scholar] [CrossRef]
- Fantuzzi, A.; Fairhead, M.; Gilardi, G. Direct electrochemistry of immobilized human cytochrome P450 2E1. J. Am. Chem. Soc. 2004, 126, 5040–5041. [Google Scholar] [CrossRef]
- Mhaske, S.D.; Ray, M.; Mazumdar, S. Covalent linkage of CYP101 with the electrode enhances the electrocatalytic activity of the enzyme: Vectorial electron transport from the electrode. Inorg. Chim. Acta 2010, 363, 2804–2811. [Google Scholar] [CrossRef]
- Joseph, S.; Rusling, J.F.; Lvov, Y.M.; Friedberg, T.; Fuhr, U. An amperometric biosensor with human CYP3A4 as a novel drug screening tool. Biochem. Pharmacol. 2003, 65, 1817–1826. [Google Scholar] [CrossRef]
- Sultana, N.; Schenkman, J.B.; Rusling, J.F. Protein film electrochemistry of microsomes genetically enriched in human cytochrome p450 monooxygenases. J. Am. Chem. Soc. 2005, 127, 13460–13461. [Google Scholar] [CrossRef]
- Krishnan, S.; Wasalathanthri, D.; Zhao, L.; Schenkman, J.B.; Rusling, J.F. Efficient bioelectronic actuation of the natural catalytic pathway of human metabolic cytochrome P450s. J. Am. Chem. Soc. 2011, 133, 1459–1465. [Google Scholar] [CrossRef]
- Dodhia, V.R.; Sassone, C.; Fantuzzi, A.; Di Nardo, G.D.; Sadeghi, S.J.; Gilardi, G. Modulating the coupling efficiency of human cytochrome P450 CYP3A4 at electrode surfaces through protein engineering. Electrochem. Commun. 2008, 10, 1744–1747. [Google Scholar] [CrossRef]
- Rhieu, S.Y.; Ludwig, D.R.; Siu, V.S.; Palmore, G.T.R. Direct electrochemistry of cytochrome P450 27B1 in surfactant films. Electrochem. Commun. 2009, 11, 1857–1860. [Google Scholar] [CrossRef]
- Rudakov, Y.O.; Shumyantseva, V.V.; Bulko, T.V.; Suprun, E.V.; Kuznetsova, G.P.; Samenkova, N.F.; Archakov, A.I. Stoichiometry of electrocatalytic cycle of cytochrome P450 2B4. J. Inorg. Biochem. 2008, 102, 2020–2025. [Google Scholar] [CrossRef]
- Shumyantseva, V.V.; Bulko, T.V.; Suprun, E.V.; Chalenko, Y.M.; Vagin, M.Y.; Rudakov, Y.O.; Shatskaya, M.A.; Archakov, A.I. Electrochemical investigations of cytochrome P450. Biochim. Biophys. Acta 2011, 1, 94–101. [Google Scholar] [CrossRef]
- Liu, S.; Peng, L.; Yang, X.; Wu, Y.; He, L. Electrochemistry of cytochrome P450 enzyme on nanoparticle-containing membrane-coated electrode and its applications for drug sensing. Anal. Biochem. 2008, 375, 209–216. [Google Scholar] [CrossRef]
- Alonso-Lomillo, M.A.; Gonzalo-Ruiz, J.; Domínguez-Renedo, O.; Muñoz, F.J.; Arcos-Martínez, M.J. CYP450 biosensors based on gold chips for antiepileptic drugs determination. Biosens. Bioelectron. 2008, 23, 1733–1737. [Google Scholar] [CrossRef]
- Holtmann, D.; Mangold, K.M.; Schrader, J. Entrapment of cytochrome P450 BM-3 in polypyrrole for electrochemically-driven biocatalysis. Biotechnol. Lett. 2009, 31, 765–770. [Google Scholar] [CrossRef]
- Yang, M.; Kabulski, J.L.; Wollenberg, L.; Chen, X.; Subramanian, M.; Tracy, T.S.; Lederman, D.; Gannett, P.M.; Wu, N. Electrocatalytic drug metabolism by CYP2C9 bonded to a self-assembled monolayer-modified electrode. Drug Metab. Dispos. 2009, 37, 892–899. [Google Scholar] [CrossRef]
- Fantuzzi, A.; Capria, E.; Mak, L.H.; Dodhia, V.R.; Sadeghi, S.J.; Collins, S.; Somers, G.; Huq, E.; Gilardi, G. An electrochemical microfluidic platform for human P450 drug metabolism profiling. Anal. Chem. 2010, 82, 10222–10227. [Google Scholar] [CrossRef]
- Mak, L.H.; Sadeghi, S.J.; Fantuzzi, A.; Gilardi, G. Control of human cytochrome P450 2E1 electrocatalytic response as a result of unique orientation on gold electrodes. Anal. Chem. 2010, 82, 5357–5362. [Google Scholar] [CrossRef] [PubMed]
- Schneider, E.; Clark, D.S. Cytochrome P450 (CYP) enzymes and the development of CYP biosensors. Biosens. Bioelectron. 2013, 39, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Cunningham, M.; Kim, S.; Burn, T.; Lin, J.; Sinz, M.; Hamilton, G.; Rizzo, C.; Jolley, S.; Gilbert, D.; et al. CYP3A4 induction by drugs: Correlation between a pregnane X receptor reporter gene assay and CYP3A4 expression in human hepatocytes. Drug Metab. Dispos. 2002, 30, 795–804. [Google Scholar] [CrossRef]
- Mie, Y.; Suzuki, M.; Komatsu, Y. Electrochemically driven drug metabolism by membranes containing human cytochrome P450. J. Am. Chem. Soc. 2009, 131, 6646–6647. [Google Scholar] [CrossRef]
- Xue, Q.; Kato, D.; Kamata, T.; Guo, Q.; You, T.; Niwa, O. Human cytochrome P450 3A4 and a carbon nanofiber modified film electrode as a platform for the simple evaluation of drug metabolism and inhibition reactions. Analyst 2013, 138, 6463–6468. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, K.; Kato, D.; Kamata, T.; Niwa, O. Cytochrome P450 Modified Polycrystalline indium tin oxide Film asa Drug Metabolizing Electrochemical Biosensor with a Simple Configuration. Anal. Chem. 2013, 85, 9996–9999. [Google Scholar] [CrossRef]
- Shumyantseva, V.V.; Ivanov, Y.D.; Bistolas, N.; Scheller, F.W.; Archakov, A.I.; Wollenberger, U. Direct electron transfer of cytochrome P450 2B4 at electrodes modified with nonionic detergent and colloidal clay nanoparticles. Anal. Chem. 2004, 76, 6046–6052. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, X.; Zhang, L.; Wang, C. An amperometric biosensor based on rat cytochrome p450 1A1 for benzo[a]pyrene determination. Biosens. Bioelectron. 2011, 26, 2177–2182. [Google Scholar] [CrossRef]
- Antonini, M.; Ghisellini, P.; Pastorino, L.; Paternolli, C.; Nicolini, C. Preliminary electrochemical characterisation of cytochrome P4501A2-clozapine interaction. IEE Proc. Nanobiotechnol. 2003, 150, 31–34. [Google Scholar] [CrossRef]
- Shumyantseva, V.V.; Bulko, T.V.; Rudakov, Y.O.; Kuznetsova, G.P.; Samenkova, N.F.; Lisitsa, A.V.; Karuzina, I.I.; Archakov, A.I. Electrochemical properties of cytochroms P450 using nanostructured electrodes: Direct electron transfer and electro catalysis. J. Inorg. Biochem. 2007, 101, 859–865. [Google Scholar] [CrossRef]
- Sugihara, N.; Ogoma, Y.; Abe, K.; Kondo, Y.; Akaike, T. Immobilization of cytochrome P-450 and electrochemical control of its activity. Polym. Adv. Technol. 1998, 9, 307–313. [Google Scholar] [CrossRef]
- Dai, C.; Ding, Y.; Li, M.; Fei, J. Direct electrochemistry of cytochrome P450 in a biocompatible film composed of an epoxy polymer and acetylene black. Microchim. Acta 2012, 176, 397–404. [Google Scholar] [CrossRef]
- Carrara, S.; Cavallini, A.; Erokhin, V.; De Micheli, G. Multi-panel drugs detection in human serum for personalized therapy. Biosens. Bioelectron. 2011, 26, 3914–3919. [Google Scholar] [CrossRef]
- Mie, Y.; Ikegami, M.; Komatsu, Y. Gold sputtered electrode surfaces enhance direct electron transfer reactions of human cytochrome P450s. Electrochem. Commun. 2010, 12, 680–683. [Google Scholar] [CrossRef]
- Ikegami, M.; Mie, Y.; Hirano, Y.; Suzuki, M.; Komatsu, Y. Size-controlled fabrication of gold nanodome arrays and its application to enzyme electrodes. Colloids Surf. A Physicochem. Eng. Asp. 2011, 384, 388–392. [Google Scholar] [CrossRef]
- Ndangili, P.M.; Jijana, A.M.; Baker, P.G.L.; Iwuoha, E.I. 3-mercaptopropionic acid capped ZnSe quantum dot-cytochrome P450 3A4 enzyme biotransducer for 17β-estradiol. J. Electroanal. Chem. 2011, 653, 67–74. [Google Scholar] [CrossRef]
- Niwa, O.; Ohta, S.; Takahashi, S.; Zhang, Z.; Kamata, T.; Kato, D.; Shiba, S. Hybrid carbon film electrodes for electroanalysis. Anal. Sci. 2021, 37, 37–47. [Google Scholar] [CrossRef]
- Mostafa, I.M.; Meng, C.; Dong, Z.; Lou, B.; Xu, G. Potentiometric sensors for the determination of pharmaceutical drugs. Anal. Sci. 2022, 38, 23–37. [Google Scholar] [CrossRef]
- Liu, Y.; Xue, Q.; Chang, C.; Wang, R.; Liu, Z.; He, L. Recent progress regarding electrochemical sensors for the detection of typical pollutants in water environments. Anal. Sci. 2022, 38, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Troudt, B.K.; Rousseau, C.R.; Dong, X.I.N.; Anderson, E.L.; Bühlmann, P. Recent progress in the development of improved reference electrodes for electrochemistry. Anal. Sci. 2022, 38, 71–83. [Google Scholar] [CrossRef]
- Si, Y.; Li, J.; Jhung, S.H.; Lee, H.J. Recent research trends in voltammetric sensing platforms for hormones and their applications to human serum analyses. Anal. Sci. 2022, 38, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Zambrano-Intriago, L.A.; Amorim, C.G.; Rodríguez-Díaz, J.M.; Araújo, A.N.; Montenegro, M.C.B.S.M. Challenges in the design of electrochemical sensor for glyphosate-based on new materials and biological recognition. Sci. Total Environ. 2021, 793, 148496. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).