Uncovering the Resistome of a Peruvian City through a Metagenomic Analysis of Sewage Samples
Abstract
:1. Introduction
2. Methods
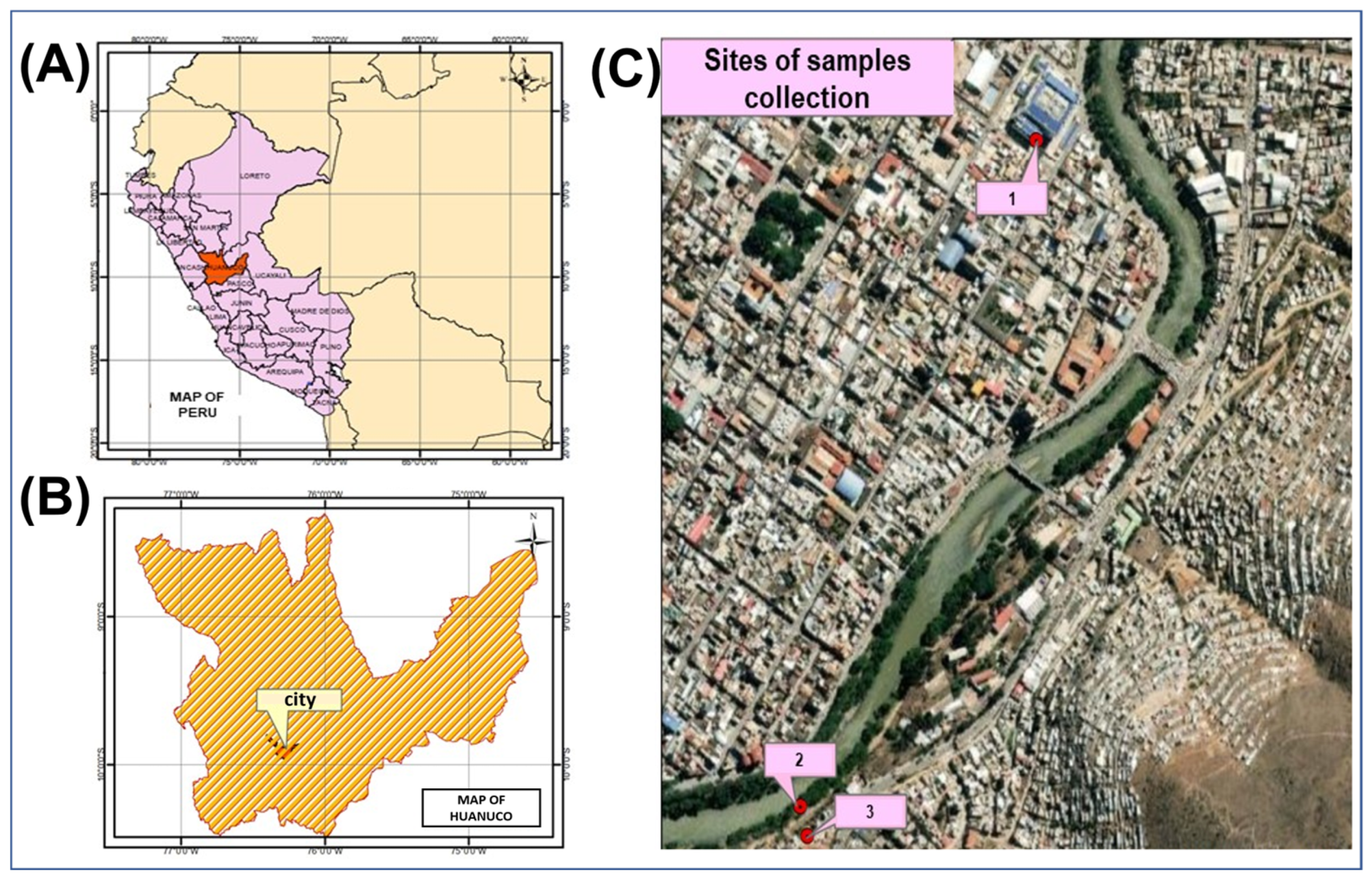
2.1. Location and Samples
2.2. Sample, DNA Extraction, and Sequencing
2.3. 16S rRNA Gene Amplification
2.4. Shotgun Metagenomics Sequencing
2.5. Antibiotic Resistance Analysis
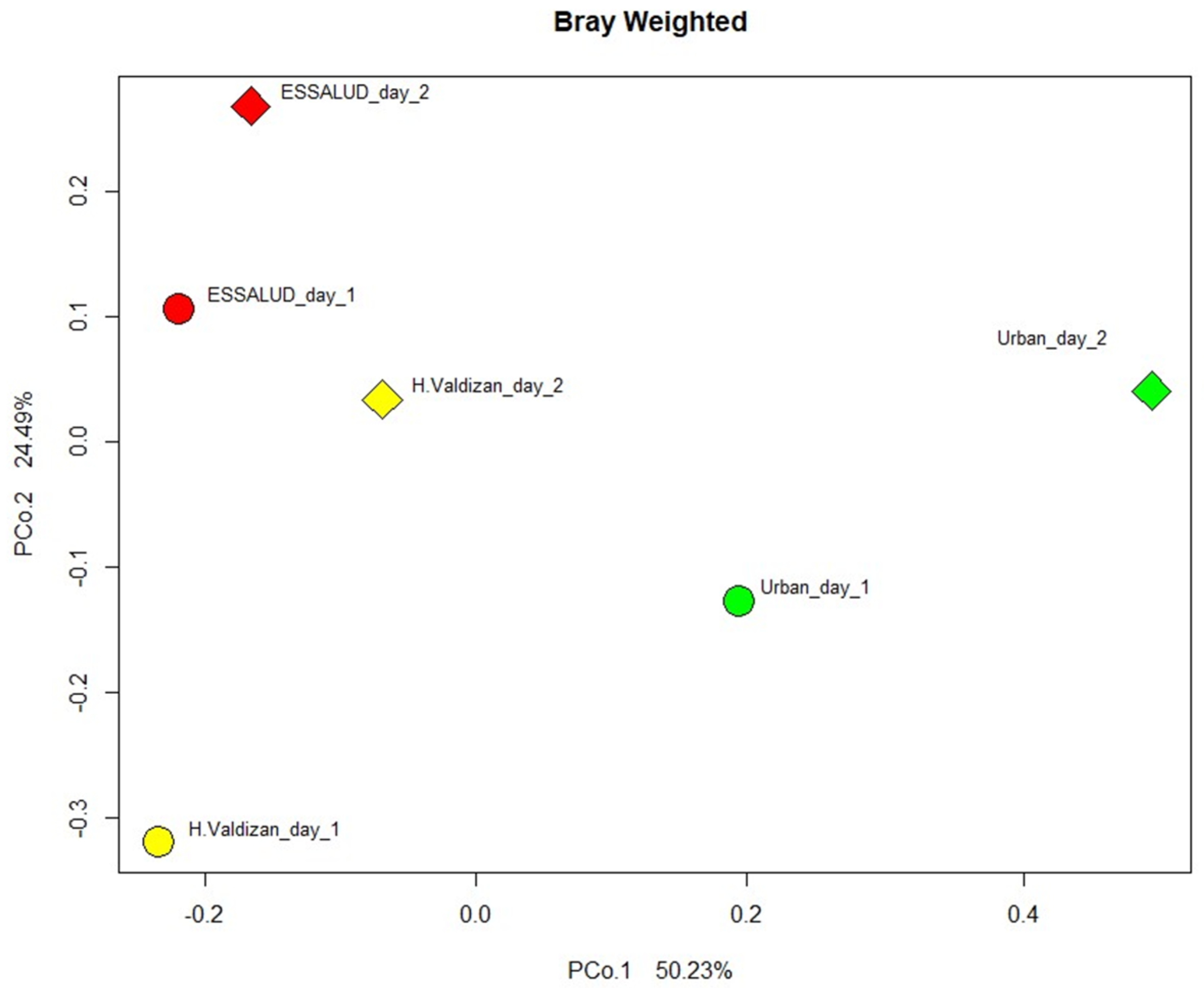
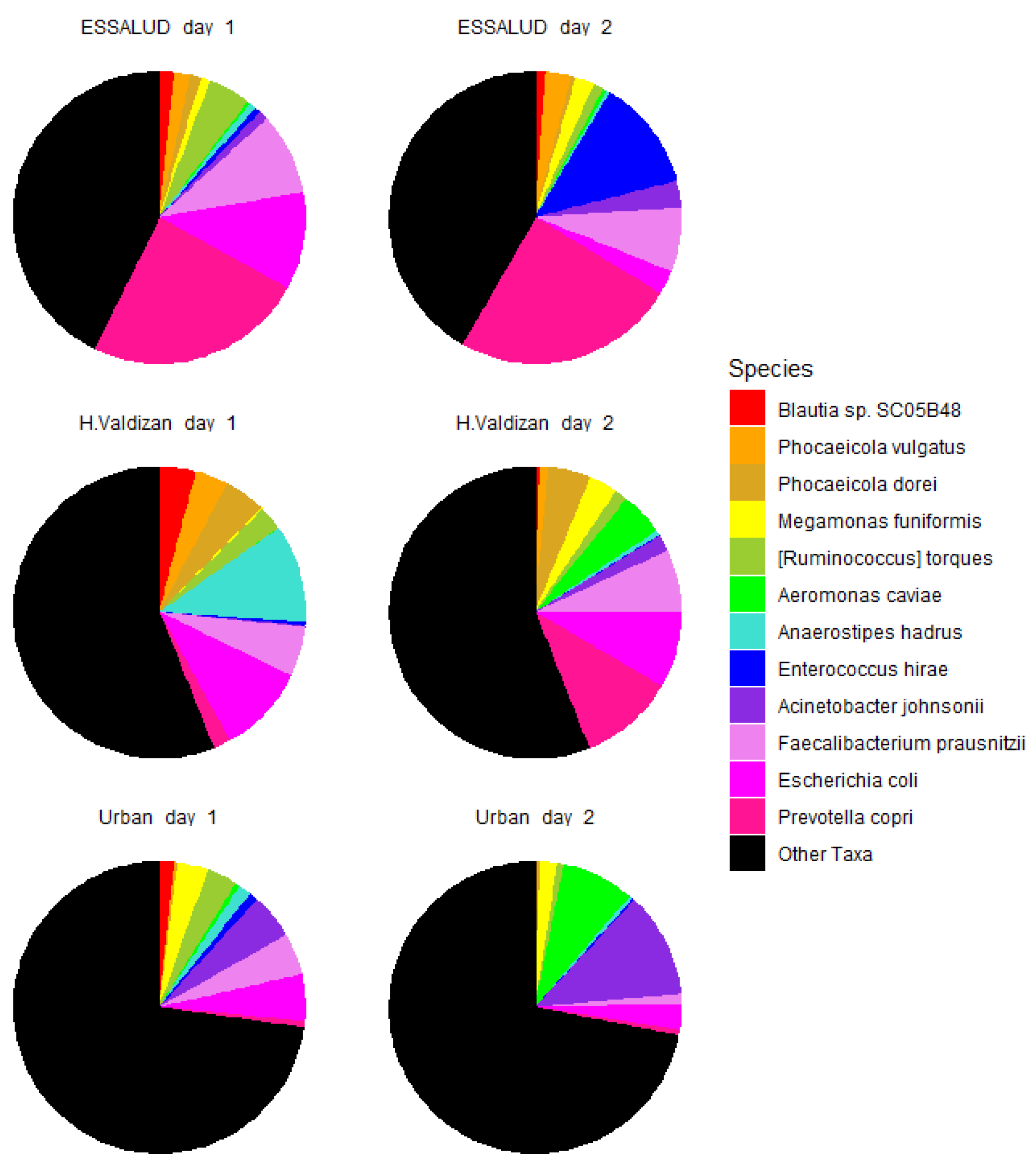
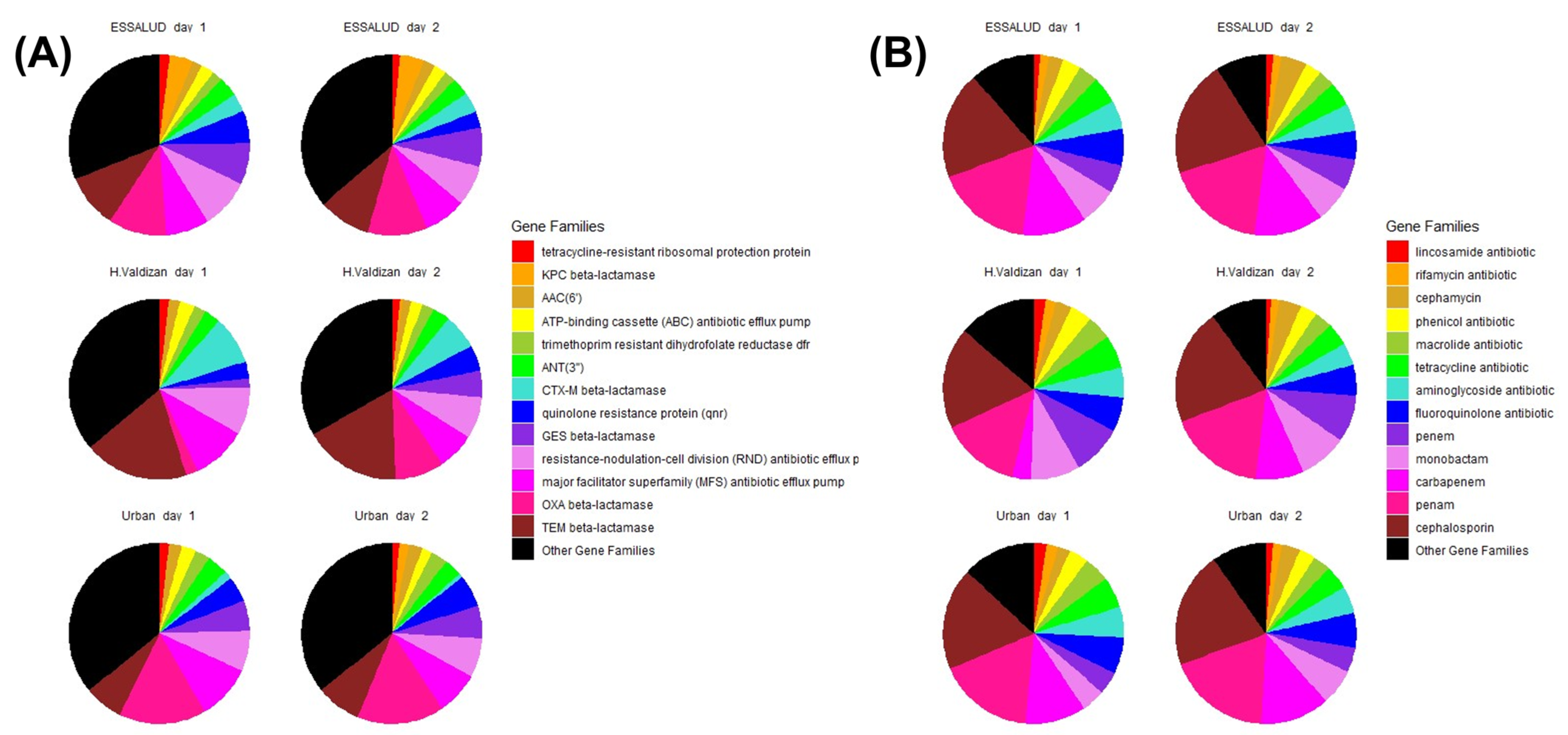
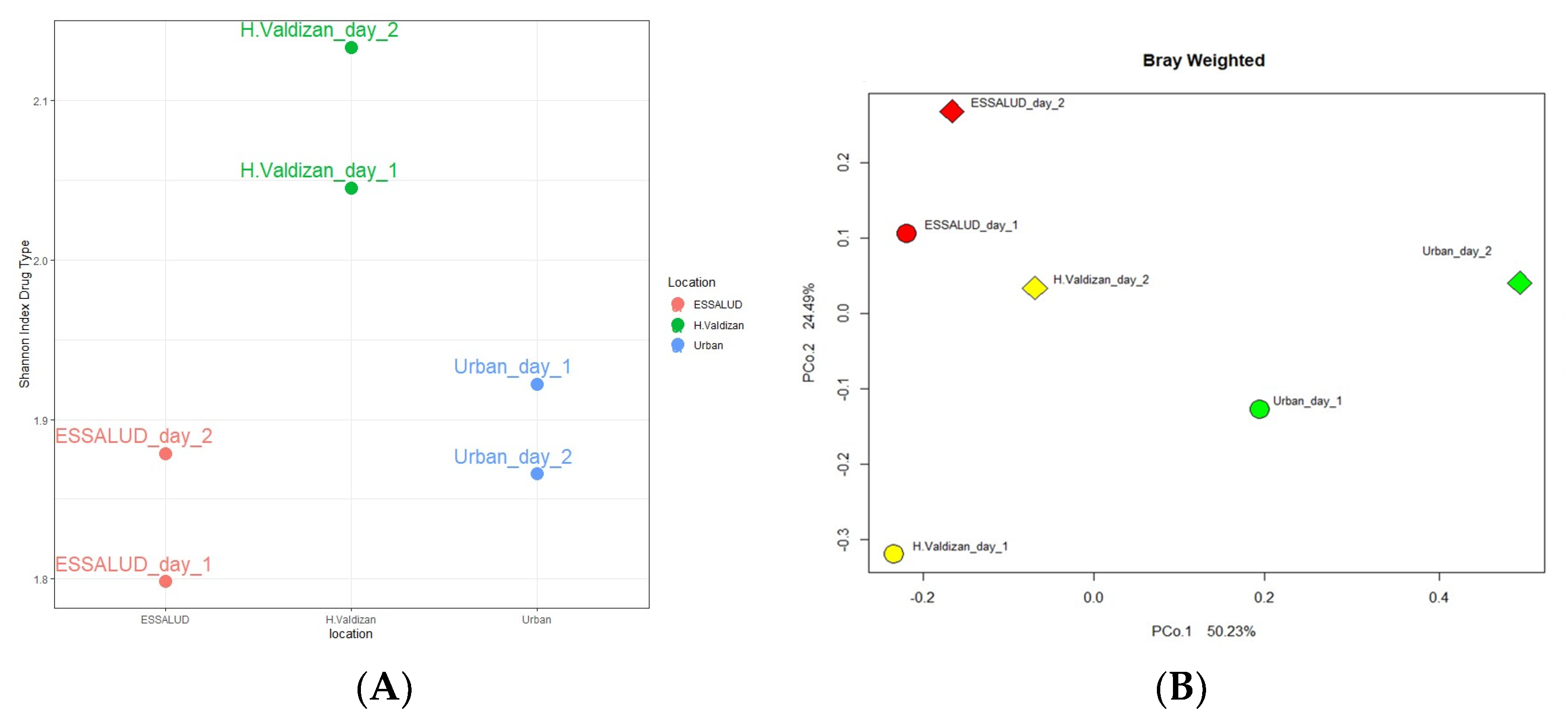
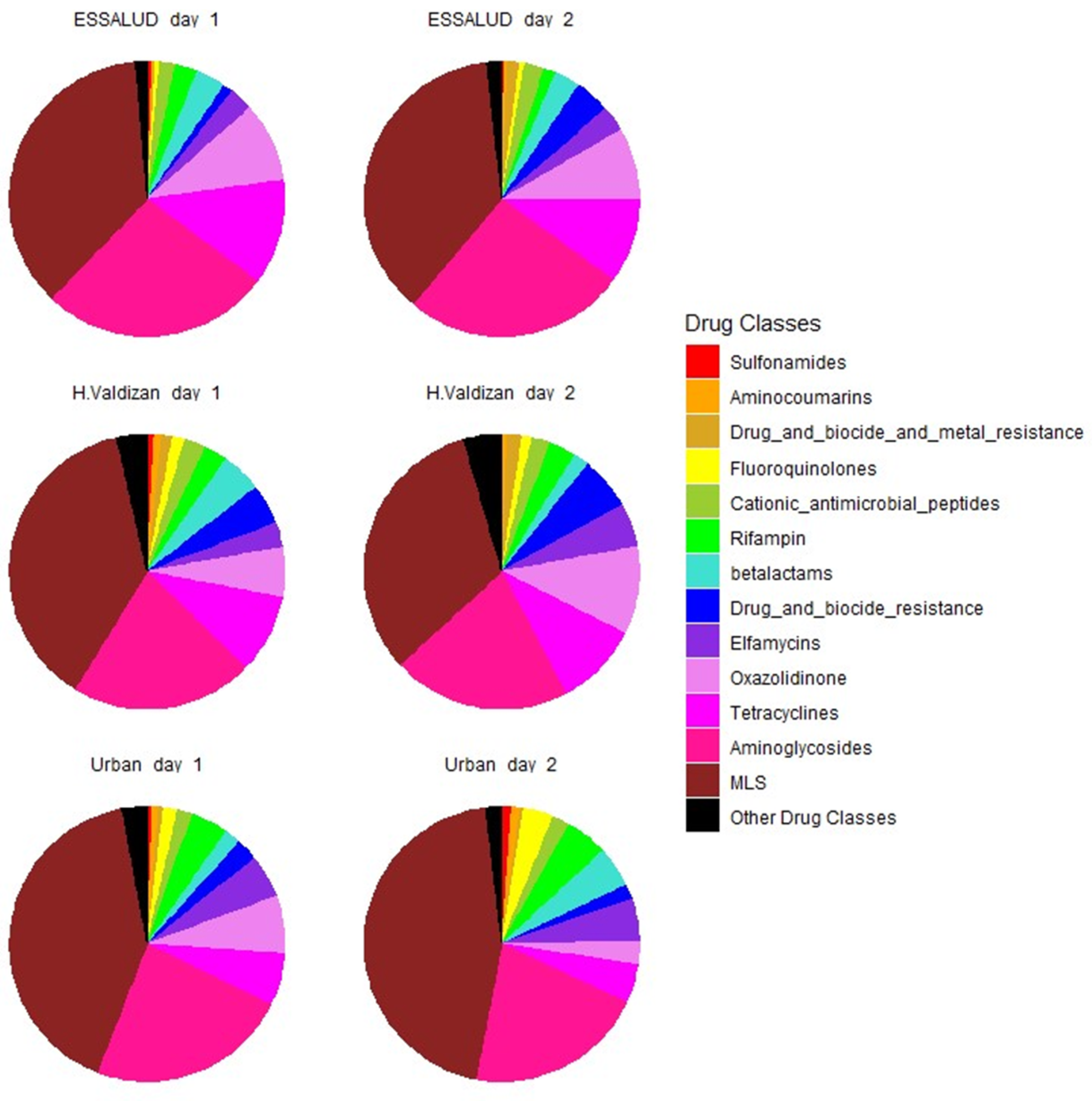
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mackenzie, J.S.; Jeggo, M. The One Health Approach—Why Is It So Important? Trop. Med. Infect. Dis. 2019, 4, 88. [Google Scholar] [CrossRef]
- Vikesland, P.; Garner, E.; Gupta, S.; Kang, S.; Maile-Moskowitz, A.; Zhu, N. Differential Drivers of Antimicrobial Resistance across the World. Acc. Chem. Res. 2019, 52, 916–924. [Google Scholar] [CrossRef]
- Booton, R.D.; Meeyai, A.; Alhusein, N.; Buller, H.; Feil, E.; Lambert, H.; Mongkolsuk, S.; Pitchforth, E.; Reyher, K.K.; Sakcamduang, W.; et al. One Health drivers of antibacterial resistance: Quantifying the relative impacts of human, animal and environmental use and transmission. One Health 2021, 12, 100220. [Google Scholar] [CrossRef]
- Albarqouni, L.; Palagama, S.; Chai, J.; Sivananthajothy, P.; Pathirana, T.; Bakhit, M.; Arab-Zozani, M.; Ranakusuma, R.; Cardona, M.; Scott, A.; et al. Overuse of medications in low- and middle-income countries: A scoping review. Bull. World Health Organ. 2023, 101, 36–61D. [Google Scholar] [CrossRef]
- Samreen; Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental antimicrobial resistance and its drivers: A potential threat to public health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016; Available online: https://wellcomecollection.org/works/thvwsuba (accessed on 1 March 2023).
- Hay, S.I.; Rao, P.C.; Dolecek, C.; Day, N.P.J.; Stergachis, A.; Lopez, A.D.; Murray, C.J.L. Measuring and mapping the global burden of antimicrobial resistance. BMC Med. 2018, 16, 78. [Google Scholar] [CrossRef]
- Baker, S.; Thomson, N.; Weill, F.-X.; Holt, K.E. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science 2018, 360, 733–738. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Woolhouse, M.E.J. Using sewage for surveillance of antimicrobial resistance. Science 2020, 367, 630–632. [Google Scholar] [CrossRef]
- The Global Sewage Surveillance Project Consortium; Hendriksen, R.S.; Munk, P.; Njage, P.; van Bunnik, B.; McNally, L.; Lukjancenko, O.; Röder, T.; Nieuwenhuijse, D.; Pedersen, S.K.; et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 2019, 10, 1124. [Google Scholar] [CrossRef]
- Munk, P.; Brinch, C.; Møller, F.D.; Petersen, T.N.; Hendriksen, R.S.; Seyfarth, A.M.; Kjeldgaard, J.S.; Svendsen, C.A.; van Bunnik, B.; Berglund, F.; et al. Genomic analysis of sewage from 101 countries reveals global landscape of antimicrobial resistance. Nat. Commun. 2022, 13, 7251. [Google Scholar] [CrossRef] [PubMed]
- Hamadeh, N.; Van Rompaey, C.; Metreau, E.; Eapen, G. New World Bank Country Classifications by Income Level: 2022–2023. The World Bank Blogs. 2022. Available online: https://blogs.worldbank.org/opendata/new-world-bank-country-classifications-income-level-2022-2023 (accessed on 1 March 2023).
- Primary Health Care Systems (PRIMASYS): Case Study from Peru, abridged version; World Health Organization (WHO): Geneva, Switzerland, 2017.
- Instituto Nacional de Estadística e Informática (INEI). Huanuco: Resultados Definitivos.; Report No.: Tomo I. 2018. Available online: https://www.inei.gob.pe/media/MenuRecursivo/publicaciones_digitales/Est/Lib1570/10TOMO_01.pdf (accessed on 1 March 2023).
- Gohl, D.M.; Vangay, P.; Garbe, J.; MacLean, A.; Hauge, A.; Becker, A.; Gould, T.J.; Clayton, J.B.; Johnson, T.J.; Hunter, R.; et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol. 2016, 34, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating species abundance in metagenomics data. PeerJ Comput. Sci. 2017, 3, e104. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Doster, E.; Lakin, S.M.; Dean, C.J.; Wolfe, C.; Young, J.G.; Boucher, C.; Belk, K.E.; Noyes, N.R.; Morley, P.S. MEGARes 2.0: A database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 2020, 48, D561–D569. [Google Scholar] [CrossRef]
- Carrillo-Larco, R.M.; Guzman-Vilca, W.C.; Leon-Velarde, F.; Bernabe-Ortiz, A.; Jimenez, M.M.; Penny, M.E.; Gianella, C.; Leguía, M.; Tsukayama, P.; Hartinger, S.M.; et al. Peru—Progress in health and sciences in 200 years of independence. Lancet Reg. Health Am. 2022, 7, 100148. [Google Scholar] [CrossRef]
- Zürcher, K.; Reichmuth, M.L.; Ballif, M.; Loiseau, C.; Borrell, S.; Reinhard, M.; Skrivankova, V.; Hömke, R.; Sander, P.; Avihingsanon, A.; et al. Mortality from drug-resistant tuberculosis in high-burden countries comparing routine drug susceptibility testing with whole-genome sequencing: A multicentre cohort study. Lancet Microbe 2021, 2, e320–e330. [Google Scholar] [CrossRef]
- Ministerio de Salud (MINSA). Modificatoria de la NTS N° 104—MINSA/DGSP V.01. Norma Técnica de Salud para la Atención Integral de las Personas Afectadas por Tuberculosis; Ministerio de Salud (MINSA): Lima, Peru, 2018; Available online: http://www.tuberculosis.minsa.gob.pe/portaldpctb/recursos/20190404114640.pdf (accessed on 4 April 2023).
- Schwalb, A.; Cachay, R.; Meza, E.; Cáceres, T.; Blackman, A.; Maruri, F.; Sterling, T.R.; Gotuzzo, E. Fluoroquinolone susceptibility in first-line drug-susceptible M. tuberculosis isolates in Lima, Peru. BMC Res. Notes 2021, 14, 413. [Google Scholar] [CrossRef]
- Torres Ortiz, A.; Coronel, J.; Vidal, J.R.; Bonilla, C.; Moore, D.A.J.; Gilman, R.H.; Balloux, F.; Kon, O.M.; Didelot, X.; Grandjean, L. Genomic signatures of pre-resistance in Mycobacterium tuberculosis. Nat. Commun. 2021, 12, 7312. [Google Scholar] [CrossRef] [PubMed]
- Villavicencio Saque, R.; Sánchez Pérez, G.; Chávez Cruz, C.; Loza Munarriz, C.; Espinoza Ríos, J. Antibiotic resistance of Helicobacter pylori in the Peruvian population: A systematic review and meta-analysis of its prevalence in the general population. Rev. Gastroenterol. Peru 2022, 42, 151–162. [Google Scholar]
- Soriano-Moreno, D.R.; Fernandez-Guzman, D.; Sangster-Carrasco, L.; Quispe-Vicuña, C.; Grados-Espinoza, P.; Ccami-Bernal, F.; Morocho-Alburqueque, N.; Coba-Villan, N.; Velasquez-Fernandez, R.; Nieto-Gutierrez, W. Factors Associated with Drug Consumption Without Scientific Evidence in Patients with Mild COVID-19 in Peru. J. Patient Saf. 2022, 18, e1189–e1195. [Google Scholar] [CrossRef] [PubMed]
- Soto, G.; Bautista, C.T.; Roth, D.E.; Gilman, R.H.; Velapatiño, B.; Ogura, M.; Dailide, G.; Razuri, M.; Meza, R.; Katz, U.; et al. Helicobacter pylori Reinfection Is Common in Peruvian Adults after Antibiotic Eradication Therapy. J. Infect. Dis. 2003, 188, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Flores, J.A.; Vargas, S.K.; Leon, S.R.; Perez, D.G.; Ramos, L.B.; Chow, J.; Konda, K.A.; Calvo, G.M.; Salvatierra, H.J.; Klaussner, J.D.; et al. Treponema pallidum pallidum Genotypes and Macrolide Resistance Status in Syphilitic Lesions among Patients at 2 Sexually Transmitted Infection Clinics in Lima, Peru. Sex. Transm. Dis. 2016, 43, 465–466. [Google Scholar] [CrossRef]
- Jorge-Berrocal, A.; Vargas-Herrera, N.; Benites, C.; Salazar-Quispe, F.; Mayta-Barrios, M.; Barrios-Cárdenas, Y.J.; Melano, R.G.; Yagui, M.; Neisseria gonorrhoeae Surveillance Working Group. Antimicrobial Susceptibility of Neisseria gonorrhoeae Isolates From Peru, 2018 and 2019. Sex. Transm. Dis. 2022, 49, 682–686. [Google Scholar] [CrossRef]
- Resurrección-Delgado, C.; Chiappe-Gonzalez, A.; Bolarte-Espinoza, J.; Martínez-Dionisio, L.; Muñante-Meneses, R.; Vicente-Lozano, Y.; Rondan-Guerrero, P.; Chávarri-Velásquez, W.; Álvarezcano-Berroa, J.; Montenegro-Idrogo, J. Uso de antibióticos en pacientes internados en un hospital nacional de Lima, Perú. Rev. Peru. Med. Exp. Salud Pública 2020, 37, 620–626. [Google Scholar] [CrossRef]
- Ymaña, B.; Luque, N.; Ruiz, J.; Pons, M.J. Worrying levels of antimicrobial resistance in Gram-negative bacteria isolated from cell phones and uniforms of Peruvian intensive care unit workers. Trans. R. Soc. Trop. Med. Hyg. 2022, 116, 676–678. [Google Scholar] [CrossRef]
- Nieto-Juárez, J.I.; Torres-Palma, R.A.; Botero-Coy, A.M.; Hernández, F. Pharmaceuticals and environmental risk assessment in municipal wastewater treatment plants and rivers from Peru. Environ. Int. 2021, 155, 106674. [Google Scholar] [CrossRef] [PubMed]
- Nadimpalli, M.L.; Marks, S.J.; Montealegre, M.C.; Gilman, R.H.; Pajuelo, M.J.; Saito, M.; Tsukayama, P.; Njenga, S.M.; Kiiru, J.; Swarthout, J.; et al. Urban informal settlements as hotspots of antimicrobial resistance and the need to curb environmental transmission. Nat. Microbiol. 2020, 5, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Pradier, L.; Bedhomme, S. Ecology, more than antibiotics consumption, is the major predictor for the global distribution of aminoglycoside-modifying enzymes. eLife 2023, 12, e77015. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poterico, J.A.; Jaramillo-Valverde, L.; Pablo-Ramirez, N.; Roa-Linares, V.C.; Martinez-Jaramillo, C.; Alvites-Arrieta, S.; Ubillus, M.; Palma-Lozano, D.; Castrejon-Cabanillas, R.; Davison, S.; et al. Uncovering the Resistome of a Peruvian City through a Metagenomic Analysis of Sewage Samples. Environments 2023, 10, 191. https://doi.org/10.3390/environments10110191
Poterico JA, Jaramillo-Valverde L, Pablo-Ramirez N, Roa-Linares VC, Martinez-Jaramillo C, Alvites-Arrieta S, Ubillus M, Palma-Lozano D, Castrejon-Cabanillas R, Davison S, et al. Uncovering the Resistome of a Peruvian City through a Metagenomic Analysis of Sewage Samples. Environments. 2023; 10(11):191. https://doi.org/10.3390/environments10110191
Chicago/Turabian StylePoterico, Julio A., Luis Jaramillo-Valverde, Nelis Pablo-Ramirez, Vicky C. Roa-Linares, Catalina Martinez-Jaramillo, Sandra Alvites-Arrieta, Milward Ubillus, Diana Palma-Lozano, Rony Castrejon-Cabanillas, Samuel Davison, and et al. 2023. "Uncovering the Resistome of a Peruvian City through a Metagenomic Analysis of Sewage Samples" Environments 10, no. 11: 191. https://doi.org/10.3390/environments10110191
APA StylePoterico, J. A., Jaramillo-Valverde, L., Pablo-Ramirez, N., Roa-Linares, V. C., Martinez-Jaramillo, C., Alvites-Arrieta, S., Ubillus, M., Palma-Lozano, D., Castrejon-Cabanillas, R., Davison, S., Gomez, A., & Guio, H. (2023). Uncovering the Resistome of a Peruvian City through a Metagenomic Analysis of Sewage Samples. Environments, 10(11), 191. https://doi.org/10.3390/environments10110191






