Study of Expression of MST3 in Myeloid Leukaemia
Abstract
1. Introduction
2. Materials and Methods
2.1. Counting of White Blood Corpuscles and Red Blood Corpuscles in Patients with Leukaemia
2.2. Expression of MST3 Using Real-Time Polymerase Chain Reaction
2.3. Statistical Analysis
2.4. Correlation Analysis
3. Results
3.1. White Blood Corpuscle and Red Blood Corpuscle Counting
3.2. Expression of MST3
| Samples | Expression of MST3 | Expression (Fold Change) |
|---|---|---|
| Control Sample | 1.33 | |
| Sample 1 | 8.81 | 5.62 |
| Sample 2 | 10.97 | 7.25 |
| Sample 3 | 11.23 | 7.44 |
| Sample 4 | 10.1 | 6.59 |
| Sample 5 | 5.2 | 2.90 |
| Sample 6 | 11.04 | 7.3 |
| Sample 7 | 11.11 | 7.35 |
| Sample 8 | 12.83 | 8.65 |
| Sample 9 | 10.9 | 7.19 |
| Sample 10 | 12.78 | 8.60 |
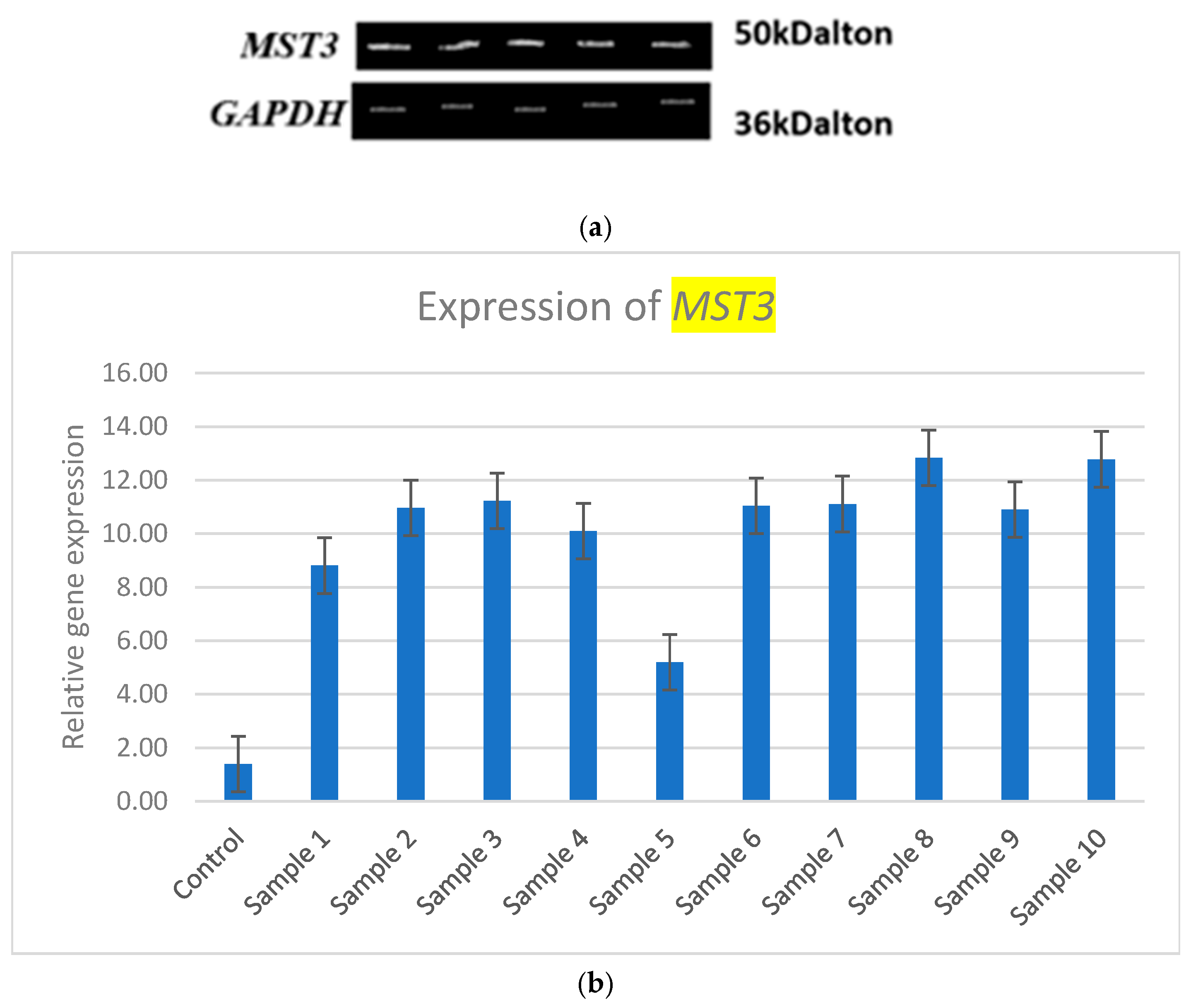
3.3. Statistical Analysis of MST3 Expression
3.4. Correlation Analysis with MST3
| Genes | r | p-Value |
|---|---|---|
| KRAS | 0.41 | 2.1 × 10−8 |
| NRAS | 0.48 | 4 × 10−11 |
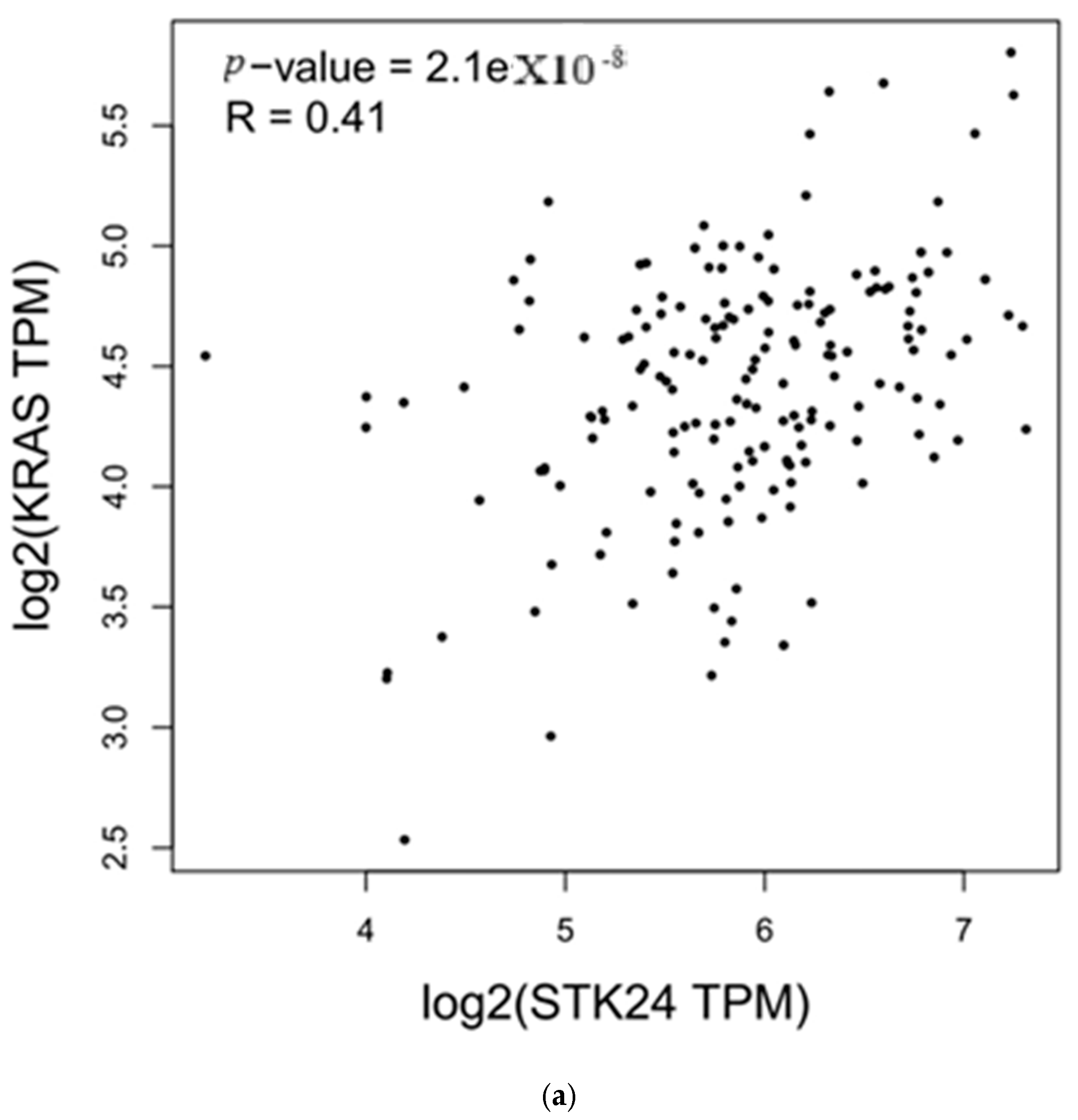
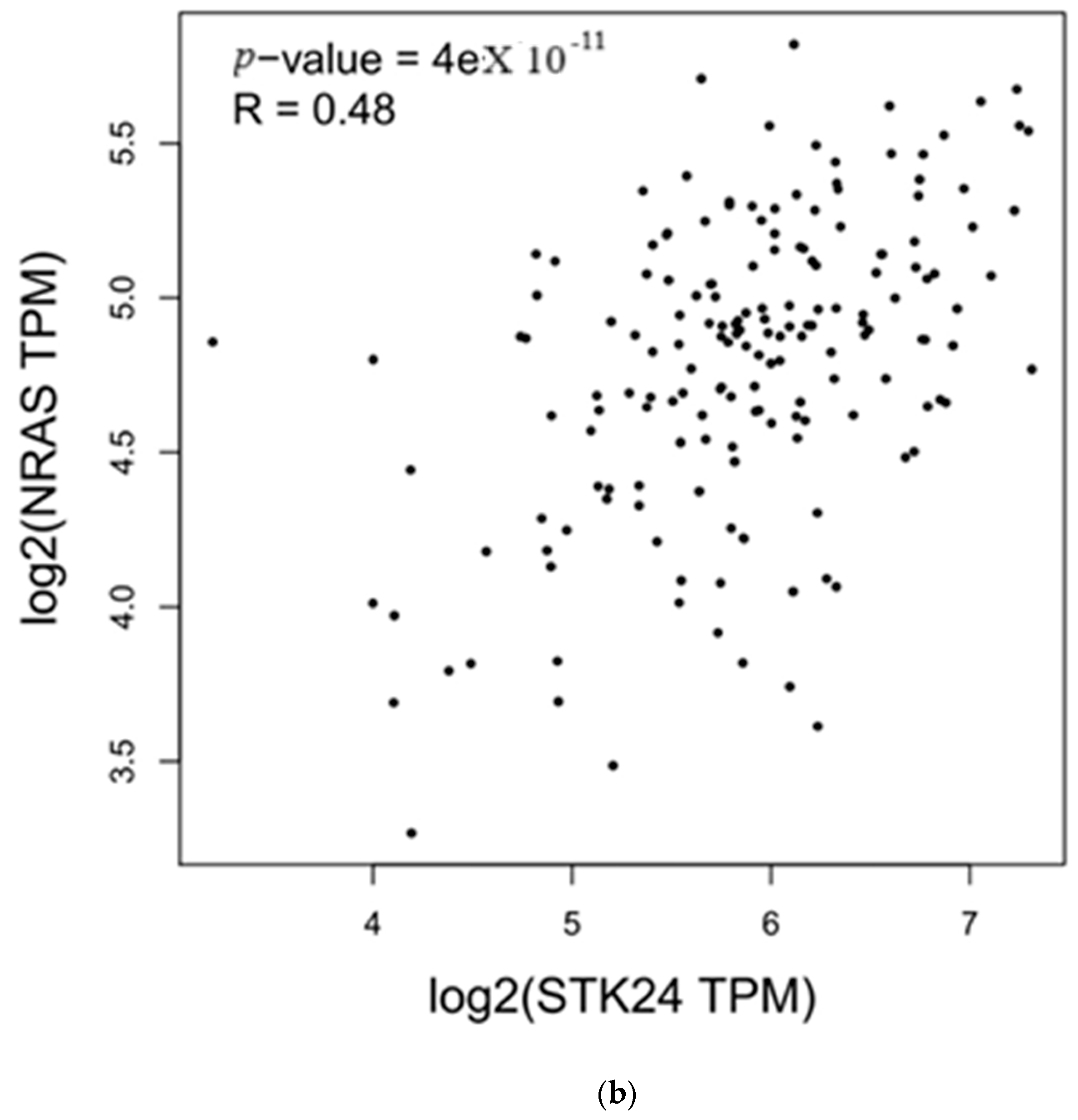
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ML | Myeloid leukaemia |
| ELN | European Leukaemia Net |
| GCK | Germinal centre kinase |
| MAPK | Mitogen-activated protein kinase |
| RBC | Red blood corpuscle |
| WBC | White blood corpuscle |
| RT-PCR | Real-time polymerase chain reaction |
| ROCK2 | Rho-associated coiled-coil kinase 2 |
| Ste20 | Sterile 20 kinase |
References
- Wachter, F.; Pikman, Y. Pathophysiology of Acute Myeloid Leukemia. Acta Haematol. 2024, 147, 229–246. [Google Scholar] [PubMed]
- Chen, Q.; Hong, Y.; Chen, W.; Lin, F.; Zeng, J.; Huang, Y.; Zhang, L.; Yao, J.; Xu, B. Prognostic implications of cGAS and STING gene expression in acute myeloid leukemia. Exp. Biol. Med. 2024, 249, 10108. [Google Scholar] [CrossRef] [PubMed]
- Stefańczyk, S.A.; Hayn, C.; Heitmann, J.; Jung, S.; Zekri, L.; Märklin, M. Expression and Prognostic Value of a Novel B7-H3 (CD276) Antibody in Acute Myeloid Leukemia. Cancers 2024, 16, 2455. [Google Scholar] [CrossRef]
- Pelcovits, A.; Niroula, R. Acute Myeloid Leukemia: A Review. Rhode Isl. Med. J. 2020, 103, 38–40. [Google Scholar]
- Shimony, S.; Stahl, M.; Stone, R.M. Acute myeloid leukemia: 2023 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2023, 98, 502–526. [Google Scholar]
- Kayser, S.; Levis, M.J. The clinical impact of the molecular landscape of acute myeloid leukemia. Haematologica 2023, 108, 308–320. [Google Scholar] [CrossRef]
- Yin, H.; Shi, Z.; Jiao, S.; Chen, C.; Wang, W.; Greene, M.I.; Zhou, Z. Germinal center kinases in immune regulation. Cell. Mol. Immunol. 2012, 9, 439–445. [Google Scholar] [CrossRef]
- Ceccarelli, D.F.; Laister, R.C.; Mulligan, V.K.; Kean, M.J.; Goudreault, M.; Scott, I.C.; Derry, W.B.; Chakrabartty, A.; Gingras, A.C.; Sicheri, F. CCM3/PDCD10 Heterodimerizes with Germinal Center Kinase III (GCKIII) Proteins Using a Mechanism Analogous to CCM3 Homodimerization. J. Biol. Chem. 2011, 286, 25056–25064. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Wang, K.; Xue, D.; Huang, Y.; Tan, Z.; Chen, Y. MST3 Promotes Progression of LUAD and Modulates the Immune Microenvironment. Mediat. Inflamm. 2023, 2023, 8646088. [Google Scholar] [CrossRef]
- Thakral, D.; Singh, V.K.; Gupta, R.; Jha, N.; Khan, A.; Kaur, G.; Rai, S.; Kumar, V.; Supriya, M.; Bakhshi, S.; et al. Integrated single-cell transcriptome analysis of CD34+ enriched leukemic stem cells revealed intra-and inter-patient transcriptional heterogeneity in pediatric acute myeloid leukemia. Ann. Hematol. 2023, 102, 73–87. [Google Scholar] [CrossRef]
- Stirewalt, D.L.; Meshinchi, S.; Kopecky, K.J.; Fan, W.; Pogosova-Agadjanyan, E.L.; Engel, J.H.; Cronk, M.R.; Dorcy, K.S.; McQuary, A.R.; Hockenbery, D.; et al. Identification of genes with abnormal expression changes in acute myeloid leukemia. Genes Chromosomes Cancer 2008, 47, 8–20. [Google Scholar] [PubMed]
- Bullinger, L.; Döhner, K.; Bair, E.; Fröhling, S.; Schlenk, R.F.; Tibshirani, R.; Döhner, H.; Pollack, J.R. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N. Engl. J. Med. 2004, 350, 1605–1616. [Google Scholar] [PubMed]
- Li, C.; Xin, H.; Hao, J.; Miao, Y. Decreasing of serine/threonine kinase 39 has tumour inhibiting effects on acute myeloid leukaemia by impacting the PI3K/AKT and Wnt/β-catenin signalling cascades. Toxicol. Appl. Pharmacol. 2024, 489, 116982. [Google Scholar] [PubMed]
- Yan, M.; Luo, X.; Han, H.; Qiu, J.; Ye, Q.; Zhang, L.; Wang, Y. ROCK2 increases drug resistance in acute myeloid leukemia via metabolic reprogramming and MAPK/PI3K/AKT signaling. Int. Immunopharmacol. 2024, 140, 112897. [Google Scholar]
- Wang, N.; Jiang, Y.; Li, M.; Wang, H.; Pan, J.; Tang, Y.; Xie, S.; Xu, Y.; Li, X.; Zhou, X.; et al. Protein Kinase MST3 Promotes Tumor Immune Evasion via the AKT-PD-L1 Axis. Adv. Sci. 2024, 11, 2304342. [Google Scholar]
- Huang, N.; Lin, W.; Shi, X.; Tao, T. MST3 expression is modulated by DNA copy number/methylation in lung adenocarcinoma and predicts poor survival. Future Oncol. 2018, 14, 2253–2263. [Google Scholar]
- Lai, S.; Wang, D.; Sun, W.; Cao, X. Serine/threonine-protein kinase MST3 induces tumorigenesis by regulating the STAT3/VEGFA signaling pathway. J. Biol. Chem. 2023, 299, 102961. [Google Scholar]
- Chen, Y.L.; Wang, C.Y.; Fang, J.H.; Hsu, H.P. Serine/threonine-protein kinase 24 is an inhibitor of gastric cancer metastasis through suppressing CDH1 gene and enhancing stemness. Am. J. Cancer Res. 2021, 11, 4277–4293. [Google Scholar]
- Suryani, E.; Wiharto, W.; Polvonov, N. Identification and counting white blood cells and red blood cells using image processing case study of leukemia. arXiv 2015, arXiv:1511.04934. [Google Scholar]
- Greenwood, M.J.; Seftel, M.D.; Richardson, C.; Barbaric, D.; Barnett, M.J.; Bruyere, H.; Forrest, D.L.; Horsman, D.E.; Smith, C.; Song, K.; et al. Leukocyte count as a predictor of death during remission induction in acute myeloid leukemia. Leuk. Lymphoma 2006, 47, 1245–1252. [Google Scholar]
- Imad, I.; Ghaloub, A.N.; Ozaslan, M.; Alwan, A.F. Comparison Between WBCs, RBCs and HGHB Levels for Iraqi AML Patients Pre and Post 3-and 7-Treatment Within Four Age Groups Segmented Based on Growth Levels. Indian J. Public Health Res. Dev. 2019, 10, 2643–2647. [Google Scholar]
- Blumberg, N.; Heal, J.M.; Gettings, K.F. WBC reduction of RBC transfusions is associated with a decreased incidence of RBC alloimmunization. Transfusion 2003, 43, 945–952. [Google Scholar] [PubMed]
- Chen, W.; Wang, H.; Hu, J. Incidence of myelosuppression in AML is higher compared with that in ALL. Mol. Clin. Oncol. 2024, 21, 95. [Google Scholar] [PubMed]
- Găman, M.A.; Dugăeşescu, M.; Popescu, D.C. Applications of Artificial Intelligence in Acute Promyelocytic Leukemia: An Avenue of Opportunities? A Systematic Review. J. Clin. Med. 2025, 14, 1670. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, S.; Vallati, M.; Gatta, R. Artificial Intelligence-based management of adult chronic myeloid leukemia: Where are we and where are we going? Cancers 2024, 16, 848. [Google Scholar] [CrossRef]
- Salah, H.T.; Muhsen, I.N.; Salama, M.E.; Owaidah, T.; Hashmi, S.K. Machine learning applications in the diagnosis of leukemia: Current trends and future directions. Int. J. Lab. Hematol. 2019, 41, 717–725. [Google Scholar]
- Radakovich, N.; Cortese, M.; Nazha, A. Acute myeloid leukemia and artificial intelligence, algorithms and new scores. Best Pract. Res. Clin. Haematol. 2020, 33, 101192. [Google Scholar]
- Alhajahjeh, A.; Nazha, A. Unlocking the potential of artificial intelligence in acute myeloid leukemia and myelodysplastic syndromes. Curr. Hematol. Malig. Rep. 2024, 19, 9–17. [Google Scholar]
| Samples | WBC (cells/μL) | RBC (cells/μL) |
|---|---|---|
| Control | 5000 | 4,692,000 |
| ML samples | 18,000 | 3,930,000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arthi, B.; Sujatha, K.; Gopal, S.; Balamuralikrishnan, B.; Arun, M.; Manikantan, P.; Sampathkumar, P.; Anand, A.V. Study of Expression of MST3 in Myeloid Leukaemia. Med. Sci. 2025, 13, 33. https://doi.org/10.3390/medsci13020033
Arthi B, Sujatha K, Gopal S, Balamuralikrishnan B, Arun M, Manikantan P, Sampathkumar P, Anand AV. Study of Expression of MST3 in Myeloid Leukaemia. Medical Sciences. 2025; 13(2):33. https://doi.org/10.3390/medsci13020033
Chicago/Turabian StyleArthi, Boro, Krishnaswamy Sujatha, Sridhar Gopal, Balasubramanian Balamuralikrishnan, Meyyazhagan Arun, Pappuswamy Manikantan, Palanisamy Sampathkumar, and Arumugam Vijaya Anand. 2025. "Study of Expression of MST3 in Myeloid Leukaemia" Medical Sciences 13, no. 2: 33. https://doi.org/10.3390/medsci13020033
APA StyleArthi, B., Sujatha, K., Gopal, S., Balamuralikrishnan, B., Arun, M., Manikantan, P., Sampathkumar, P., & Anand, A. V. (2025). Study of Expression of MST3 in Myeloid Leukaemia. Medical Sciences, 13(2), 33. https://doi.org/10.3390/medsci13020033







