Transferred Bacterial Community on the Potentially Pathogenic Bacteria among Aquatic Water, Plant Root, and Sediment When Planting with Chinese Herbs
Abstract
:1. Introduction
2. Materials and Methods
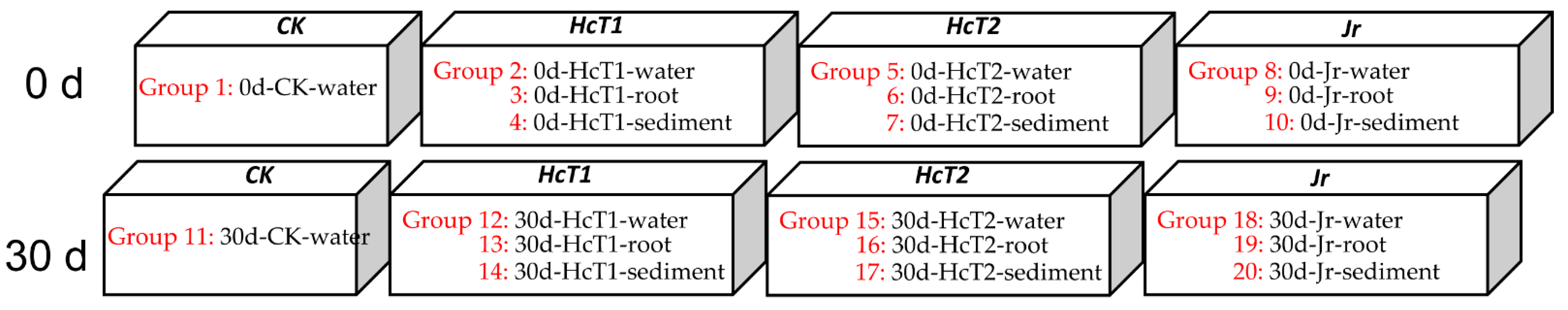
2.1. Experiment Design and Sample Collection
2.2. DNA Extraction, PCR Amplification of 16S rDNA, Amplicon Sequencing, and Sequence Data Processing
2.3. The Possible Transformation and Beneficial Influence for Prevention of Pathogenic Bacteria
2.4. Data Analysis
3. Results
3.1. OTUs Collection, α- and β-Diversity
3.2. Specific OTU Abundances with Chinese Herbs Planting
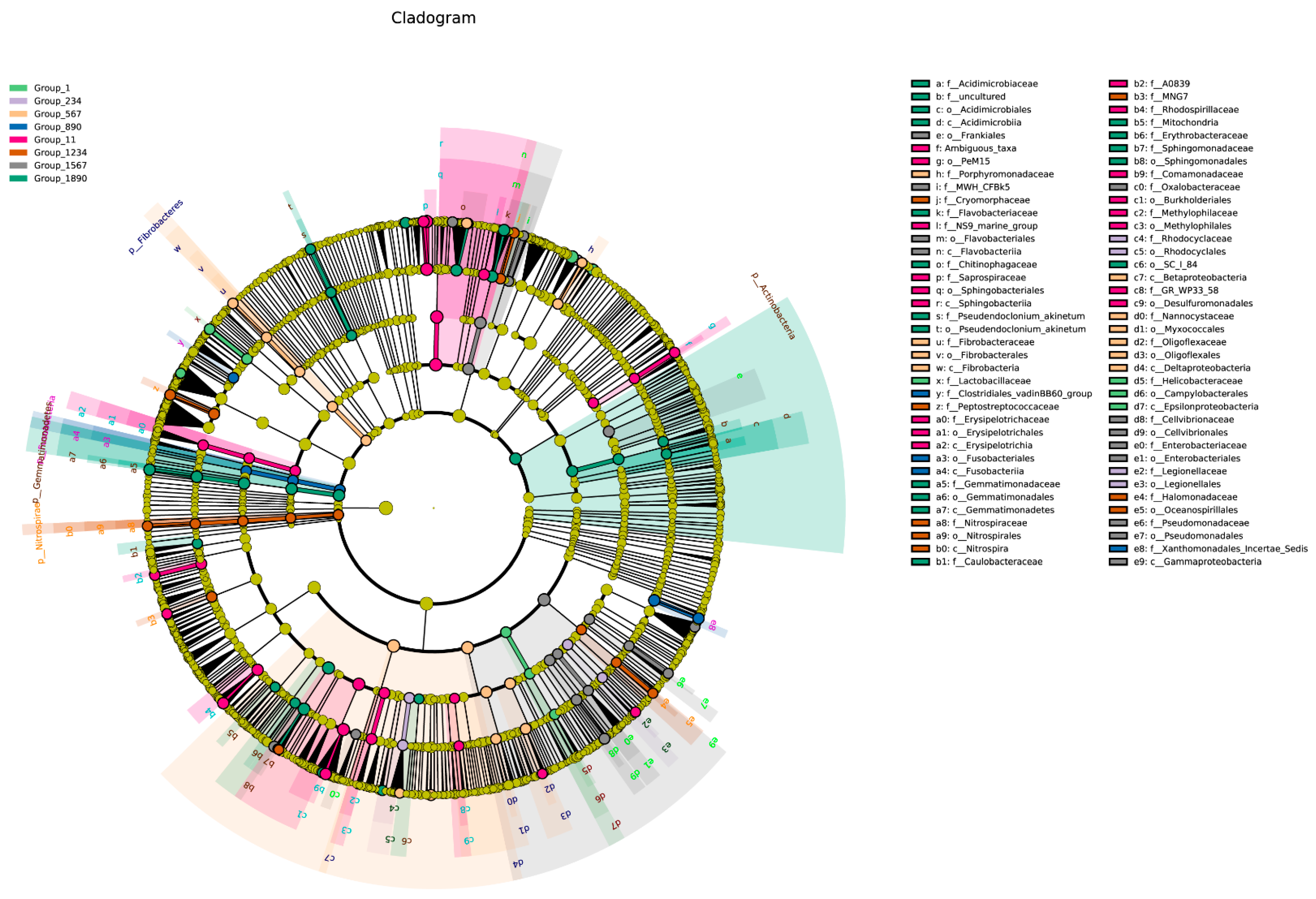
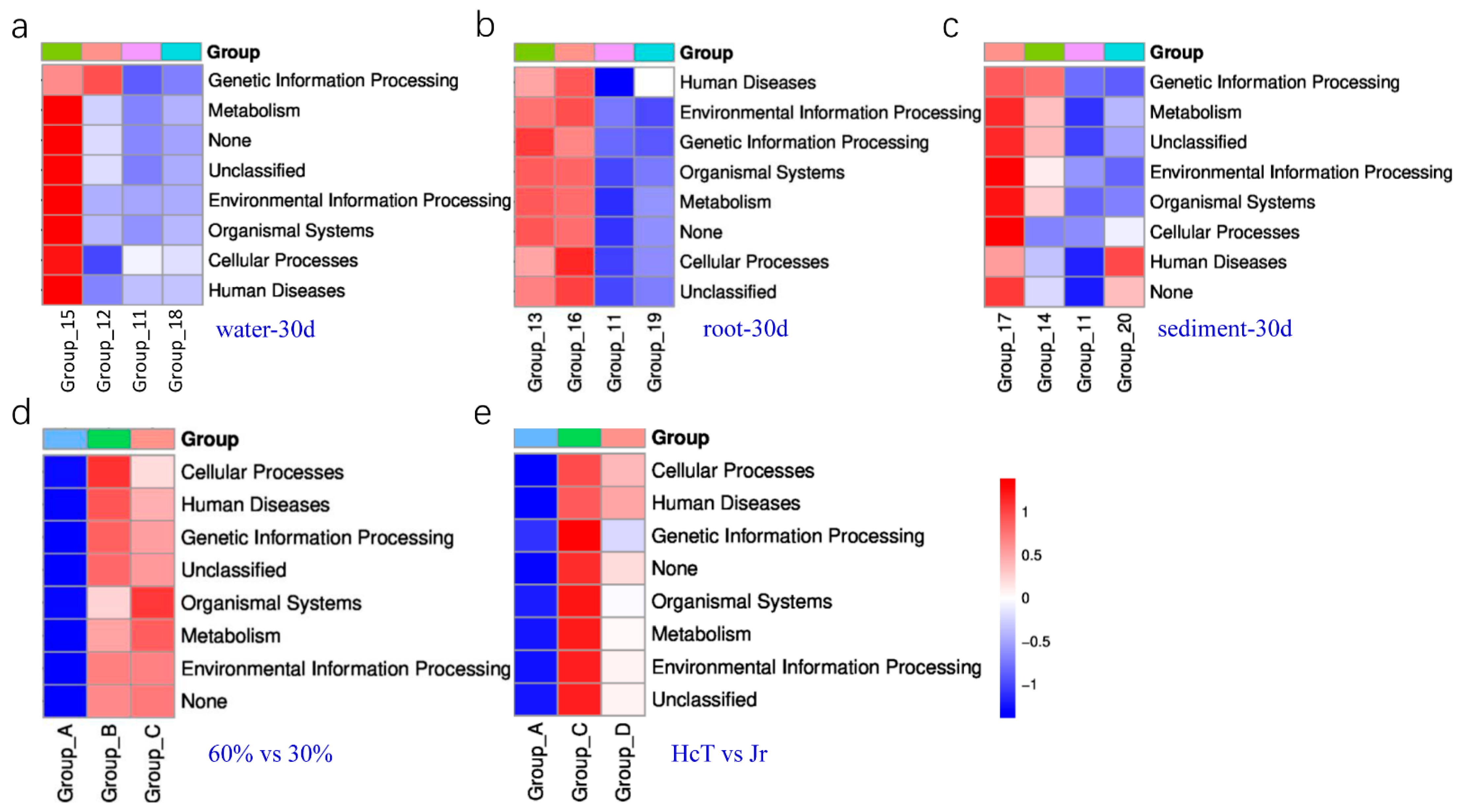
3.3. Biomarker Bacteria with Different Planting Area
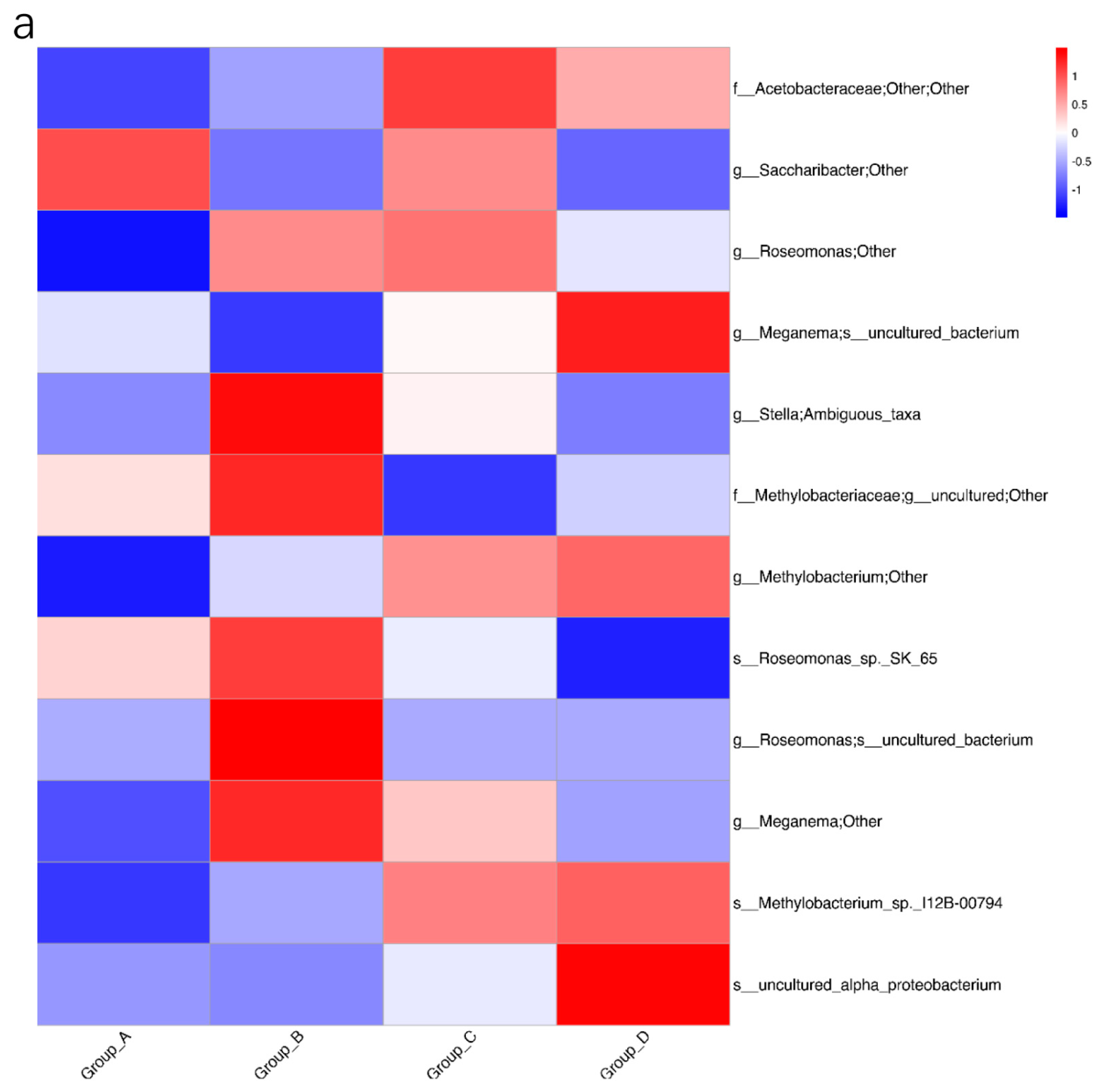
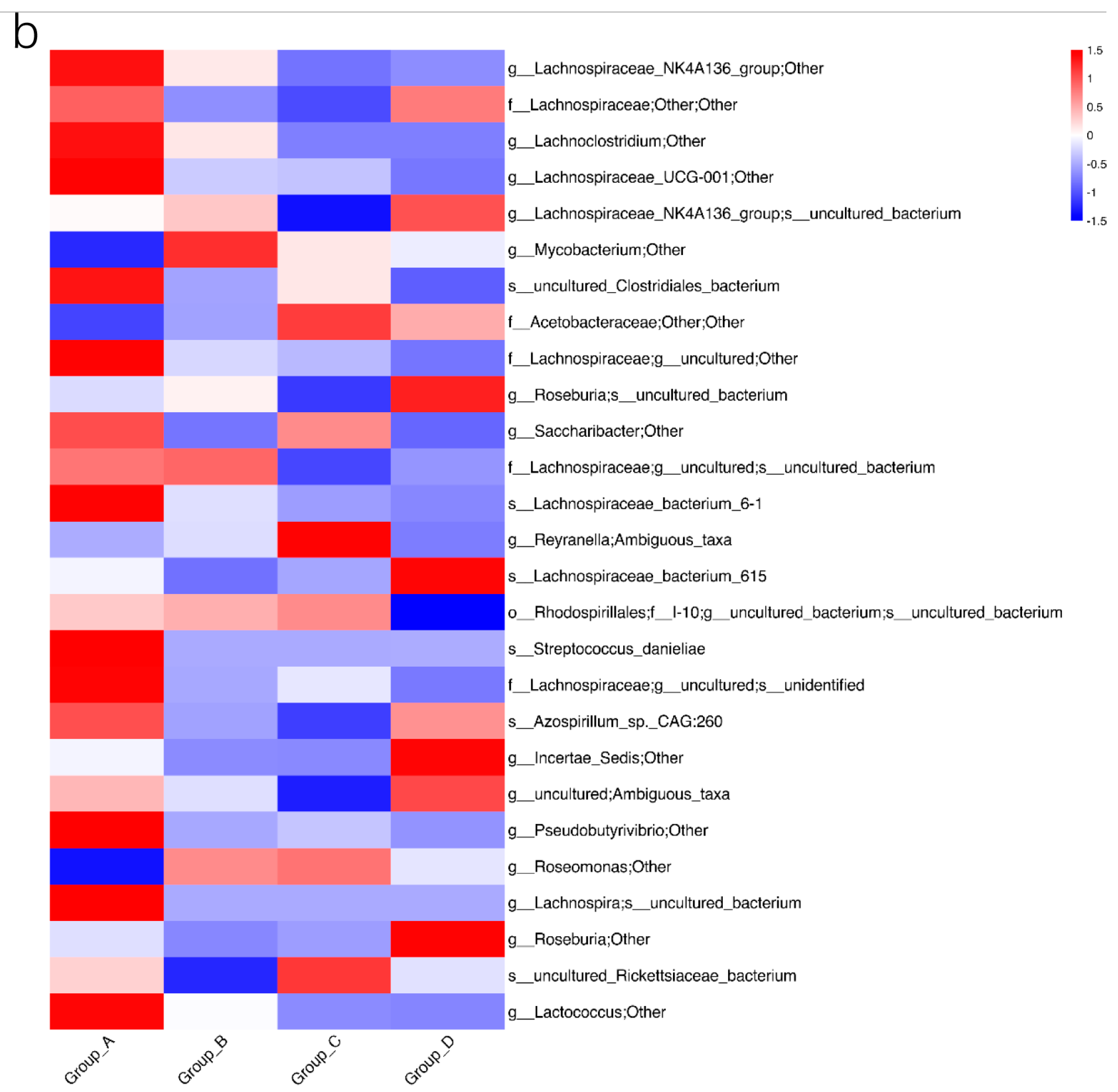
3.4. Biomarker Bacteria with Different Media Possibly Related to Transformation
3.5. Beneficial and Harmful Bacteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ananda Raja, R.; Sridhar, R.; Balachandran, C.; Palanisammi, A.; Ramesh, S.; Nagarajan, K. Pathogenicity profile of Vibrio parahaemolyticus in farmed Pacific white shrimp, Penaeus vannamei. Fish Shellfish Immunol. 2017, 67, 368–381. [Google Scholar] [CrossRef]
- Zheng, Y.; Hu, G.; Wu, W.; Qiu, L.; Li, D.; Bing, X.; Chen, J. Reshaping faecal gut microbiota composition by floating bed cultivation of Polygonum cuspidatum, Houttuynia cordata Thunb and Ipomoea aquatica Forsk. Can. J. Microbiol. 2019, 65, 522–529. [Google Scholar] [CrossRef]
- Zheng, Y.; Wu, W.; Hu, G.; Qiu, L.; Bing, X.; Chen, J. Varieties of immunity activities and gut contents in tilapia with seasonal changes. Fish Shellfish Immunol. 2019, 90, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zheng, Y.; Bing, X.; Yuan, J. Experimental study on the inhibition of algae and bacteria by the extract of Jussiaea stipulacea Ohwi. Nat. Prod. Commun. 2022, 17, 1–10. [Google Scholar] [CrossRef]
- Aniya; Nomura, Y.; Appiah, K.S.; Fuerdeng; Suzuki, Y.; Fujii, Y.; Xia, Q. Relationship between the antioxidant activity and allelopathic activities of 55 Chinese pharmaceutical plants. Plants 2022, 11, 2481. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, B.; Xing, K.; Zhang, X.; Tian, X.; Dai, W. Inhibitory effects of golden thread (Coptis chinensis) and berberine on Microcystis aeruginosa. Water Sci. Technol. 2010, 61, 763–769. [Google Scholar] [CrossRef]
- Jing, X.; Zheng, Y.; Mulbah, J.P.; Chen, J.; Xu, G. Effect of methomyl on growth, antioxidant system of GIFT (Oreochromis niloticus) and residue in the presence of water spinach (Ipomoea aquatica forsk). Int. J. Anal. Chem. 2022, 2022, 7434426. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Benkhelifa, F.; Xu, G. Combined analysis of the fruit metabolome and transcriptome reveals candidate genes involved in flavonoid biosynthesis in Bohe (Mentha haplocalyx Briq.). Aquat. Toxicol. 2023, 263, 106675. [Google Scholar] [CrossRef] [PubMed]
- Futagawa, M.; Iwasaki, T.; Murata, H.; Ishida, M.; Sawada, K. A miniature integrated multimodal sensor for measuring pH, EC and temperature for precision agriculture. Sensors 2012, 12, 8338–8354. [Google Scholar] [CrossRef]
- Ptaszek, M.; Orlikowski, L.B.; Trzewik, A.; Orlikowska, T.; Sadowski, C. Relationship between occurrence of Phytophthora cambivora on plants in hardy ornamental nursery stocks and detection of the pathogen from water ponds. Commun. Agric. Appl. Biol. Sci. 2010, 75, 659–663. [Google Scholar]
- Makovcova, J.; Slany, M.; Babak, V.; Slana, I.; Kralik, P. The water environment as a source of potentially pathogenic mycobacteria. J. Water Health 2014, 12, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Willmon, E.; Burgos, F.A.; Ray, C.L.; Hanson, T.; Arias, C.R. Biofilm and sediment are major reservoirs of virulent Aeromonas hydrophila (vAh) in catfish production ponds. J. Aquat. Anim. Health 2019, 31, 112–120. [Google Scholar] [CrossRef]
- Klanicova, B.; Slany, M.; Slana, I. Analysis of sediments and plants from the system of five fishponds in the Czech Republic using culture and PCR methods. Sci. Total Environ. 2014, 472, 851–854. [Google Scholar] [CrossRef]
- Xiong, W.; Sun, Y.; Zhang, T.; Ding, X.; Li, Y.; Wang, M.; Zeng, Z. Antibiotics, antibiotic resistance genes, and bacterial community composition in fresh water aquaculture environment in China. Microb. Ecol. 2015, 70, 425–432. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Li, H.; Gatesoupe, F.J.; She, R.; Lin, Q.; Yan, X.; Li, J.; Li, X. Bacterial signatures of “red-operculum” disease in the gut of crucian carp (Carassius auratus). Microb. Ecol. 2017, 74, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Minniti, G.; Hagen, L.H.; Porcellato, D.; Jørgensen, S.M.; Pope, P.B.; Vaaje-Kolstad, G. The skin-mucus microbial community of farmed Atlantic salmon (Salmo salar). Front. Microbiol. 2017, 8, 2043. [Google Scholar] [CrossRef]
- Shi, H.; Lu, L.; Ye, J.; Shi, L. Effects of two Bacillus velezensis microbial inoculants on the growth and rhizosphere soil environment of Prunus davidiana. Int. J. Mol. Sci. 2022, 23, 13639. [Google Scholar] [CrossRef]
- Yan, B.; Liu, N.; Liu, M.; Du, X.; Shang, F.; Huang, Y. Soil actinobacteria tend to have neutral interactions with other co-occurring microorganisms, especially under oligotrophic conditions. Environ. Microbiol. 2021, 23, 4126–4140. [Google Scholar] [CrossRef]
- Yang, X.; He, Q.; Guo, F.; Sun, X.; Zhang, J.; Chen, M.; Vymazal, J.; Chen, Y. Nanoplastics disturb nitrogen removal in constructed wetlands: Responses of microbes and macrophytes. Environ. Sci. Technol. 2020, 54, 14007–14016. [Google Scholar] [CrossRef]
- Gao, T.; Cui, B.; Kong, X.; Bai, Z.; Zhuang, X.; Qian, Z. Investigation of bacterial diversity and pathogen abundances in gibel carp (Carassius auratus gibelio) ponds during a cyprinid herpesvirus 2 outbreak. Microbiologyopen 2019, 8, e907. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lambrides, C.J.; Li, J.; Xu, Q.; Toh, R.; Tian, S.; Yang, P.; Yang, H.; Ryder, M.; Denton, M.D. Nitrifying microbes in the rhizosphere of perennial grasses are modified by biological nitrification inhibition. Microorganisms 2020, 8, 1687. [Google Scholar] [CrossRef] [PubMed]
- Waajen, G.W.; Faassen, E.J.; Lürling, M. Eutrophic urban ponds suffer from cyanobacterial blooms: Dutch examples. Environ. Sci. Pollut. Res. Int. 2014, 21, 9983–9994. [Google Scholar] [CrossRef]
- Gugliandolo, C.; Michaud, L.; Lo Giudice, A.; Lentini, V.; Rochera, C.; Camacho, A.; Maugeri, T.L. Prokaryotic community in lacustrine sediments of byers peninsula (livingston island, maritime antarctica). Microb. Ecol. 2016, 71, 387–400. [Google Scholar] [CrossRef]
- Petersen, K.S.; Anderson, S.; Chen See, J.R.; Leister, J.; Kris-Etherton, P.M.; Lamendella, R. Herbs and spices modulate gut bacterial composition in adults at risk for cardiovascular disease: Results of a pre-specified exploratory analysis from a randomized, crossover, controlled-feeding study. J. Nutr. 2022, 152, 2461–2470. [Google Scholar] [CrossRef]
- Bi, X.; Dai, W.; Wang, X.; Dong, S.; Zhang, S.; Zhang, D.; Wu, M. Microcystins distribution, bioaccumulation, and Microcystis genotype succession in a fish culture pond. Sci. Total Environ. 2019, 688, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Lodhi, M.A.; Killingbeck, K.T. Effects of pine-produced chemicals on selected understory species in a Pinus ponderosa community. J. Chem. Ecol. 1982, 8, 275–283. [Google Scholar] [CrossRef]
- Wang, N.; Wang, X.; Liu, H.; Zheng, Y.; Zhang, Y.; Xiong, J.; Pan, P.; Liu, Y. Speciation, distribution and risk assessment of metals in sediments from a water body replenished by effluent from a wastewater treatment plant. Bull. Environ. Contam. Toxicol. 2019, 102, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Suo, Y.; Li, E.; Li, T.; Jia, Y.; Qin, J.G.; Gu, Z.; Chen, L. Response of gut health and microbiota to sulfide exposure in Pacific white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 2017, 63, 87–96. [Google Scholar] [CrossRef]
- Corrochano-Monsalve, M.; González-Murua, C.; Estavillo, J.M.; Estonba, A.; Zarraonaindia, I. Impact of dimethylpyrazole-based nitrification inhibitors on soil-borne bacteria. Sci. Total Environ. 2021, 792, 148374. [Google Scholar] [CrossRef]
- Díaz, E.E.; Stams, A.J.; Amils, R.; Sanz, J.L. Phenotypic properties and microbial diversity of methanogenic granules from a full-scale upflow anaerobic sludge bed reactor treating brewery wastewater. Appl. Environ. Microbiol. 2006, 72, 4942–4949. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, S.; Chen, Y.; Guo, J.; Li, C.; Zhang, J. Metatranscriptomic approach reveals the functional and enzyme dynamics of core microbes during noni fruit fermentation. Food Res. Int. 2021, 141, 109999. [Google Scholar] [CrossRef] [PubMed]
- Pei, L.P.; Song, J.Z.; Liu, W.; Wu, E.Q.; Ling, Y.Q. Effect of water extracts from Cynanchum thesioides (Freyn) K. Schum. on visceral hypersensitivity and gut microbiota profile in maternally separated rats. J. Ethnopharmacol. 2021, 264, 113352. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Hu, J.; Xu, G. Transferred Bacterial Community on the Potentially Pathogenic Bacteria among Aquatic Water, Plant Root, and Sediment When Planting with Chinese Herbs. Environments 2023, 10, 200. https://doi.org/10.3390/environments10120200
Zheng Y, Hu J, Xu G. Transferred Bacterial Community on the Potentially Pathogenic Bacteria among Aquatic Water, Plant Root, and Sediment When Planting with Chinese Herbs. Environments. 2023; 10(12):200. https://doi.org/10.3390/environments10120200
Chicago/Turabian StyleZheng, Yao, Jiawen Hu, and Gangchun Xu. 2023. "Transferred Bacterial Community on the Potentially Pathogenic Bacteria among Aquatic Water, Plant Root, and Sediment When Planting with Chinese Herbs" Environments 10, no. 12: 200. https://doi.org/10.3390/environments10120200
APA StyleZheng, Y., Hu, J., & Xu, G. (2023). Transferred Bacterial Community on the Potentially Pathogenic Bacteria among Aquatic Water, Plant Root, and Sediment When Planting with Chinese Herbs. Environments, 10(12), 200. https://doi.org/10.3390/environments10120200





