Abstract
Background: Breast and ovarian cancers (BC and OC) are prevalent malignancies in women globally, with germline variants in the BRCA2 gene significantly increasing the risk of developing these cancers. Despite extensive studies, the frequency and impact of BRCA2 variants in women from Jalisco, Mexico, remain underexplored. Objective: The aim of this study was to identify and characterize BRCA2 gene variants in Mexican women diagnosed with BC and OC and to assess their functional and structural consequences using computational analyses. Methodology: Genomic DNA from 140 Mexican women with BC and/or OC, selected based on clinical criteria suggestive of BRCA2 variants, was sequenced using NGS targeting BRCA2 coding regions. Functional effects were predicted with Ensembl VEP, SIFT, and PolyPhen-2. Structural impacts of missense variants were assessed using HOPE and AlphaFold models. Results: BRCA2 variants were identified in 12.86% of patients, with higher frequency in OC (21.05%) than BC (12%). Several mapped to key functional domains, including BRC repeats and the DNA-binding domain. Many were predicted as deleterious or probably damaging, though clinical classifications were often conflicting. Structural analysis indicated potential disruptions in protein stability or interactions for most missense variants. Clinically, BRCA2-positive BC patients were younger at diagnosis and showed a trend toward lower complete response. Conclusion: BRCA2 variants were found in 12.86% of patients, including six VUSs not reported in other populations. Several affected key functional domains with predicted deleterious effects. Findings support the need for genetic panels tailored to the Mexican population.
1. Introduction
Breast and ovarian cancer (BC and OC) are among the most common and deadly malignancies affecting women worldwide [1]. Both environmental and genetic factors influence their development. Notably, carriers of germline variants in the BRCA2 gene have a 55% increased risk of developing BC compared to non-carriers [2]. Furthermore, BRCA2 variant carriers face a lifetime risk of OC estimated between 11% and 27%, significantly higher than the general population [3,4]. Variants in BRCA2 can also increase the risk of developing prostate, pancreatic, and colorectal cancers [5,6,7].
The BRCA2 gene, located on chromosome 13q12.3, plays a crucial role in maintaining genomic stability by regulating DNA repair and cell cycle progression. Its primary function is the repair of DNA double-strand breaks (DSBs) through homologous recombination (HR), a high-fidelity repair pathway essential for maintaining the genome. The BRCA2 protein mediates the recruitment and loading of RAD51 recombinase onto single-stranded DNA at sites of damage, a critical step for the formation and stabilization of the RAD51 nucleoprotein filament. This process involves specific protein domains, including the eight BRC repeats that directly bind RAD51, and the C-terminal domain that further stabilizes the RAD51–DNA complex. BRCA2 also contains a nuclear localization signal (NLS) that ensures proper transport into the nucleus, where DNA repair occurs. Pathogenic variants in BRCA2 can disrupt these interactions, impairing RAD51 filament assembly, compromising HR efficiency, and ultimately leading to genomic instability and increased cancer predisposition [8,9].
Genetic sequencing has enabled the identification of numerous germline variants in the BRCA2 gene, classified as benign, probably benign, of uncertain significance, probably pathogenic, and pathogenic [10,11]. However, many variants described in the ClinVar database have conflicting classifications, as their clinical relevance lacks clear consensus among different laboratories, databases, or scientific studies. Large international consortia, such as CIMBA [12] and ENIGMA [13], have conducted extensive analyses linking specific BRCA2 variants to the risk of BC and OC, providing strong evidence for the pathogenicity of certain variants through case–control studies, segregation analyses, and functional assays. Functional studies have further demonstrated that pathogenic BRCA2 variants can disrupt RAD51 binding, impair homologous recombination efficiency, and lead to genomic instability [14]. The integration of bioinformatic tools to evaluate the impact of variants on protein stability, domain integrity, and molecular interactions provides valuable complementary evidence to refine variant classification and assess their functional consequences [15].
Although studies describing the frequency and spectrum of clinically relevant BRCA2 variants have been conducted in different regions of Mexico [16,17,18], none have specifically focused on the population from the state of Jalisco. This regional focus is relevant because Mexico is characterized by marked genetic heterogeneity and admixture patterns that vary by state [19]; for example, Jalisco exhibits a distinctive profile with roughly balanced European and Native American ancestry [20]. In addition, disparities in healthcare access and cancer diagnostic services across the country, including documented delays in rural areas of Jalisco [21], may influence both variant detection and clinical outcomes. These factors underscore the importance of a population-specific analysis to better capture the genetic and clinical landscape of BRCA2 in this region.
Furthermore, most previous reports have not incorporated computational analyses to predict the functional effects of these variants on the gene or protein, particularly in the case of VUS. Therefore, analyzing this population using both genetic and in silico approaches provides valuable insights into the genetic diversity within the country and may reveal clinically relevant variants not previously characterized in other Mexican cohorts.
In this study, we analyzed BRCA2 gene variants identified in Mexican women with BC and OC. Our focus was to characterize the frequency of these variants within the Mexican BC and OC population and to perform computational analyses assessing their functional significance, structural stability, and potential impact on the BRCA2 protein.
2. Materials and Methods
2.1. Patients
This study included Mexican women over 18 years of age with a confirmed diagnosis of BC and/or OC, with clinical suspicion of being carriers of germline pathogenic variants in the BRCA2 gene, from the population of Jalisco, Mexico. Patient selection comprised women diagnosed with BC and/or OC who met at least one of the following criteria: early disease onset (diagnosis before 50 years of age), a significant family history of related cancers (such as breast, ovarian, prostate, or pancreatic cancer in first- or second-degree relatives), or a diagnosis of multiple primary neoplasms (bilateral or multiple cancers). Additionally, relevant tumor characteristics were considered, including the HER2-negative subtype in BC, due to its known association with BRCA2 pathogenic variants.
The guidelines provided in the Declaration of Helsinki were followed to ensure the welfare and rights of the study participants. All patients were informed about the objectives and procedures of the study, and their written informed consent was obtained before sample collection. The study protocol was approved by the ethics committee under registration number R-2022-1305-114 at the Centro de Investigación Biomédica de Occidente, Instituto Mexicano del Seguro Social (CLIES #1305), ensuring compliance with all relevant ethical and legal regulations.
2.2. Identification of Variants in the BRCA2 Gene
2.2.1. Genomic DNA Extraction
Genomic DNA was extracted from peripheral blood samples using the AmoyDx Blood/Bone Marrow Spin Column kit (AmoyDX, Singapore), strictly following the manufacturer’s protocol. The quality and concentration of the extracted DNA were assessed using spectrophotometry (Qubit 4, ThermoFisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis to ensure its integrity and purity before proceeding with downstream analyses.
2.2.2. BRCA2 Sequencing
The complete coding regions of the BRCA2 gene were targeted for analysis. Selective capture and amplification of these regions were performed using amplicon enrichment technology with the commercial AmpliSeq for Illumina BRCA Panel. This panel is specifically designed to comprehensively cover the coding exons and splice site boundaries of BRCA1 and BRCA2, enabling accurate detection of single nucleotide variants, insertions, deletions, and splice-affecting alterations. DNA libraries were prepared following the standard AmpliSeq protocol, which includes amplification, purification, and quantification steps. Subsequently, libraries were sequenced on the Illumina MiSeq next-generation sequencing platform (Illumina, Inc., San Diego, CA, USA) using transcripts of the variants BRCA2 (NM_000059.3) with a compatible sequencing kit that generated high-quality, appropriately long reads for detailed analysis.
2.2.3. Bioinformatic Analysis and Variant Annotation
Sequence reads were processed and variants identified using the ANDAS-Amoy platform, a specialized tool that performs sequence alignment, variant calling, and functional annotation. Variants were filtered based on quality metrics and a minor allele frequency (MAF) of ≤0.01, as per the gnomAD and ClinVar databases. Potentially damaging variants were prioritized based on their predicted pathogenicity by in silico tools integrated into the platform. Additional annotations included genomic position, predicted protein effect, and allele frequency in public databases. The analysis focused on coding variants; no splice site variants meeting these criteria were identified in the cohort.
2.2.4. Variant Classification and Evaluation
Clinical interpretation of detected variants was conducted according to the guidelines established by the ACMG/AMP consortium [22]. This framework categorizes variants as pathogenic, likely pathogenic, variants of uncertain significance, likely benign, or benign.
2.3. Computational Analysis of Variants
2.3.1. Structural Identification of Functional Domains Using UniProt
To characterize the structural localization of a set of variants in the BRCA2 gene, the UniProt database [23] (https://www.uniprot.org, accessed on 20 June 2025) was utilized through its official web platform. The entry corresponding to the identifier P51587, representing the BRCA2 protein encoded by the NM_000059.4 transcript, which was selected as part of the MANE Select set, was consulted. From this entry, functional domains and structural regions of the protein were identified, along with their corresponding annotations.
UniProt [23] is a comprehensive database that provides detailed protein sequence and functional information, including annotations on protein function, structure, and involvement in diseases. It is a key resource for researchers in the fields of genomics and bioinformatics.
For each analyzed variant, the corresponding position of the amino acid change in the protein sequence was located and manually compared with the annotated region ranges. This allowed for determining whether the variant resided within a functional domain, an unstructured region, or in areas with no known annotation. This approach enabled the classification of each variant based on its relative location to critical domains of the BRCA2 protein, providing an initial assessment of its potential functional relevance.
2.3.2. Functional Analysis and Impact Prediction Using Ensembl VEP
Subsequently, a more detailed functional annotation was performed using the Variant Effect Predictor (VEP) tool, version 115, from Ensembl [24] (https://useast.ensembl.org/info/docs/tools/vep/index.html, accessed on 26 June 2025), which is available online via its web interface. The reference transcript NM_000059.4, corresponding to the BRCA2 gene and recognized as the MANE Select transcript, was used. Variants were analyzed in rsID format when available, and in HGVS cDNA format when the polymorphism was not registered in databases.
VEP version 115 [24] is a tool used to predict the functional effects of genetic variants. It provides detailed annotations for variants, including their impact on protein-coding regions, regulatory regions, and noncoding sequences. VEP integrates data from multiple sources, including gene models, regulatory features, and known pathogenic variants, to help researchers assess the potential consequences of genetic variations on gene function and disease.
For each entered variant, VEP version 115 [24] provided information on the type of molecular consequence (such as missense_variant, frameshift_variant, or stop_gained), the estimated functional impact (classified as high, moderate, or low), codon changes, and alignment with the selected transcript. Additionally, computational predictions generated by the SIFT [25] and PolyPhen version 2 (PolyPhen-2) [26] algorithms, which estimate whether a nonsynonymous variant could alter protein function or structure, were included. SIFT and PolyPhen-2 predictions are only available for missense variants.
SIFT [25] classifies variants as deleterious or tolerated based on the evolutionary conservation of the affected residues. At the same time, PolyPhen-2 [26] categorizes them as probably damaging, possibly damaging, or benign, based on structural and functional criteria. Both tools also provide a quantitative score between 0 and 1, indicating the confidence in the prediction. The results were recorded in a comparative table for each variant, integrating information about their localization in functional domains with their functional predictions, to facilitate biological and clinical interpretation.
2.4. Structural Impact Prediction and Local Visualization of Variants
2.4.1. Structural Impact Prediction
To explore the possible structural consequences of the identified missense variants in BRCA2, the HOPE tool (Have (y)Our Protein Explained) version 1.1.1 [27] was used (https://www3.cmbi.umcn.nl/hope/, accessed on 19 April 2025). This resource enables automated analysis based on the physicochemical properties of amino acids, evolutionary conservation information, and annotations from UniProt. Due to the lack of a resolved three-dimensional structure for the complete BRCA2 protein, the analysis was performed based on computational predictions and sequence alignments, using the UniProt identifier P51587 as a reference for the protein encoded by the NM_000059.4 transcript (MANE Select).
For each missense variant, HOPE [27] provided a comparison between the wild-type and mutant amino acids in terms of size, charge, and hydrophobicity, as well as indicating whether the affected residue was evolutionarily conserved. Additionally, when possible, annotations on proximity to functional regions or previously reported variants in databases were provided. The tool generated an automated diagnosis of the potential structural or functional disruption caused by the change, based on the biochemical differences between the residues.
2.4.2. Fragment Modeling
To illustrate the precise localization of selected missense variants in the BRCA2 protein, three-dimensional models of local fragments of approximately 20 amino acids centered on the position of each amino acid change were generated. This modeling was performed exclusively for missense variants, regardless of their clinical classification, in order to visualize their immediate structural environment. The corresponding sequences were obtained from the UniProt entry ID P51587, corresponding to the canonical isoform of BRCA2. The fragments were submitted to the AlphaFold Server, using the AlphaFold version 3 structure prediction model (AlphaFold3) [28] (https://alphafoldserver.com/, accessed on 30 June 2025). Models were generated for both the wild-type and mutated sequences, with the residue corresponding to each variant manually modified. The resulting models were visualized using UCSF ChimeraX version 1.9 [29] (https://www.cgl.ucsf.edu/chimerax/, accessed on 30 June 2025), where mutated residues were highlighted using various representation styles, such as “stick” and “sphere,” along with text labels. This structural analysis was performed solely for illustrative purposes to show the immediate environment of each variant, without making functional or protein stability inferences; therefore, confidence metrics such as pLDDT values were not considered in this work.
AlphaFold [28] is an artificial intelligence–based computational tool developed by DeepMind that enables the high-accuracy prediction of protein three-dimensional structures from their amino acid sequences. It utilizes deep learning models trained on known structures to infer the most probable conformation of the polypeptide chain. Its use has become widespread in structural bioinformatics studies due to its ability to generate reliable models even in the absence of experimental data. ChimeraX [29], on the other hand, is a molecular visualization software developed by the University of California, San Francisco (UCSF), designed for the analysis, editing, and graphical representation of biomolecular structures. Its intuitive interface and capacity to work with both predicted models and experimental structures make it a key tool for exploring molecular interactions, locating variants, and generating high-quality figures for scientific publications.
It is important to note that the Structural Impact Prediction and Local Visualization of Variants analysis was applied only to missense variants, as stop-gained variants are assumed to disrupt protein formation.
3. Results
3.1. Sociodemographic and Clinicopathological Characteristics of the Study Patients
A total of 140 patients were included in this study, comprising 116 women with BC, 19 with ovarian cancer (OC), and 5 with combined BC and OC diagnoses (Table 1 and Table 2). Regarding sociodemographic characteristics (Table 1), the mean ages were 46.9 ± 13.3 years (range, 20–79) for BC patients, 56.6 ± 11.1 years (range, 35–77) for OC patients, and 49.0 ± 10.7 years (range, 33–61) for the combined group. A statistically significant age difference was observed between the BC and OC groups (p = 0.028), with patients diagnosed with OC being, on average, older. The combined group was excluded from the comparative analysis due to its small sample size. The age at menarche averaged 12.2 ± 1.56 years (range, 8–16) in BC, 11.8 ± 0.89 years (range, 10–13) in OC, and 12.4 ± 0.54 years (range, 12–13) in the combined group. Regarding BMI, normal weight was predominant in BC (76%) and OC (58%), while all patients in the combined group were obese (100%). Alcohol, tobacco consumption, and hormone therapy were reported as negative in most groups. Pre-menopause status (53%) was more frequent in the BC, in contrast to the OC group (84%) and the combined group (60%). Breastfeeding and the family history of cancer in first or second degree relatives with breast, ovarian, prostate, or pancreatic cancer were reported as positive in most groups. The autodetection was characteristic of the BC, and sonogram, ultrasonogram in the OC, and the combined group.

Table 1.
Sociodemographic characteristics of the cohort of patients.

Table 2.
Clinicopathological characteristics of the cancer shown by the patients.
Clinicopathological characteristics (Table 2) revealed that the study groups were predominantly diagnosed between 1 and 4 years. The tumors were predominantly unilateral. The most frequent clinical stages in BC were II and III. Histologically, invasive ductal carcinoma was the predominant type in BC (94%), whereas high-grade serous carcinoma was universal in OC and the combined group (100%). Regarding molecular subtypes, 44% of breast tumors were triple negative; all ovarian and combined cases were high-grade serous, with combined cases showing serous with Luminal A and serous with triple negative subtypes. In the three study groups, most patients had a Ki-67 proliferation index of 20% or greater. Lymph node involvement was observed in 40% of BC patients and 21% of OC patients. Treatment response was complete in 63% of BC cases, partial in 42% of OC cases, and complete in 80% of the combined group.
3.2. Frequency and Distribution of BRCA2 Variants in Patient Cohorts
All 140 patients included in this study underwent successful BRCA2 gene sequencing. Variants were identified in 18 patients (12.86%), including 14 women with BC (12%) and four patients with OC (21.05%). The variants detected in the studied patients are detailed in Table 3.

Table 3.
Genetic BRCA2 variants detected among BC and OC cases.
3.3. Comparison of Patients Carrying and Not Carrying BRCA2 Variants
When the clinical characteristics of the study groups were compared, stratified by BRCA2-positive and BRCA2-negative status for each cancer type, the cohort included 102 BRCA2-negative and 14 BRCA2-positive BC cases, as well as 15 BRCA2-negative and 4 BRCA2-positive OC cases (Table S1). In BC, statistically significant differences were observed for overweight status (29% vs. 6%, p = 0.019), family history of first-degree relatives with breast/ovarian/pancreatic cancer (100% vs. 57%, p = 0.004), and chemotherapy response (complete response 36% vs. 69%, p = 0.032). In OC, significant associations were found for abortion history (0% vs. 80%, p = 0.018) and laterality (bilateral cases 75% vs. 13%, p = 0.037) (Table 4).

Table 4.
Summary of Clinical and Pathological Characteristics by BRCA2 Variant Status in Breast and Ovarian Cancer Patients.
It is worth noting that the variants rs80359380, rs587780646, rs397507422, and rs11571658 have been previously reported in the Mexican population among patients with BC. All of these previously reported variants showed no statistically significant differences in frequency compared to the population included in this study (p > 0.05), except for rs11571658. In contrast, the variants rs587782313, c.3481_3482dup, and rs80359479 have been previously reported in populations from Portugal and Brazil, respectively. Significant differences in allele frequencies were observed when compared to our study population (p < 0.05). Finally, the variants rs398122715, rs1329182873, c.9812T>C, rs775030825, rs1064795067, and rs80359219 have not been reported in other cohorts. However, we observed significant differences in their frequencies compared to those reported in the gnomAD and dbSNP databases (Table 5).

Table 5.
BRCA2 variant in women with BC and OC from Jalisco population.
3.4. Computational Analysis
3.4.1. Structural Localization of Variants in Functional Domains of BRCA2
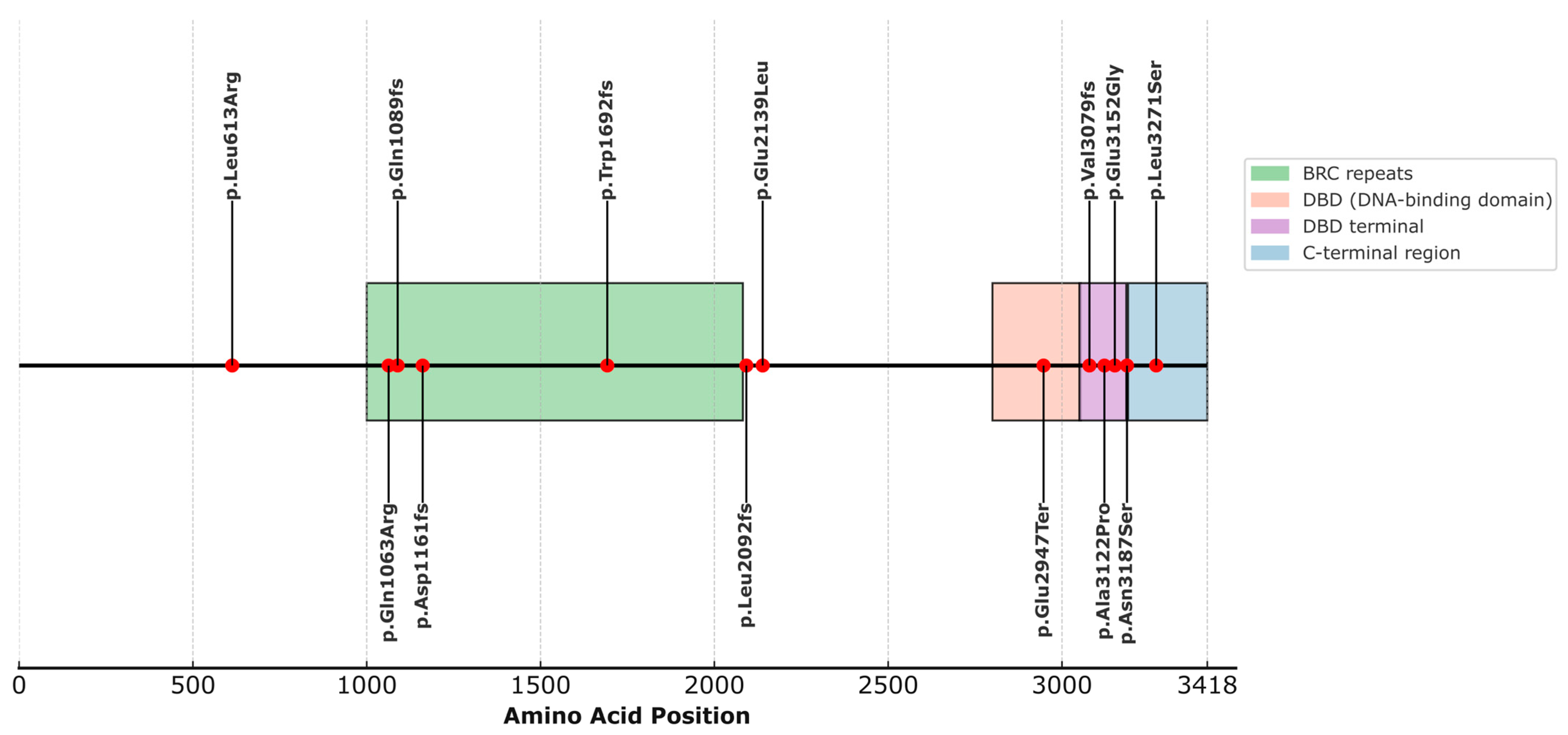
When analyzing the structural localization of the selected variants in the BRCA2 protein, we observed that most of them fall within well-defined functional domains, as indicated by the available annotation in UniProt (ID: P51587) (Figure 1, Table S2). Specifically, four variants are located within the domain known as BRC repeats (1003–2082), which play essential roles in RAD51 binding for the formation of nucleofilaments during homologous recombination repair. These include p.Gln1089fs (rs80359380), p.Gln1063Arg (rs775030825), p.Asp1161fs (c.3481_3482dup), and p.Trp1692fs (rs80359479). The variant p.Glu2947Ter (rs398122715) is located within the DNA-binding domain (DBD) (2804–3054), a crucial domain for the interaction of BRCA2 with single-stranded and double-stranded DNA, as well as facilitating binding to the regulatory protein DSS1. Variants p.Ala3122Pro (rs587782313), p.Val3079Phefs (rs397507422), and p.Glu3152Gly (c.9455A>G) are located in the terminal portion of the DBD, within a structurally recognized region known as the terminal DBD (3052–3185), with essential functions in the stability of the repair complex. The variant p.Asn3187Ser (rs1329182873) is located in the C-terminal region/DBD boundary (3190–3418) and is involved in phosphorylation processes, nuclear localization, and interaction with RAD51. On the other hand, variants p.Leu613Arg (rs587780646), p.Leu2171Ser (c.9812T>C), p.Leu2092fs (rs11571658), and p.Glu2139Leu (c.6415_6416delinsAT) are not localized within any currently described functional domain (Figure 1).
Figure 1.
Distribution of BRCA2 variants across annotated protein domains.
3.4.2. Functional Analysis via VEP Annotation
The analysis of selected BRCA2 variants using the VEP revealed a range of molecular consequences, computational predictions, and relevant clinical annotations, summarized in Table 6. A large proportion of the variants were classified as missense variants, involving single amino acid substitutions. Most of these missense changes were predicted to be deleterious by SIFT, with scores close to zero, indicating a likely detrimental effect on protein function. PolyPhen predictions showed greater variability, with some alleles classified as benign, while others were predicted to be possibly damaging or probably damaging, reflecting context-dependent structural effects. Similarly, variants like rs80359380, rs80359479, and rs11571658 were identified as frameshift variants, disrupting the reading frame and associated with clinical annotations ranging from pathogenic to uncertain significance. The clinical significance annotations from ClinVar were heterogeneous, with many variants annotated as of uncertain significance or displaying conflicting interpretations of pathogenicity. This highlights the ongoing challenges in the clinical interpretation of BRCA2 variants. For example, rs775030825 and rs587780646 share missense consequences but differ in their computational predictions and clinical classifications, ranging from uncertain significance to likely benign. Variants c.9812T>C (p.Leu3271Ser) and c.3481_3482dup (p.Asp1161fs) are not annotated in VEP; however, they can be manually classified as missense and frameshift variants, respectively.

Table 6.
Functional and Clinical Annotations for BRCA2 Variants (VEP Analysis).
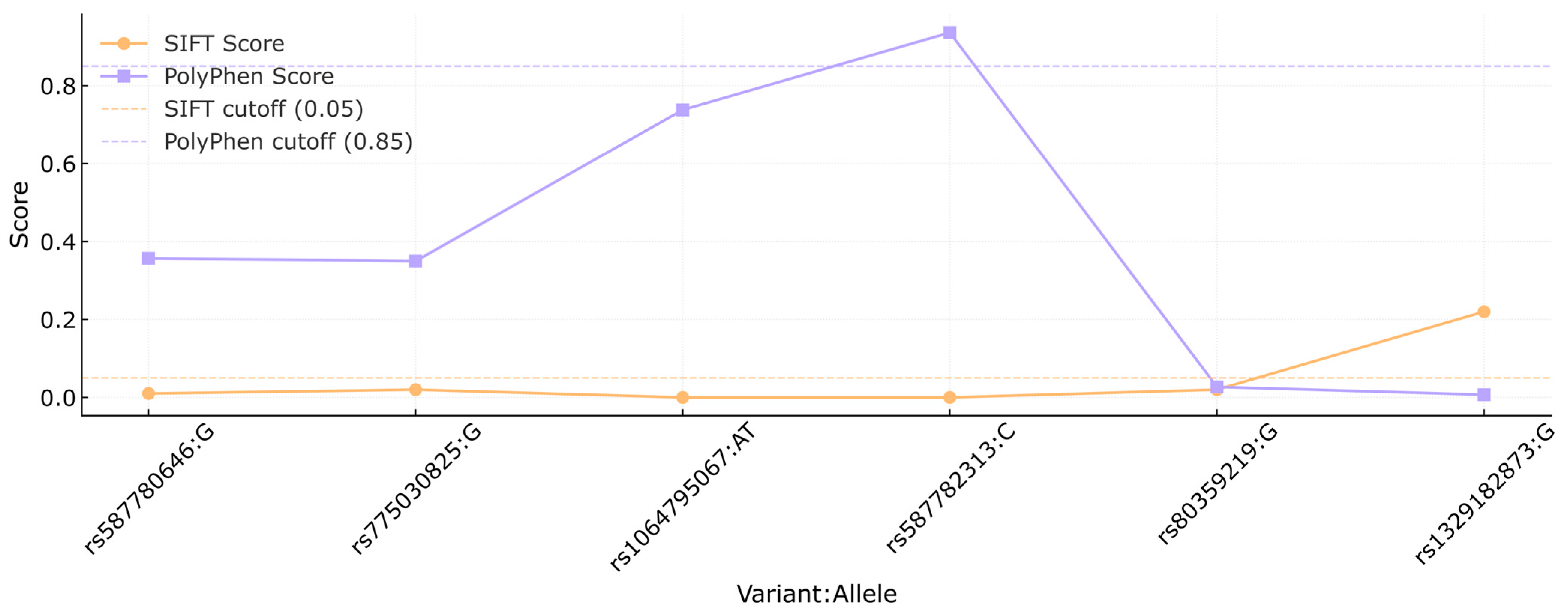
In Figure 2, the SIFT and PolyPhen scores are compared for each variant. The orange line represents the SIFT scores, while the PolyPhen scores are shown in purple. Cutoff thresholds for SIFT (0.05) and PolyPhen (0.85) are marked by the orange and purple dashed lines, respectively, to indicate the functional impact ranges for each score. Variants rs80359380, rs80359479, rs11571658, rs398122715, rs397507422, c.9812T>C, and c.3481_3482dup had no reports available in the previously mentioned platforms.
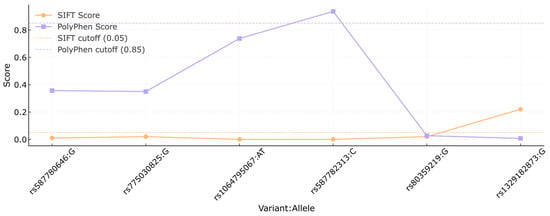
Figure 2.
Comparison of SIFT and PolyPhen scores of the BRCA2 variants. Each dot represents the score assigned to a specific variant (shown as rsID:allele) by the SIFT (orange) and PolyPhen (purple) algorithms. Dashed lines indicate the cutoff thresholds for predicting deleterious effects: 0.05 for SIFT and 0.85 for PolyPhen. This analysis was conducted to assess the potential functional impact of missense variants based on in silico predictions.
3.4.3. Structural Consequences of the Variants
The seven missense variants in the BRCA2 gene were analyzed using the HOPE tool (Table 7), which focuses on predicting structural consequences based on amino acid properties. Four variants (rs587780646, rs1329182873, rs587782313, and c.9812T>C) affect highly conserved residues, with predictions indicating alterations in charge, size, and/or hydrophobicity, potentially compromising protein architecture or its molecular interactions. In contrast, the variant rs775030825 (p.Gln1063Arg) does not affect a conserved residue, and its substitution is observed in other species, making HOPE less likely to predict a detrimental effect.

Table 7.
Structural Consequences of Missense Variants in BRCA2 Predicted by HOPE.
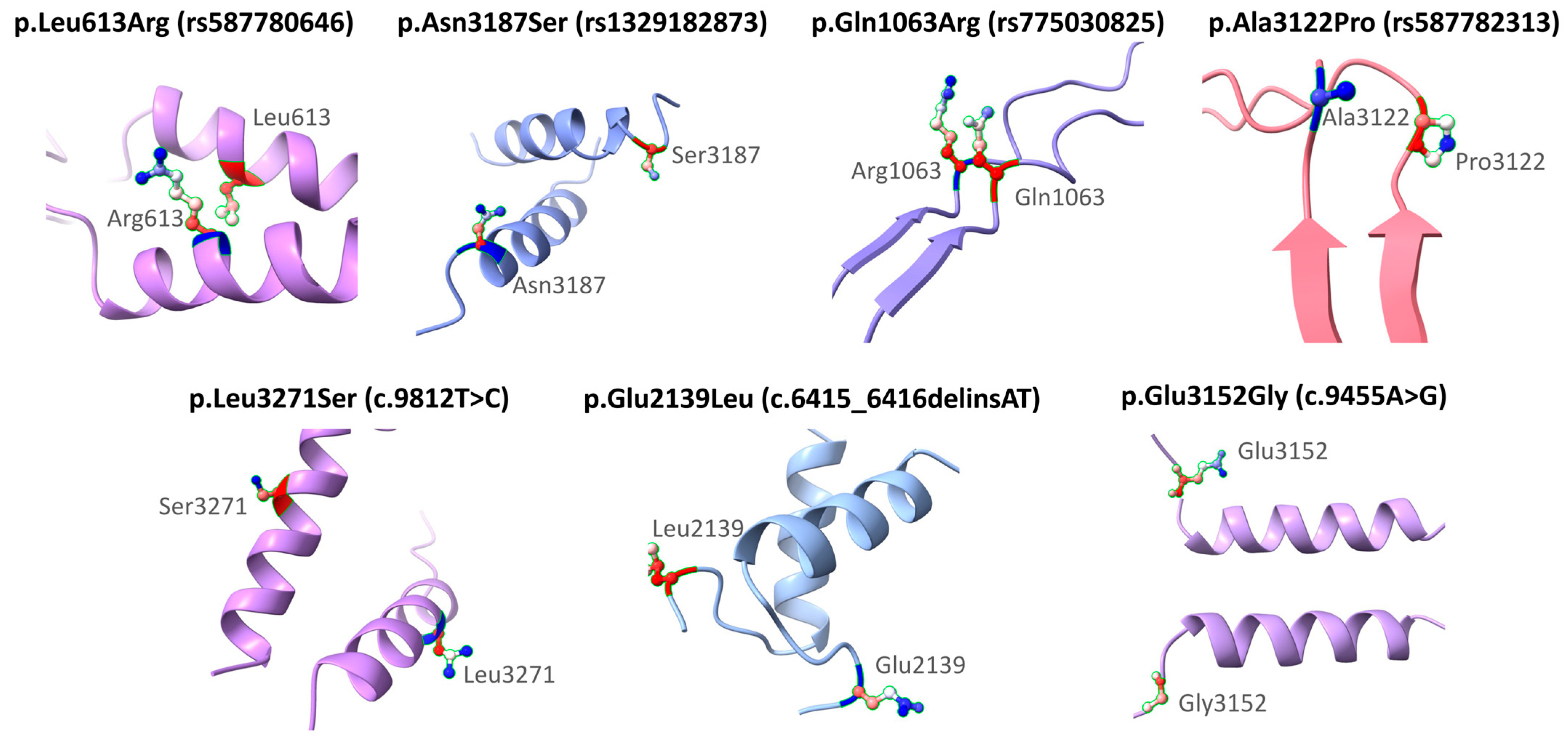
Figure 3 shows the structural modeling of seven missense variants in the BRCA2 gene using AlphaFold. The original and substituted residues are displayed within their local three-dimensional structural context. The variants included are p.Leu613Arg (rs587780646), p.Asn3187Ser (rs1329182873), p.Gln1063Arg (rs775030825), p.Ala3122Pro (rs587782313), p.Leu3271Ser (c.9812T>C), p.Glu2139Leu (c.6415_6416delinsAT), and p.Glu3152Gly (c.9455A>G). Each substitution is mapped onto the corresponding secondary structure model to visualize its position and the nature of the amino acid change. This modeling was performed solely for illustrative purposes and does not aim to predict functional or clinical consequences. The clinical interpretations of these variants are based on ACMG classification and other computational analyses described in previous sections. Frameshift variants (rs80359380, rs80359479, rs11571658, and rs397507422) were not modeled because they are predicted by VEP to generate truncated proteins (Table 6).
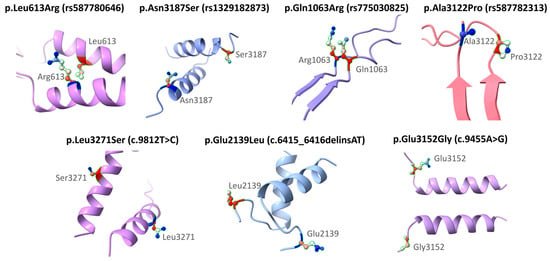
Figure 3.
Structural representation of missense variants in the BRCA2 gene.
3.5. Proposed Prioritization of BRCA2 Variants
The BRCA2 variants identified in this study were organized according to their domain localization, computational predictions, and reported clinical classification. Based on these parameters, we propose a prioritization that may help to guide subsequent functional or clinical investigations. Table 8 summarizes the integration of these data for all variants analyzed.

Table 8.
Integrated summary and suggested priority level of BRCA2 variants based on domain localization, computational predictions and clinical classification.
4. Discussion
In Mexico, as in other regions of the world, BC and OC represent a significant public health issue due to their increasing incidence and mortality rates. BC ranks first, and OC second, in terms of incidence and mortality among gynecological cancers in the country [1,37]. The frequency of BC is higher in women around 50 years of age, an age considered productive, which is also reflected in a higher mortality rate within this age group [38]. On the other hand, the average age at diagnosis for OC is estimated to be around 63 years [39]. The prevalence and impact of both types of cancer in Mexico and worldwide depend on multiple factors, including genetic predisposition, lifestyle, and access to healthcare services [5,39,40].
Regarding the sociodemographic characteristics observed in the groups analyzed in this study (Table 1), the average age of patients was similar to that reported in the literature [38,39]: 49.0 ± 10.7 years in the group with both BC and OC, 46.9 ± 13.3 years in the BC group, and 56.6 ± 11.1 years in the OC group. Menopausal status was more frequent in the OC and combined groups, while premenopausal status predominated in the BC group. Clinically (Table 2), BC was unilateral in most cases, recently diagnosed (1 to 4 years), of invasive ductal type, and notably presented a triple-negative profile in 44% of cases. This overrepresentation of TNBC, consistent with the study’s inclusion criteria emphasizing HER2-negative subtypes, should be acknowledged as a potential source of selection bias in the clinical profile of our BC cohort. OC cases were predominantly high-grade serous, also characterized by elevated Ki-67 levels, and were diagnosed at advanced stages; however, the small number of OC cases in our series limits the generalizability of these clinicopathological observations. Likewise, the subgroup of patients diagnosed with both BC and OC was very small, and the clinical patterns observed in this group should be interpreted as anecdotal.
Advances in the understanding of BC and OC have led to improvements in health policies, resulting in better quality of care and earlier detection for patients with these conditions. However, there is still a noticeable trend of self-detection of BC, often at advanced stages.
Since BC and OC are multifactorial diseases, various risk factors may influence their development [4,39,40]. Nevertheless, genetic contribution has been recognized as a key factor in the susceptibility to these tumors. It is estimated that 5 to 10% of BC and OC cases are hereditary, and that germline variants in genes such as BRCA1 and BRCA2 account for a significant proportion of these cases [41,42]. Specifically, variants in these genes increase the risk of developing BC by more than 50% and OC by up to 40% [43,44].
The crucial role of BRCA proteins in the cell cycle and DNA repair through homologous recombination highlights the importance of studying these genes [9,45]. Moreover, variants in these genes are considered dominant, which means they can increase cancer risk even in the heterozygous state. These variants have also been associated with other cancer types beyond BC and OC, including prostate, pancreatic, and other malignancies [44,46,47,48].
The detection of variants in the BRCA genes is a well-established and widely available method for families at risk of breast, ovarian, prostate, and pancreatic cancers in developed countries [49]. Thousands of variants associated with these cancer types have been described. Most of these variants lead to premature stop codons, caused by deletions, insertions, or base substitutions, resulting in the early termination of the protein. These so-called deleterious variants are often located in critical sequence regions that contain recognition sites for functional motifs of the protein [50,51]. However, the functional effect of many of these variants remains unknown. The reported frequency of BRCA2 variant carriers in the general population is approximately 1 in 800 individuals [52], with a higher prevalence observed in the Ashkenazi Jewish population [53].
The management of patients carrying pathogenic or likely pathogenic BRCA2 is individualized and may include increased surveillance, chemoprevention with tamoxifen, bilateral prophylactic oophorectomy, and/or bilateral prophylactic mastectomy [54,55]. This approach encompasses the etiology, epidemiology, pathophysiology, screening, evaluation, and management of BRCA2 variants, and highlights the importance of an interprofessional team in educating patients about cancer risk and appropriate management strategies.
In our analysis (Table 3), thirteen BRCA2 variants were identified in Mexican women with BC (n = 14) and OC (n = 4), resulting in a carrier frequency of 12.86%. The frequency was higher in the OC group (27.8%) compared to the BC group (12%). Although this difference was not statistically significant, it is consistent with studies showing that some BRCA2 variants are more frequently associated with OC [56].
When we compared the clinical and pathological characteristics of carrier patients with non-carrier patients, we observed that, in BC, BRCA2 carriers were more frequently overweight, had a more marked family history of breast, ovarian, or pancreatic cancer, and showed a lower complete response rate to chemotherapy, suggesting both a hereditary influence and possible differences in treatment sensitivity. In OC, carriers more frequently presented with bilateral cases, consistent with hereditary patterns, while the absence of a history of miscarriages should be interpreted with caution due to the small sample size. Overall, these findings highlight the clinical relevance of BRCA2 variant status in patient presentation and outcomes.
Among the identified variants, frameshift, missense, and one truncating variant were reported. Several of these, such as rs80359380, rs80359479, rs11571658, rs397507422, rs878853569, rs80359479, and rs398122715, have been previously classified as pathogenic. The variant rs587782313 was classified as likely pathogenic. Additionally, six VUS were identified, reflecting the challenges of clinical interpretation, particularly in underrepresented populations such as the Mexican population. These VUS included rs775030825, rs1064795067, rs80359219, rs1329182873, c.9812T>C, and rs587780646. VUS are typically characterized by their low population frequency, lack of direct functional evidence, location outside well-defined functional domains, and conflicting annotations in clinical databases such as ClinVar [57,58]. This highlights the need for complementary analyses to clarify their functional impact.
The in silico analysis allowed us to characterize these variants from both structural and functional perspectives (Table 6 and Table 7). For example, rs587782313, located in the terminal region of the DNA-binding domain (DBD), showed consistent predictions of deleterious effect in SIFT (0.0), probably damaging in PolyPhen (0.936), and structural alterations according to HOPE. Its position in a critical region for DSS1 interaction and DNA binding further supports its potential functional impact. Similarly, rs1064795067, although outside a canonical domain, showed an unfavorable profile across all tools, suggesting disruption of salt bridges due to the loss of negative charge. Other variants, such as rs587780646, located outside known domains, presented mixed predictions across SIFT, PolyPhen, and HOPE. Despite not falling within classic functional regions, it has been proposed that the intermediate regions of BRCA2 may have regulatory roles that are not yet fully characterized [9]. These observations suggest that current structural annotation may still be incomplete, particularly in large and complex proteins such as BRCA2.
A critical finding was that four of the six VUS identified in our research (rs775030825, rs80359219, rs1329182873, and c.9812T>C) were located within functionally relevant regions (such as the BRC repeats and the DBD), highlighting the need to re-evaluate these classifications based on functional and structural evidence. This pattern is also observed in other Latin American populations, where variants classified as rare or uncertain in European databases appear at higher frequencies, suggesting a distinct ethnic and genomic background [16,17,32,36]. For example, variants such as rs80359380 and rs11571658 were significantly more frequent in our cohort (2.1% and 1.4%, respectively) compared to their global frequencies reported in dbSNP (0.000012% and 0.000753%, respectively).
We applied a stepwise rubric that integrates variant type, domain localization, concordant in silico predictions and ClinVar annotation. This approach organizes the thirteen BRCA2 variants into high, moderate, and low priority categories to guide follow-up.
High priority included truncating changes p.Gln1089fs (rs80359380), p.Trp1692fs (rs80359479), p.Asp1161fs (rs878853569), p.Glu2947Ter (rs398122715), p.Val3079fs (rs397507422), and p.Leu2092fs (rs11571658). All are expected to abolish essential motifs, including BRC repeats or the DNA-binding domain. ClinVar reports these variants as pathogenic, frequently with expert panel review, which is entirely consistent with their high priority in our rubric. The missense p.Ala3122Pro (rs587782313) also reached high priority due to its location in the terminal DNA-binding domain and concordant deleterious predictions. ClinVar is conflicting for this variant (uncertain significance versus likely pathogenic). Our designation, therefore, diverges from current clinical consensus but is supported by the convergence of domain and computational evidence.
Moderate priority comprised p.Glu3152Gly (rs80359219), p.Glu2139Leu (rs1064795067), p.Leu613Arg (rs587780646), and p.Gln1063Arg (rs775030825). For p.Glu3152Gly, the location of the terminal DNA-binding domain is relevant, although PolyPhen is classified as benign, and ClinVar lists uncertain significance. This aligns with a moderate assignment. For p.Glu2139Leu and p.Leu613Arg, both lie outside annotated domains but show deleterious SIFT and HOPE-suggested perturbations; ClinVar provides conflicting or likely benign signals, which support an intermediate position rather than a high or low one. For p.Gln1063Arg, ClinVar is conflicting, and HOPE suggests a limited impact; however, the location within BRC repeats and a deleterious SIFT score justify a moderate level under this rubric.
Low priority included p.Asn3187Ser (rs1329182873) and p.Leu3271Ser (no rsID). These variants show predominantly benign or non-concordant predictions and lack strong domain anchoring. ClinVar lists p.Asn3187Ser as uncertain significance with no conflicts, which is consistent with a low assignment. p.Leu3271Ser lacks VEP annotation and has no consolidated ClinVar entry in our dataset; the low level reflects limited current evidence and indicates the need for data accrual rather than immediate functional testing. Nevertheless, the absence of experimental validation or orthogonal evidence (e.g., functional assays, family segregation, or tumor sequencing) limits the robustness of these prioritization levels. Integrating such data in future studies will be essential to confirm or refine the functional effect of the variants.
It is worth noting that several of the variants detected in this study have been previously reported in other populations, which supports their potential involvement in hereditary cancer. For instance, rs587782313 has been described in patients with OC in Portugal [30], while rs80359380 has been reported in both Spanish [31] and Mexican cohorts [32], and rs80359479 and c.3481_3482dup in Brazilian patients [33,34]. Likewise, variants such as rs587780646, rs397507422, and rs11571658 have been previously reported in studies conducted in the Mexican population [16,17,36], suggesting that these variants may recur in specific population subgroups.
In our analysis, most of these previously reported variants did not show significant differences in frequency compared to the present cohort, with the exception of rs80359380 and rs11571658, which were significantly different when contrasted with Spanish and Canadian studies, respectively. Conversely, variants such as rs587782313, c.3481_3482dup, and rs80359479 did not differ from other cohorts but showed significant differences when compared with gnomAD data. In addition, several variants not previously reported in other cohorts (rs398122715, rs1329182873, c.9812T>C, rs775030825, rs1064795067, and rs80359219) also displayed significant differences relative to gnomAD (Table 5). These comparisons should be interpreted with caution, as study design, sample size, and population ancestry may influence observed frequencies; however, they provide valuable context to highlight potential population-specific effects.
Regarding the six of the thirteen variants identified in this study (rs398122715, rs1329182873, c.9812T>C, rs775030825, rs1064795067, and rs80359219) that have not been previously reported in the Mexican population or other described cohorts, this study represents the first time these variants have been documented in this population, underscoring the need for ongoing evaluation of their potential clinical significance. Their low frequency in global databases such as gnomAD, along with their absence in prior regional and international studies, supports the hypothesis that these may represent rare but potentially significant events in the Mexican genomic context.
This observation becomes even more relevant considering that, among these previously unreported variants, all except rs398122715 are currently classified as VUS. These findings underscore the importance of conducting genetic studies in underrepresented populations, such as those in Latin America, to broaden our understanding of genetic variability in susceptibility genes like BRCA2. Furthermore, they highlight the need to update international databases with variants identified in diverse ethnic contexts, as this could significantly improve the clinical interpretation of variants currently classified as VUS.
When compared with independent studies in the Mexican population, our results for BRCA2 in Jalisco complement and expand the national mutational landscape. The systematic review by Alday-Montañez et al. (2024) [59] aggregated 9026 Mexican genotypes and documented 657 pathogenic variants, reporting that while BRCA1 accounts for a larger proportion of hereditary breast and ovarian cancer cases, BRCA2 harbors recurrent and regionally enriched variants, particularly in exon 11. Similarly, Catalán et al. (2019) [60] described 12 distinct BRCA2 pathogenic variants in Mexican families, several of which were not previously reported in Latin America, underscoring the ongoing discovery of BRCA2 alleles in the country. In ovarian cancer, Gallardo-Rincón et al. (2020) [61] identified germline BRCA2 variants in approximately one-third of mutation-positive patients, highlighting clinically relevant differences in outcome compared to BRCA1 carriers. Our study contributes to this national context by characterizing BRCA2 variants in a regional (Jalisco) cohort, including six not previously reported in Mexican or other populations. These findings expand the catalog of BRCA2 diversity and provide population-specific information that can aid in clinical interpretation. Nevertheless, given the consistent evidence that BRCA1 represents a larger share of the hereditary burden in Mexico, future research in Jalisco should integrate BRCA1, particularly screening for the exon 9–12 founder deletion.
Taken together, our study, which integrated clinical, population-based, and computational data, provides a more comprehensive view of the potential functional and clinical impact of BRCA2 variants in Mexican patients with breast and ovarian cancer. This strategy enabled us to address not only the frequency and distribution of the variants but also their possible biological effects by incorporating structural predictions, localization within functional domains, and associated clinical evidence. This approach is particularly relevant for VUS, whose interpretation remains a persistent challenge in oncological and genetic practice. The multidimensional contextual analysis of these variants offers a stronger foundation for generating hypotheses about their potential pathogenicity and suggests clear directions for their future reclassification.
Moreover, our findings underscore the need to characterize genetic variants in underrepresented populations, such as the Mexican population, where ancestral backgrounds, local evolutionary dynamics, and admixture profiles may give rise to unique variants or result in different allele frequencies compared to those reported in international databases. The identification of several VUS in functionally relevant regions of the BRCA2 gene, combined with their limited representation in global cohorts, reveals a significant gap in current knowledge and constrains clinical interpretation in these populations. This supports the need for comprehensive functional studies to assess the impact of these variants on protein structure, stability, and function, as well as on DNA repair mechanisms mediated by homologous recombination.
Likewise, the importance of conducting validations in larger and genetically diverse cohorts is emphasized, as such efforts would allow for more reliable associations between specific variants and phenotypic traits such as histological type, age at diagnosis, or family history of cancer. These studies would help mitigate the interpretation bias arising from the overreliance on data from European populations and improve the predictive power of genetic panels in Latin American settings.
Finally, we propose working toward the inclusion of these variants in clinical genetic screening panels tailored to the Mexican context, in both public and private healthcare systems. This would enhance early diagnosis, improve the stratification of hereditary cancer risk, guide preventive or surgical management decisions, and expand therapeutic options based on individual molecular profiles. Integrating these findings into clinical practice would contribute to the development of a more equitable, inclusive, and ethnically contextualized precision medicine, aligned with the specific needs of the Mexican population and other Latin American communities.
5. Conclusions
BRCA2 variants were identified in 12.86% of patients, with a higher prevalence in ovarian cancer. Several affected key functional domains and showed functional and structural profiles consistent with pathogenic effects. Notably, six had not been previously reported in other populations, highlighting the distinct genomic background of the Mexican population. These findings underscore the urgent need to develop population-specific genetic screening panels that incorporate locally relevant variants to enhance diagnostic accuracy and inform clinical decision-making in hereditary cancer care.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/medsci13040248/s1, Table S1: Comparison of Clinical and Pathological Characteristics by BRCA2 Variant Status in Breast and Ovarian Cancer Patients.; Table S2: Structural Localization of BRCA2 Variants within Functional Domains and Their Associated Roles.
Author Contributions
Conceptualization, P.M.G.-V., J.E.G.-O., A.F.G.-R., I.P.D.-R., S.d.C.M.-R., M.T.M.-T., M.A.R.-R., G.M.Z.-G., B.C.G.-M., R.V.-P., S.O.M.-C. and M.P.G.-A.; Data curation, J.E.G.-O., A.F.G.-R., S.d.C.M.-R., M.A.R.-R. and M.P.G.-A.; Formal analysis, P.M.G.-V., J.E.G.-O., A.F.G.-R., I.P.D.-R., S.d.C.M.-R., M.T.M.-T., L.E.F., C.d.J.T.-J., G.M.Z.-G., B.M.T.-M. and M.P.G.-A.; Investigation, P.M.G.-V., J.E.G.-O., A.F.G.-R., I.P.D.-R., S.d.C.M.-R., M.T.M.-T., M.A.R.-R., C.d.J.T.-J., G.M.Z.-G., S.O.M.-C. and M.P.G.-A.; Methodology, P.M.G.-V., J.E.G.-O., A.F.G.-R., I.P.D.-R., S.d.C.M.-R., L.E.F., G.M.Z.-G., B.M.T.-M., R.V.-P., R.G.-C., J.C.C.V. and M.P.G.-A.; Project administration, J.E.G.-O., I.P.D.-R., R.G.-C., J.C.C.V. and M.P.G.-A.; Resources, J.E.G.-O., A.F.G.-R., S.d.C.M.-R., C.d.J.T.-J. and M.P.G.-A.; Software, J.E.G.-O., A.F.G.-R., B.C.G.-M. and M.P.G.-A.; Supervision, J.E.G.-O., I.P.D.-R., L.E.F., B.C.G.-M., B.M.T.-M., R.V.-P., R.G.-C., J.C.C.V. and M.P.G.-A.; Validation, P.M.G.-V., J.E.G.-O., A.F.G.-R., M.A.R.-R. and M.P.G.-A.; Visualization, P.M.G.-V., J.E.G.-O., A.F.G.-R., M.A.R.-R., C.d.J.T.-J., G.M.Z.-G., B.C.G.-M., B.M.T.-M., R.V.-P. and M.P.G.-A.; Writing—original draft, P.M.G.-V., J.E.G.-O., A.F.G.-R., M.T.M.-T., L.E.F., G.M.Z.-G. and M.P.G.-A.; Writing—review and editing, P.M.G.-V., J.E.G.-O., A.F.G.-R., I.P.D.-R., S.d.C.M.-R., M.T.M.-T., L.E.F., M.A.R.-R., C.d.J.T.-J., B.C.G.-M., B.M.T.-M., R.V.-P., R.G.-C., J.C.C.V., S.O.M.-C. and M.P.G.-A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and Local Ethics and Research Committees (1305, CIBO, IMSS) (protocol code R-2022-1305-114 on 22 December 2022).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patient(s) to publish this paper.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Acknowledgments
The authors thank the CIBO, IMSS, and AstraZeneca for their support in the realization of this research.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Giaquinto, A.N.; Sung, H.; Newman, L.A.; Freedman, R.A.; Smith, R.A.; Star, J.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics 2024. CA Cancer J. Clin. 2024, 74, 477–495. [Google Scholar] [CrossRef]
- Li, P.-C.; Zhu, Y.-F.; Cao, W.-M.; Li, B. ER-Positive and BRCA2-Mutated Breast Cancer: A Literature Review. Eur. J. Med. Res. 2024, 29, 30. [Google Scholar] [CrossRef]
- Walker, M.; Jacobson, M.; Sobel, M. Management of Ovarian Cancer Risk in Women with BRCA1/2 Pathogenic Variants. CMAJ 2019, 191, E886–E893. [Google Scholar] [CrossRef] [PubMed]
- Kuchenbaecker, K.B.; Hopper, J.L.; Barnes, D.R.; Phillips, K.-A.; Mooij, T.M.; Roos-Blom, M.-J.; Jervis, S.; van Leeuwen, F.E.; Milne, R.L.; Andrieu, N.; et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA 2017, 317, 2402–2416. [Google Scholar] [CrossRef] [PubMed]
- Thorat, M.A.; Balasubramanian, R. Breast Cancer Prevention in High-Risk Women. Best. Pract. Res. Clin. Obstet. Gynaecol. 2020, 65, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Sopik, V.; Phelan, C.; Cybulski, C.; Narod, S.A. BRCA1 and BRCA2 Mutations and the Risk for Colorectal Cancer. Clin. Genet. 2015, 87, 411–418. [Google Scholar] [CrossRef]
- Patel, M.M.; Adrada, B.E. Hereditary Breast Cancer: BRCA Mutations and Beyond. Radiol. Clin. N. Am. 2024, 62, 627–642. [Google Scholar] [CrossRef]
- Shahid, T.; Soroka, J.; Kong, E.; Malivert, L.; McIlwraith, M.J.; Pape, T.; West, S.C.; Zhang, X. Structure and Mechanism of Action of the BRCA2 Breast Cancer Tumor Suppressor. Nat. Struct. Mol. Biol. 2014, 21, 962–968. [Google Scholar] [CrossRef]
- Andreassen, P.R.; Seo, J.; Wiek, C.; Hanenberg, H. Understanding BRCA2 Function as a Tumor Suppressor Based on Domain-Specific Activities in DNA Damage Responses. Genes 2021, 12, 1034. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, H.; Yu, Z.; Li, L.; Zhang, J.; Liang, X.; Huang, Q. Germline Variants Profiling of BRCA1 and BRCA2 in Chinese Hakka Breast and Ovarian Cancer Patients. BMC Cancer 2022, 22, 842. [Google Scholar] [CrossRef]
- Pal, T.; Mundt, E.; Richardson, M.E.; Chao, E.; Pesaran, T.; Slavin, T.P.; Couch, F.J.; Monteiro, A.N.A. Reduced Penetrance BRCA1 and BRCA2 Pathogenic Variants in Clinical Germline Genetic Testing. npj Precis. Onc. 2024, 8, 247. [Google Scholar] [CrossRef] [PubMed]
- Chenevix-Trench, G.; Milne, R.L.; Antoniou, A.C.; Couch, F.J.; Easton, D.F.; Goldgar, D.E.; CIMBA. An International Initiative to Identify Genetic Modifiers of Cancer Risk in BRCA1 and BRCA2 Mutation Carriers: The Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA). Breast Cancer Res. 2007, 9, 104. [Google Scholar] [CrossRef] [PubMed]
- Parsons, M.T.; Tudini, E.; Li, H.; Hahnen, E.; Wappenschmidt, B.; Feliubadaló, L.; Aalfs, C.M.; Agata, S.; Aittomäki, K.; Alducci, E.; et al. Large Scale Multifactorial Likelihood Quantitative Analysis of BRCA1 and BRCA2 Variants: An ENIGMA Resource to Support Clinical Variant Classification. Hum. Mutat. 2019, 40, 1557–1578. [Google Scholar] [CrossRef]
- Jimenez-Sainz, J.; Mathew, J.; Moore, G.; Lahiri, S.; Garbarino, J.; Eder, J.P.; Rothenberg, E.; Jensen, R.B. BRCA2 BRC Missense Variants Disrupt RAD51-Dependent DNA Repair. eLife 2022, 11, e79183. [Google Scholar] [CrossRef] [PubMed]
- Stella, S.; Vitale, S.R.; Massimino, M.; Martorana, F.; Tornabene, I.; Tomarchio, C.; Drago, M.; Pavone, G.; Gorgone, C.; Barone, C.; et al. In Silico Prediction of BRCA1 and BRCA2 Variants with Conflicting Clinical Interpretation in a Cohort of Breast Cancer Patients. Genes 2024, 15, 943. [Google Scholar] [CrossRef]
- Ruiz-Flores, P.; Sinilnikova, O.M.; Badzioch, M.; Calderon-Garcidueñas, A.L.; Chopin, S.; Fabrice, O.; González-Guerrero, J.F.; Szabo, C.; Lenoir, G.; Goldgar, D.E.; et al. BRCA1 and BRCA2 Mutation Analysis of Early-Onset and Familial Breast Cancer Cases in Mexico. Human. Mutat. 2002, 20, 474–475. [Google Scholar] [CrossRef]
- Villarreal-Garza, C.; Alvarez-Gómez, R.M.; Pérez-Plasencia, C.; Herrera, L.A.; Herzog, J.; Castillo, D.; Mohar, A.; Castro, C.; Gallardo, L.N.; Gallardo, D.; et al. Significant Clinical Impact of Recurrent BRCA1 and BRCA2 Mutations in Mexico. Cancer 2015, 121, 372–378. [Google Scholar] [CrossRef]
- Villarreal-Garza, C.; Weitzel, J.N.; Llacuachaqui, M.; Sifuentes, E.; Magallanes-Hoyos, M.C.; Gallardo, L.; Alvarez-Gómez, R.M.; Herzog, J.; Castillo, D.; Royer, R.; et al. The Prevalence of BRCA1 and BRCA2 Mutations among Young Mexican Women with Triple-Negative Breast Cancer. Breast Cancer Res. Treat. 2015, 150, 389–394. [Google Scholar] [CrossRef]
- Silva-Zolezzi, I.; Hidalgo-Miranda, A.; Estrada-Gil, J.; Fernandez-Lopez, J.C.; Uribe-Figueroa, L.; Contreras, A.; Balam-Ortiz, E.; del Bosque-Plata, L.; Velazquez-Fernandez, D.; Lara, C.; et al. Analysis of Genomic Diversity in Mexican Mestizo Populations to Develop Genomic Medicine in Mexico. Proc. Natl. Acad. Sci. USA 2009, 106, 8611–8616. [Google Scholar] [CrossRef]
- Bravo-Acevedo, A.; Escobedo-Ruíz, A.; Barquera, R.; Clayton, S.; García-Arias, V.E.; Arrieta-Bolaños, E.; Goné-Vázquez, I.; Hernández-Zaragoza, D.I.; Arellano-Prado, F.P.; Rodríguez-López, M.E.; et al. Genetic Diversity of HLA System in Six Populations from Jalisco, Mexico: Guadalajara City, Tlajomulco, Tlaquepaque, Tonalá, Zapopan and Rural Jalisco. Hum. Immunol. 2020, 81, 502–505. [Google Scholar] [CrossRef]
- Herrera, I.M.R.; Castañeda, M.E.G.; Sandoval, C.G.; Sevilla, A.R.; López, R.M.V.; Fernández, A.M.; Pastrana, J.d.D.R.; Águila, S.L.; Camet, Z. Access and Availability of Diagnostic Services: The Experiences of Women with Breast Cancer in Jalisco, Mexico. JOJPH 2019, 4, 555650. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Adusumalli, S.; Leng, S.N.; Sikic, M.; Ng, P.C. SIFT Missense Predictions for Genomes. Nat. Protoc. 2016, 11, 1–9. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef]
- Venselaar, H.; te Beek, T.A.; Kuipers, R.K.; Hekkelman, M.L.; Vriend, G. Protein Structure Analysis of Mutations Causing Inheritable Diseases. An e-Science Approach with Life Scientist Friendly Interfaces. BMC Bioinform. 2010, 11, 548. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for Structure Building and Analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Peixoto, A.; Pinto, P.; Guerra, J.; Pinheiro, M.; Santos, C.; Pinto, C.; Santos, R.; Escudeiro, C.; Bartosch, C.; Canário, R.; et al. Tumor Testing for Somatic and Germline BRCA1/BRCA2 Variants in Ovarian Cancer Patients in the Context of Strong Founder Effects. Front. Oncol. 2020, 10, 1318. [Google Scholar] [CrossRef]
- Rosado-Jiménez, L.; Mestre-Terkemani, Y.; García-Aliaga, Á.; Marín-Vera, M.; Macías-Cerrolaza, J.A.; Sarabia-Meseguer, M.D.; García-Hernández, M.R.; Zafra-Poves, M.; Sánchez-Henarejos, P.; de la Peña, F.A.; et al. Recurrent Genetic Variants and Prioritization of Variants of Uncertain Clinical Significance Associated with Hereditary Breast and Ovarian Cancer in Families from the Region of Murcia. Adv. Lab. Med./Av. En Med. Lab. 2023, 4, 279–287. [Google Scholar] [CrossRef]
- Chavarri-Guerra, Y.; Villarreal-Garza, C.; Ferrigno, A.S.; Mohar, A.; Aguilar, D.; Alvarez-Gomez, R.M.; Gallardo-Alvarado, L.; del Toro-Valero, A.; Quintero-Beulo, G.; Gutierrez-Delgado, F.; et al. Germline Pathogenic Variants in Mexican Patients with Hereditary Triple-Negative Breast Cancer. Salud Pública Méx. 2022, 64, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Duarte, C.A.B.; dos Santos, C.A.; de Oliveira, C.D.D.; Spautz, C.C.; Sumita, L.M.; Nakatani, S.M. Hereditary Breast Cancer Next----generation Sequencing Panel Evaluation in the South Region of Brazil: A Novel BRCA2 Candidate Pathogenic Variant Is Reported. Mol. Genet. Genom. Med. 2024, 12, e2504. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, R.L.; Leite, A.C.R.; Barbalho, D.M.; Assad, D.X.; Barroso, R.; Polidorio, N.; dos Anjos, C.H.; de Miranda, A.D.; de Mendonça Ferreira, A.C.S.; Fernandes, G.d.S.; et al. Germline Molecular Data in Hereditary Breast Cancer in Brazil: Lessons from a Large Single-Center Analysis. PLoS ONE 2021, 16, e0247363. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Royer, R.; Li, S.; McLaughlin, J.R.; Rosen, B.; Risch, H.A.; Fan, I.; Bradley, L.; Shaw, P.A.; Narod, S.A. Frequencies of BRCA1 and BRCA2 Mutations among 1,342 Unselected Patients with Invasive Ovarian Cancer. Gynecol. Oncol. 2011, 121, 353–357. [Google Scholar] [CrossRef]
- Fernández-Lopez, J.C.; Romero-Córdoba, S.; Rebollar-Vega, R.; Alfaro-Ruiz, L.A.; Jiménez-Morales, S.; Beltrán-Anaya, F.; Arellano-Llamas, R.; Cedro-Tanda, A.; Rios-Romero, M.; Ramirez-Florencio, M.; et al. Population and Breast Cancer Patients’ Analysis Reveals the Diversity of Genomic Variation of the BRCA Genes in the Mexican Population. Hum. Genom. 2019, 13, 3. [Google Scholar] [CrossRef]
- Cancer (IARC). T.I.A. for R. on Global Cancer Observatory. Available online: https://gco.iarc.fr/ (accessed on 8 April 2025).
- Hendrick, R.E.; Monticciolo, D.L.; Biggs, K.W.; Malak, S.F. Age Distributions of Breast Cancer Diagnosis and Mortality by Race and Ethnicity in US Women. Cancer 2021, 127, 4384–4392. [Google Scholar] [CrossRef]
- Ali, A.T.; Al-Ani, O.; Al-Ani, F. Epidemiology and Risk Factors for Ovarian Cancer. Menopause Rev. 2023, 22, 93–104. [Google Scholar] [CrossRef]
- Ferris, J.S.; Morgan, D.A.; Tseng, A.S.; Terry, M.B.; Ottman, R.; Hur, C.; Wright, J.D.; Genkinger, J.M. Risk Factors for Developing Both Primary Breast and Primary Ovarian Cancer: A Systematic Review. Crit. Rev. Oncol. Hematol. 2023, 190, 104081. [Google Scholar] [CrossRef]
- Yoshida, R. Hereditary Breast and Ovarian Cancer (HBOC): Review of Its Molecular Characteristics, Screening, Treatment, and Prognosis. Breast Cancer 2021, 28, 1167–1180. [Google Scholar] [CrossRef]
- Marmolejo, D.H.; Wong, M.Y.Z.; Bajalica-Lagercrantz, S.; Tischkowitz, M.; Balmaña, J.; Patócs, A.B.; Chappuis, P.; Colas, C.; Genuardi, M.; Haanpää, M.; et al. Overview of Hereditary Breast and Ovarian Cancer (HBOC) Guidelines across Europe. Eur. J. Med. Genet. 2021, 64, 104350. [Google Scholar] [CrossRef]
- Si, S.; Li, J.; Tewara, M.A.; Li, H.; Liu, X.; Li, Y.; Chen, X.; Liu, C.; Yuan, T.; Li, W.; et al. Identifying Causality, Genetic Correlation, Priority and Pathways of Large-Scale Complex Exposures of Breast and Ovarian Cancers. Br. J. Cancer 2021, 125, 1570–1581. [Google Scholar] [CrossRef] [PubMed]
- Samuel, D.; Diaz-Barbe, A.; Pinto, A.; Schlumbrecht, M.; George, S. Hereditary Ovarian Carcinoma: Cancer Pathogenesis Looking beyond BRCA1 and BRCA2. Cells 2022, 11, 539. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Engebrecht, J. BRCA1 and BRCA2 Tumor Suppressor Function in Meiosis. Front. Cell Dev. Biol. 2021, 9, 668309. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: Important Differences with Common Interests. Nat. Rev. Cancer 2012, 12, 372. [Google Scholar] [CrossRef]
- Messina, C.; Cattrini, C.; Soldato, D.; Vallome, G.; Caffo, O.; Castro, E.; Olmos, D.; Boccardo, F.; Zanardi, E. BRCA Mutations in Prostate Cancer: Prognostic and Predictive Implications. J. Oncol. 2020, 2020, 4986365. [Google Scholar] [CrossRef]
- Wong, W.; Raufi, A.G.; Safyan, R.A.; Bates, S.E.; Manji, G.A. BRCA Mutations in Pancreas Cancer: Spectrum, Current Management, Challenges and Future Prospects. Cancer Manag. Res. 2020, 12, 2731–2742. [Google Scholar] [CrossRef]
- Pujol, P.; Barberis, M.; Beer, P.; Friedman, E.; Piulats, J.M.; Capoluongo, E.D.; Foncillas, J.G.; Ray-Coquard, I.; Penault-Llorca, F.; Foulkes, W.D.; et al. Clinical Practice Guidelines for BRCA1 and BRCA2 Genetic Testing. Eur. J. Cancer 2021, 146, 30–47. [Google Scholar] [CrossRef]
- Boussios, S.; Rassy, E.; Moschetta, M.; Ghose, A.; Adeleke, S.; Sanchez, E.; Sheriff, M.; Chargari, C.; Pavlidis, N. BRCA Mutations in Ovarian and Prostate Cancer: Bench to Bedside. Cancers 2022, 14, 3888. [Google Scholar] [CrossRef]
- Sekine, M.; Nishino, K.; Enomoto, T. Differences in Ovarian and Other Cancers Risks by Population and BRCA Mutation Location. Genes. 2021, 12, 1050. [Google Scholar] [CrossRef]
- Salazar Saez, R.; Zorrilla, M.; Sánchez, R.; Cebollero, A.; Manrique, I.; Martín, A.; de Ávila, L.; Lacalle-Emborujo, A.; Martin-Rodriguez, S.; Bernardo-González, I.; et al. Molecular Analysis of BRCA1 and BRCA2 Genes in La Rioja (Spain): Five New Variants. Hered. Cancer Clin. Pract. 2024, 22, 22. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, S.; Chen-Shtoyerman, R.; Levi, Z.; Shkedi-Rafid, S.; Zuckerman, S.; Bernstein-Molho, R.; Levi, G.R.; Shachar, S.S.; Flugelman, A.; Libman, V.; et al. Common Founder BRCA2 Pathogenic Variants and Breast Cancer Characteristics in Ethiopian Jews. Breast Cancer Res. Treat. 2022, 193, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Rachmat, R.; Enyioma, S.; Ghose, A.; Revythis, A.; Boussios, S. BRCA Mutations in Prostate Cancer: Assessment, Implications and Treatment Considerations. Int. J. Mol. Sci. 2021, 22, 12628. [Google Scholar] [CrossRef] [PubMed]
- Arun, B.; Couch, F.J.; Abraham, J.; Tung, N.; Fasching, P.A. BRCA-Mutated Breast Cancer: The Unmet Need, Challenges and Therapeutic Benefits of Genetic Testing. Br. J. Cancer 2024, 131, 1400–1414. [Google Scholar] [CrossRef]
- Fanale, D.; Fiorino, A.; Incorvaia, L.; Dimino, A.; Filorizzo, C.; Bono, M.; Cancelliere, D.; Calò, V.; Brando, C.; Corsini, L.R.; et al. Prevalence and Spectrum of Germline BRCA1 and BRCA2 Variants of Uncertain Significance in Breast/Ovarian Cancer: Mysterious Signals From the Genome. Front. Oncol. 2021, 11, 682445. [Google Scholar] [CrossRef]
- Sullivan, K.E. The Scary World of Variants of Uncertain Significance (VUS): A Hitchhiker’s Guide to Interpretation. J. Allergy Clin. Immunol. 2021, 147, 492–494. [Google Scholar] [CrossRef]
- Lucci-Cordisco, E.; Amenta, S.; Panfili, A.; del Valle, J.; Capellá, G.; Pineda, M.; Genuardi, M. Variants of Uncertain Significance (VUS) in Cancer Predisposing Genes: What Are We Learning from Multigene Panels? Eur. J. Med. Genet. 2022, 65, 104400. [Google Scholar] [CrossRef]
- Alday-Montañez, F.D.; Dickens-Terrazas, D.; Mejia-Carmona, G.E.; Robles-Escajeda, E.; Kirken, R.A.; Bencomo-Alvarez, A.E.; Carrasco-Urrutia, V.J.; Lobo-Galo, N.; Plenge-Tellechea, L.F.; Diaz-Sanchez, Á.G.; et al. Pathogenic Variants in BRCA1 and BRCA2 Genes Associated with Female Breast and Ovarian Cancer in the Mexican Population. J. Med. Life 2025, 18, 38–47. [Google Scholar] [CrossRef]
- Millan Catalan, O.; Campos-Parra, A.D.; Vázquez-Romo, R.; Cantú de León, D.; Jacobo-Herrera, N.; Morales-González, F.; López-Camarillo, C.; Rodríguez-Dorantes, M.; López-Urrutia, E.; Pérez-Plasencia, C. A Multi-Center Study of BRCA1 and BRCA2 Germline Mutations in Mexican-Mestizo Breast Cancer Families Reveals Mutations Unreported in Latin American Population. Cancers 2019, 11, 1246. [Google Scholar] [CrossRef]
- Gallardo-Rincón, D.; Álvarez-Gómez, R.M.; Montes-Servín, E.; Toledo-Leyva, A.; Montes-Servín, E.; Michel-Tello, D.; Alamilla-García, G.; Bahena-González, A.; Hernández-Nava, E.; Fragoso-Ontiveros, V.; et al. Clinical Evaluation of BRCA1/2 Mutation in Mexican Ovarian Cancer Patients. Transl. Oncol. 2020, 13, 212–220. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).