Artificial Intelligence as a Diagnostic Tool in Non-Invasive Imaging in the Assessment of Coronary Artery Disease
Abstract
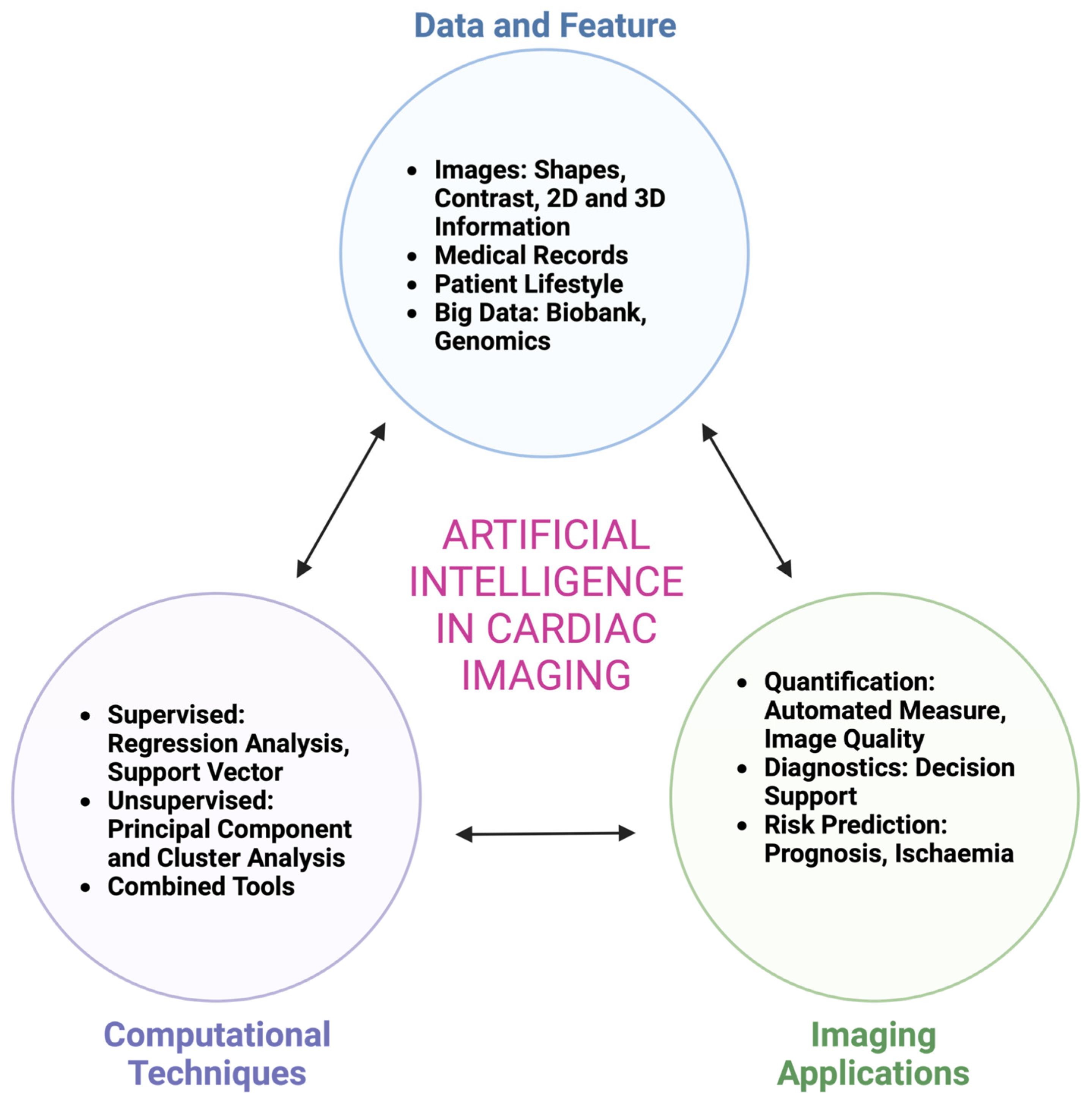
1. Introduction
2. Echocardiography
3. Coronary CT
3.1. Role of Coronary Computed Tomographic Angiography (CCTA) in Prognostication
3.2. Fat Attenuation Index
3.3. Plaque Feature and Fractional Flow Reserve-CT (CT-FFR)
4. Myocardial Perfusion Imaging
5. Cardiac MRI
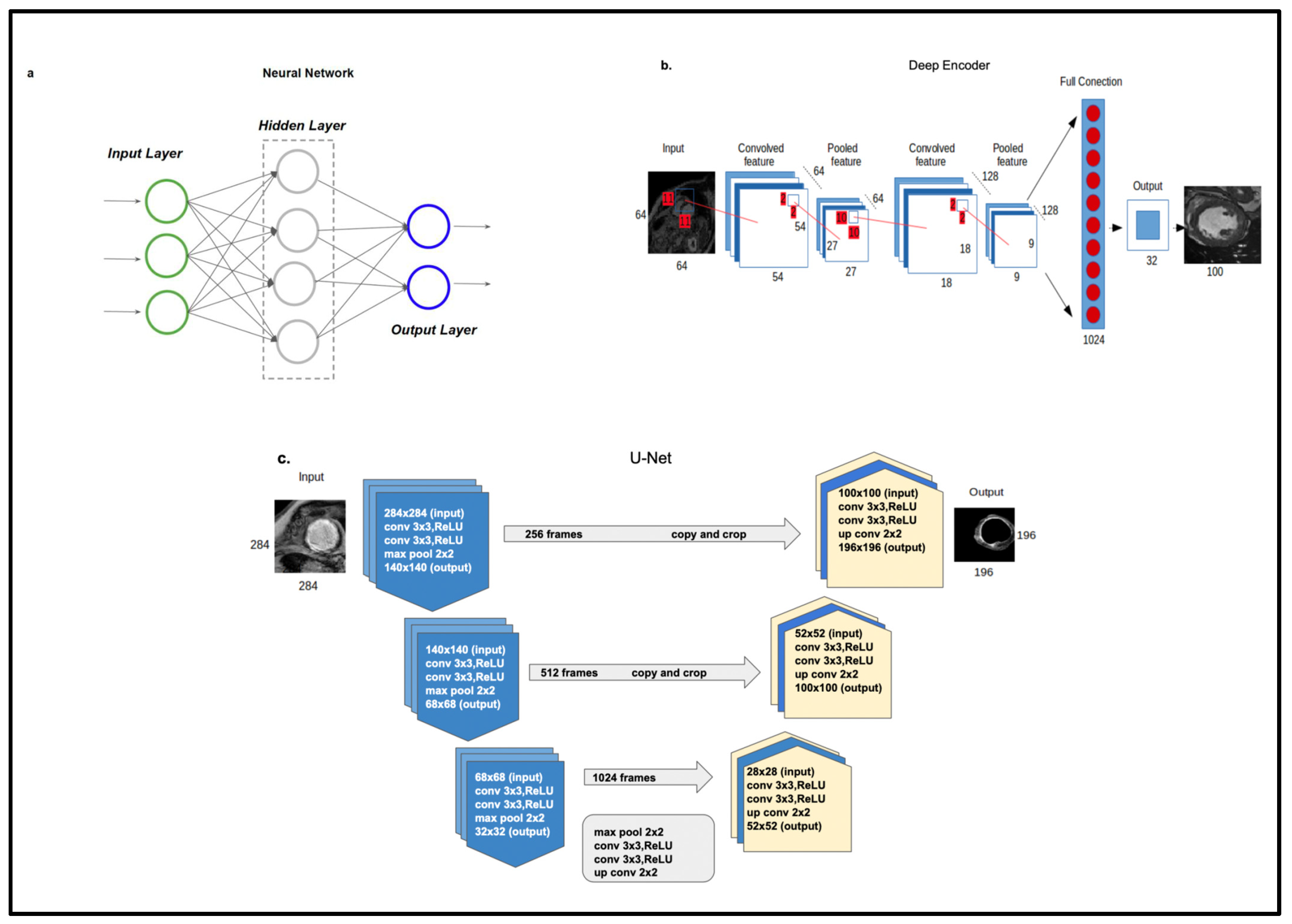
5.1. Supervised and Unsupervised Techniques for Automatic Cardiac Scar Segmentation
5.1.1. Cardiac Scar Tissue Physiology
5.1.2. Automatic Segmentation
5.1.3. Review of Scar Automatic Segmentation Techniques
5.2. Cardiac MRI in Prognostication
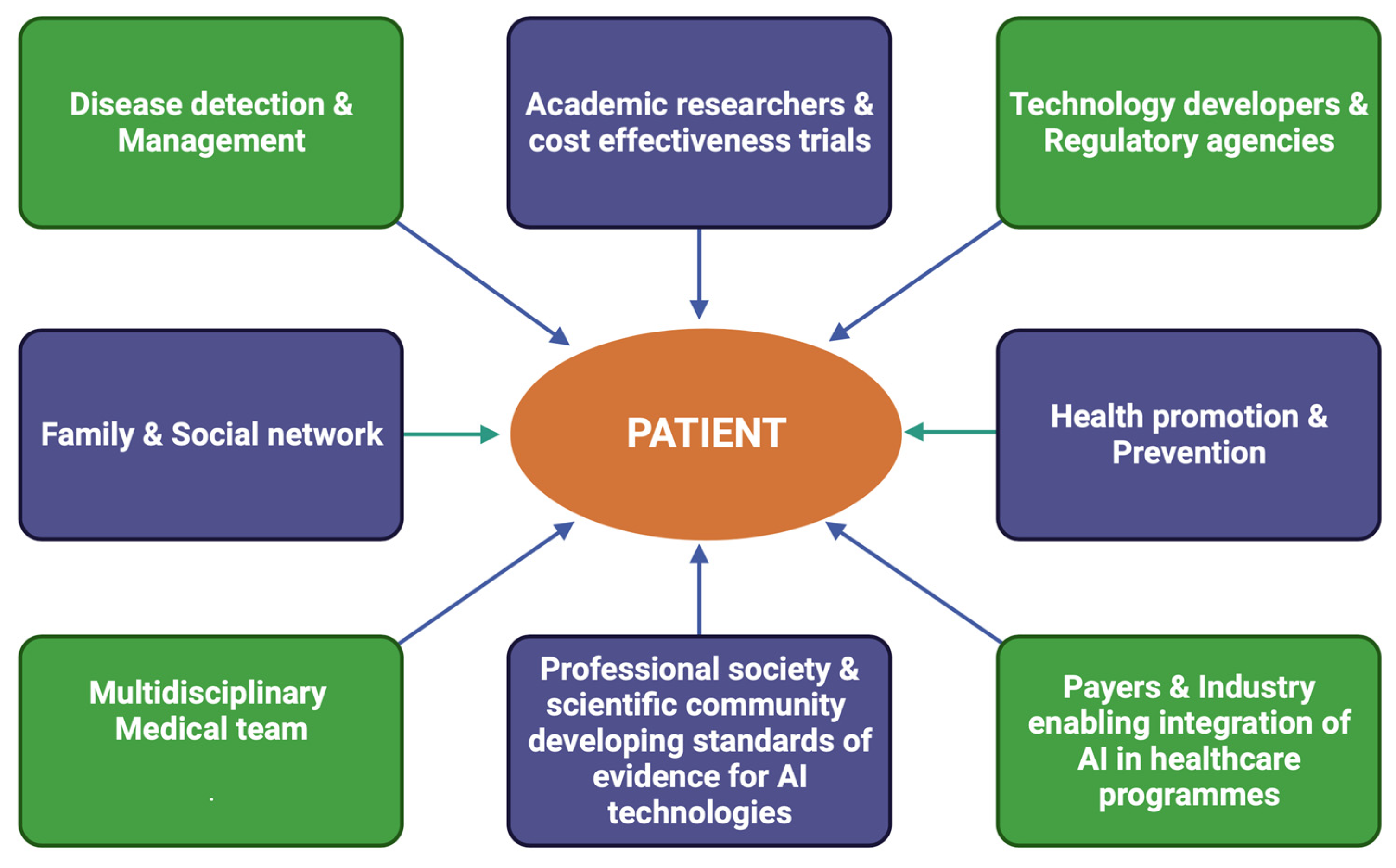
6. Clinical Context of AI Application as a Digital Diagnostic Tool
7. What Is the Future of AI in Cardiovascular Imaging?
8. Limitations and Challenges
9. Cost-Benefit Implications
10. Conclusions
11. Future Perspectives: Highlights
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fenech, M.; Buston, O. AI in Cardiac Imaging: A UK-Based Perspective on Addressing the Ethical, Social, and Political Challenges. Front. Cardiovasc. Med. 2020, 7, 54. [Google Scholar] [CrossRef]
- Royal College of Physicians. Artificial Intelligence (AI) in Health. 2018. Available online: https://www.rcplondon.ac.uk/projects/outputs/artificial-intelligence-ai-health (accessed on 3 June 2021).
- News. Eric Topol Pens Book on Artificial Intelligence in Medicine. Scripps Research. 2019. Available online: https://www.scripps.edu/news-and-events/press-room/2019/20190312-topol-deep-medicine.html (accessed on 3 June 2021).
- Turing, A.M. Computing machinery and intelligence. Mind 1950, LIX, 433–460. [Google Scholar] [CrossRef]
- McCarthy, J.; Minsky, M.L.; Rochester, N.; Shannon, C.E. A Proposal for the Dartmouth Summer Research Project on Artificial Intelligence, August 31, 1955. AI Mag. 2006, 27, 12. [Google Scholar]
- Dey, D.; Slomka, P.J.; Leeson, P.; Comaniciu, D.; Shrestha, S.; Sengupta, P.P.; Marwick, T. Artificial Intelligence in Cardiovascular Imaging: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 2019, 73, 1317–1335. [Google Scholar] [CrossRef]
- Brownlee, J. A Tour of Machine Learning Algorithm. Machine Learning Mastery. 2020. Available online: https://machinelearningmastery.com/a-tour-of-machine-learning-algorithms/ (accessed on 12 December 2022).
- Goldstein, B.A.; Navar, A.M.; Carter, R.E. Moving beyond regression techniques in cardiovascular risk prediction: Applying machine learning to address analytic challenges. Eur. Heart J. 2017, 38, 1805–1814. [Google Scholar] [CrossRef]
- Leiner, T.; Rueckert, D.; Suinesiaputra, A.; Baeßler, B.; Nezafat, R.; Išgum, I.; Young, A. Machine learning in cardiovascular magnetic resonance: Basic concepts and applications. J. Cardiovasc. Magn. Reson. 2019, 21, 61. [Google Scholar] [CrossRef]
- Heidmann, L. Unsupervised Machine Learning: Use Cases & Examples. 2020. Available online: https://blog.dataiku.com/unsupervised-machine-learning-use-cases-examples (accessed on 12 December 2022).
- IBM Cloud Education. Neural Networks. 2020. Available online: https://www.ibm.com/uk-en/cloud/learn/neural-networks (accessed on 23 October 2022).
- Yan, Y.; Zhang, J.W.; Zang, G.Y.; Pu, J. The primary use of artificial intelligence in cardiovascular diseases: What kind of potential role does artificial intelligence play in future medicine? J. Geriatr. Cardiol. 2019, 16, 585–591. [Google Scholar]
- British Heart Foundation. BHF Statistics Factsheet-UK. 2020. Available online: https://www.bhf.org.uk/what-we-do/our-research/heart-statistic (accessed on 6 January 2022).
- Maragna, R.; Giacari, C.M.; Guglielmo, M.; Baggiano, A.; Fusini, L.; Guaricci, A.I.; Rossi, A.; Rabbat, M.; Pontone, G. Artificial Intelligence Based Multimodality Imaging: A New Frontier in Coronary Artery Disease Management. Front. Cardiovasc. Med. 2021, 8, 736223. [Google Scholar] [CrossRef]
- Madani, A.; Arnaout, R.; Mofrad, M.; Arnaout, R. Fast and accurate view classification of echocardiograms using deep learning. npj Digit. Med. 2018, 1, 6. [Google Scholar] [CrossRef]
- Knackstedt, C.; Bekkers, S.C.; Schummers, G.; Schreckenberg, M.; Muraru, D.; Badano, L.P.; Franke, A.; Bavishi, C.; Omar, A.M.; Sengupta, P.P. Fully automated versus standard tracking of left ventricular ejection fraction and longitudinal strain: The FAST-EFs multicenter study. J. Am. Coll. Cardiol. 2015, 66, 1456–1466. [Google Scholar] [CrossRef]
- Lancaster, M.C.; Salem Omar, A.M.; Narula, S.; Kulkarni, H.; Narula, J.; Sengupta, P.P. Phenotypic Clustering of Left Ventricular Diastolic Function Parameters: Patterns and Prognostic Relevance. JACC Cardiovasc. Imaging 2019, 12 Pt 1, 1149–1161. [Google Scholar] [CrossRef]
- Pandey, A.; Kagiyama, N.; Yanamala, N.; Segar, M.W.; Cho, J.S.; Tokodi, M.; Sengupta, P.P. Deep-Learning Models for the Echocardiographic Assessment of Diastolic Dysfunction. JACC Cardiovasc. Imaging 2021, 14, 1887–1900. [Google Scholar] [CrossRef]
- Upton, R.; Mumith, A.; Beqiri, A.; Parker, A.; Hawkes, W.; Gao, S.P.; Porumb, M.; Sarwar, R.; Marques, P.; Markham, D.; et al. Automated Echocardiographic Detection of Severe Coronary Artery Disease Using Artificial Intelligence. JACC Cardiovasc. Imaging 2022, 15, 715–727. [Google Scholar] [CrossRef]
- Motwani, M.; Dey, D.; Berman, D.S.; Germano, G.; Achenbach, S.; Al-Mallah, M.H.; Andreini, D.; Budoff, M.J.; Cademartiri, F.; Callister, T.Q.; et al. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: A 5-year multicentre prospective registry analysis. Eur. Heart J. 2017, 38, 500–507. [Google Scholar] [CrossRef]
- Nakanishi, R.; Dey, D.; Commandeur, F.; Slomka, P.; Betancur, J.; Gransar, H.; Dailing, C.; Osawa, K.; Berman, D.; Budoff, M. Machine Learning in Predicting Coronary Heart Disease and Cardiovascular Disease Events: Results from the Multi-Ethnic Study of Atherosclerosis (Mesa). J. Am. Coll. Cardiol. 2018, 71, A1483. [Google Scholar] [CrossRef]
- Lin, A.; Kolossváry, M.; Motwani, M.; Išgum, I.; Maurovich-Horvat, P.; Slomka, P.J.; Dey, D. Artificial Intelligence in Cardiovascular Imaging for Risk Stratification in Coronary Artery Disease. Radiol. Cardiothorac. Imaging 2021, 3, e200512. [Google Scholar] [CrossRef]
- Klüner, L.V.; Oikonomou, E.K.; Antoniades, C. Assessing Cardiovascular Risk by Using the Fat Attenuation Index in Coronary CT Angiography. Radiol. Cardiothorac. Imaging 2021, 3, e200563. [Google Scholar] [CrossRef]
- Margaritis, M.; Antonopoulos, A.S.; Digby, J.; Lee, R.; Reilly, S.; Coutinho, P.S.; Shirodaria, C.; Sayeed, R.; Petrou, M.; De Silva, R.; et al. Interactions between vascular wall and perivascular adipose tissue reveal novel roles for adiponectin in the regulation of endothelial nitric oxide synthase function in human vessels. Circulation 2013, 127, 2209–2221. [Google Scholar] [CrossRef]
- Antoniades, C.; Kotanidis, C.P.; Berman, D.S. State-of-the-art review article. Atherosclerosis affecting fat: What can we learn by imaging perivascular adipose tissue? J. Cardiovasc. Comput. Tomogr. 2019, 13, 288–296. [Google Scholar] [CrossRef]
- Antonopoulos, A.S.; Sanna, F.; Sabharwal, N.; Thomas, S.; Oikonomou, E.K.; Herdman, L.; Margaritis, M.; Shirodaria, C.; Kampoli, A.-M.; Akoumianakis, I.; et al. Detecting human coronary inflammation by imaging perivascular fat. Sci. Transl. Med. 2017, 9, eaal2658. [Google Scholar] [CrossRef]
- Oikonomou, E.K.; West, H.W.; Antoniades, C. Cardiac Computed Tomography: Assessment of Coronary Inflammation and Other Plaque Features. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 2207–2219. [Google Scholar] [CrossRef]
- Oikonomou, E.K.; Antoniades, C. Detection of Coronary Inflammation Using CT: The CRISP-CT Study, American College of Cardiology. 2019. Available online: https://www.acc.org/latest-in-cardiology/articles/2019/07/29/08/34/detection-of-coronary-inflammation-using-ct (accessed on 16 February 2023).
- Oikonomou, E.K.; Marwan, M.; Desai, M.Y.; Mancio, J.; Alashi, A.; Hutt Centeno, E.; Thomas, S.; Herdman, L.; Kotadinis, C.; Thomas, K.; et al. Non-invasive detection of coronary inflammation using computed tomography and prediction of residual cardiovascular risk (the CRISP CT study): A post-hoc analysis of prospective outcome data. Lancet 2018, 392, 929–939. [Google Scholar] [CrossRef]
- Graby, J.; Khavandi, A.; Thompson, D.; Downie, P.; Antoniades, C.; Rodrigues, J.C.L. CT coronary angiography-guided cardiovascular risk screening in asymptomatic patients: Is it time? Clin. Radiol. 2021, 76, 801–811. [Google Scholar] [CrossRef]
- Oikonomou, E.K.; Williams, M.C.; Kotanidis, C.P.; Desai, M.Y.; Marwan, M.; Antonopoulos, A.S.; Thomas, K.; Thomas, S.; Akoumianakis, I.; Fan, L.; et al. A novel machine learning-derived radiotranscriptomic signature of perivascular fat improves cardiac risk prediction using coronary CT angiography. Eur. Heart J. 2019, 40, 3529–3543. [Google Scholar] [CrossRef]
- SCOT-HEART Investigators; Newby, D.E.; Adamson, P.D.; Berry, C.; Boon, N.A.; Dweck, M.R.; Flather, M.; Forbes, J.; Hunter, A.; Lewis, S.; et al. Coronary CT Angiography and 5-Year Risk of Myocardial Infarction. N. Engl. J. Med. 2018, 379, 924–933. [Google Scholar]
- Motoyama, S.; Ito, H.; Sarai, M.; Kondo, T.; Kawai, H.; Nagahara, Y.; Harigaya, H.; Kan, S.; Anno, H.; Takahashi, H.; et al. Plaque Characterization by Coronary Computed Tomography Angiography and the Likelihood of Acute Coronary Events in Mid-Term Follow-Up. J. Am. Coll. Cardiol. 2015, 66, 337–346. [Google Scholar] [CrossRef]
- Narula, J.; Nakano, M.; Virmani, R.; Kolodgie, F.D.; Petersen, R.; Newcomb, R.; Malik, S.; Fuster, V.; Finn, A.V. Histopathologic characteristics of atherosclerotic coronary disease and implications of the findings for the invasive and noninvasive detection of vulnerable plaques. J. Am. Coll. Cardiol. 2013, 61, 1041–1051. [Google Scholar] [CrossRef]
- Motoyama, S.; Kondo, T.; Anno, H.; Sugiura, A.; Ito, Y.; Mori, K.; Ishii, J.; Sato, T.; Inoue, K.; Sarai, M.; et al. Atherosclerotic plaque characterization by 0.5-mm-slice multislice computed tomographic imaging. Circ. J. 2007, 71, 363–366. [Google Scholar] [CrossRef]
- Al’Aref, S.J.; Singh, G.; Choi, J.W.; Xu, Z.; Maliakal, G.; van Rosendael, A.R.; Lee, B.C.; Fatima, Z.; Andreini, D.; Bax, J.J.; et al. A Boosted Ensemble Algorithm for Determination of Plaque Stability in High-Risk Patients on Coronary CTA. JACC Cardiovasc. Imaging 2020, 13, 2162–2173. [Google Scholar] [CrossRef]
- Lee, J.M.; Choi, G.; Koo, B.K.; Hwang, D.; Park, J.; Zhang, J.; Kim, K.J.; Tong, Y.; Kim, H.J.; Grady, L.; et al. Identification of High-Risk Plaques Destined to Cause Acute Coronary Syndrome Using Coronary Computed Tomographic Angiography and Computational Fluid Dynamics. JACC Cardiovasc. Imaging 2019, 12, 1032–1043. [Google Scholar] [CrossRef]
- Dey, D.; Gaur, S.; Ovrehus, K.A.S.; Slomka, P.J.; Betancur, J.; Goeller, M.; Hell, M.M.; Gransar, H.; Berman, D.S.; Achenbach, S.; et al. Integrated prediction of lesion-specific ischaemia from quantitative coronary CT angiography using machine learning: A multicentre study. Eur. Radiol. 2018, 28, 2655–2664. [Google Scholar] [CrossRef]
- Nagumo, S.; Collet, C.; Norgaard, B.L.; Otake, H.; Ko, B.; Koo, B.K.L.; Ko, B.; Koo, B.K.; Leipsic, J.; Andreini, D.; et al. Rationale and design of the precise percutaneous coronary intervention plan (P3) study: Prospective evaluation of a virtual computed tomography-based percutaneous intervention planner. Clin. Cardiol. 2021, 44, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Sonck, J.; Nagumo, S.; Norgaard, B.L.; Otake, H.; Ko, B.; Zhang, J.; Mizukami, T.; Maeng, M.; Andreini, D.; Takahashi, Y.; et al. Clinical Validation of a Virtual Planner for Coronary Interventions Based on Coronary CT Angiography. JACC Cardiovasc. Imaging 2022, 15, 1242–1255. [Google Scholar] [CrossRef]
- Slomka, P.J.; Xu, Y.; Berman, D.S.; Germano, G. Quantitative Analysis of Perfusion Studies: Strengths and Pitfalls. J. Nucl. Cardiol. 2012, 19, 338–346. [Google Scholar] [CrossRef]
- Betancur, J.; Commandeur, F.; Motlagh, M.; Sharir, T.; Einstein, A.J.; Bokhari, S.; Fish, M.B.; Ruddy, T.D.; Kaufmann, P.; Sinusas, A.J.; et al. Deep Learning for Prediction of Obstructive Disease from Fast Myocardial Perfusion SPECT: A Multicenter Study. JACC Cardiovasc. Imaging 2018, 11, 1654–1663. [Google Scholar] [CrossRef]
- Arsanjani, R.; Xu, Y.; Dey, D.; Vahistha, V.; Shalev, A.; Nakanishi, R.; Hayes, S.; Fish, M.; Berman, D.; Germano, G.; et al. Improved accuracy of myocardial perfusion SPECT for detection of coronary artery disease by machine learning in a large population. J. Nucl. Cardiol. 2013, 20, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Arsanjani, R.; Dey, D.; Shalev, A.; Khachatryan, T.; Hayes, S.; Fish, M.; Berman, D.; Germano, G.; Slomka, P.J. Improved Accuracy of Myocardial Perfusion Spect for Prediction of Revascularization by Machine Learning in a Large Population. J. Am. Coll. Cardiol. 2014, 63, A1229. [Google Scholar] [CrossRef]
- De Luna, A.B.; Coumel, P.; Leclercq, J.F. Ambulatory sudden cardiac death: Mechanisms of production of fatal arrhythmia on the basis of data from 157 cases. Am. Heart J. 1989, 117, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Olshausen, K.V.; Witt, T.; Pop, T.; Treese, N.; Bethge, K.-P.; Meyer, J. Sudden cardiac death while wearing a Holter monitor. Am. J. Cardiol. 1991, 67, 381–386. [Google Scholar] [CrossRef]
- Martin, R.; Maury, P.; Bisceglia, C.; Wong, T.; Estner, H.; Meyer, C.; Dallet, C.; Martin, C.A.; Shi, R.; Takigawa, M.; et al. Characteristics of Scar-Related Ventricular Tachycardia Circuits Using Ultra-High-Density Mapping. Circ. Arrhythm. Electrophysiol. 2018, 11, e006569. [Google Scholar] [CrossRef]
- Perry, D.; Morris, A.; Burgon, N.; McGann, C.; Macleod, R.; Cates, J. Automatic classification of scar tissue in late gadolinium enhancement cardiac MRI for the assessment of left-atrial wall injury after radiofrequency ablation. Proc. SPIE-Int. Soc. Opt. Eng. 2012, 8315, 406–414. [Google Scholar]
- Pashakhanloo, F.; Herzka, D.A.; Halperin, H.; McVeigh, E.R.; Trayanova, N.A. Role of 3-Dimensional Architecture of Scar and Surviving Tissue in Ventricular Tachycardia. Circ. Arrhythm. Electrophysiol. 2018, 11, e006131. [Google Scholar] [CrossRef]
- Mamalakis, M.; Garg, P.; Nelson, T.; Lee, J.; Swift, A.J.; Wild, J.M.; Clayton, R.H. Automatic development of 3D anatomical models of border zone and core scar regions in the left ventricle. Comput. Med. Imaging Graph. 2023, 103, 102152. [Google Scholar] [CrossRef]
- Tao, Q.; Milles, J.; Zeppenfeld, K.; Lamb, H.J.; Bax, J.J.; Reiber, J.H.C.; Van der Geest, R.J. Automated segmentation of myocardial scar in late enhancement MRI using combined intensity and spatial information. Magn. Reson. Med. 2010, 64, 586–594. [Google Scholar] [CrossRef]
- Top, A.; Hamarneh, G.; Abugharbieh, R. Active Learning for Interactive 3D Image Segmentation BT—Medical Image Computing and Computer-Assisted Intervention—MICCAI 2011; Fichtinger, G., Martel, A., Peters, T., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 603–610. [Google Scholar]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef]
- Detsky, J.S.; Paul, G.; Dick, A.J.; Wright, G.A. Reproducible Classification of Infarct Heterogeneity Using Fuzzy Clustering on Multicontrast Delayed Enhancement Magnetic Resonance Images. IEEE Trans. Med. Imaging 2009, 28, 1606–1614. [Google Scholar] [CrossRef]
- Karim, R.; Arujuna, A.; Housden, R.J.; Gill, J.; Cliffe, H.; Matharu, K.; Gill, J.; Rindaldi, C.A.; O’Neill, M.; Rueckert, D.; et al. A method to standardize quantification of left atrial scar from delayed-enhancement MR images. IEEE J. Transl. Eng. Health Med. 2014, 2, 1–15. [Google Scholar] [CrossRef]
- Karim, R.; Housden, R.J.; Balasubramaniam, M.; Chen, Z.; Perry, D.; Uddin, A.; Al-Beyatti, Y.; Palkhi, E.; Acheampong, P.; Obom, S.; et al. Evaluation of current algorithms for segmentation of scar tissue from late Gadolinium enhancement cardiovascular magnetic resonance of the left atrium: An open-access grand challenge. J. Cardiovasc. Magn. Reson. 2013, 15, 105. [Google Scholar] [CrossRef]
- Oakes, R.S.; Badger, T.J.; Kholmovski, E.G.; Akoum, N.; Burgon, N.S.; Fish, E.N.; Blauer, J.J.; Rao, S.N.; DiBella, E.V.; Segerson, N.M.; et al. Detection and quantification of left atrial structural remodeling with delayed-enhancement magnetic resonance imaging in patients with atrial fibrillation. Circulation 2009, 119, 1758–1767. [Google Scholar] [CrossRef]
- Lu, Y.; Yang, Y.; Connelly, K.A.; Wright, G.A.; Radau, P.E. Automated quantification of myocardial infarction using graph cuts on contrast delayed enhanced magnetic resonance images. Quant. Imaging Med. Surg. 2012, 2, 81. [Google Scholar]
- Avendi, M.R.; Kheradvar, A.; Jafarkhani, H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med. Image Anal. 2016, 30, 108–119. [Google Scholar] [CrossRef]
- Zabihollahy, F.; White, J.A.; Ukwatta, E. Myocardial scar segmentation from magnetic resonance images using convolutional neural network. In Proceedings of the SPIE, Houston, TX, USA, 12–15 February 2018. [Google Scholar]
- Kolipaka, A.; Chatzimavroudis, G.P.; White, R.D.; O’Donnell, T.P.; Setser, R.M. Segmentation of non-viable myocardium in delayed enhancement magnetic resonance images. Int. J. Cardiovasc. Imaging 2005, 21, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Ukwatta, E.; Arevalo, H.; Li, K.; Yuan, J.; Qiu, W.; Malamas, P.; Wu, K.C.; Trayanova, N.A.; Vadakkumpadan, F. Myocardial Infarct Segmentation from Magnetic Resonance Images for Personalized Modeling of Cardiac Electrophysiology. IEEE Trans. Med. Imaging. 2016, 35, 1408–1419. [Google Scholar] [CrossRef]
- Bilchick, K.C. Integration of CMR scar imaging and electroanatomic mapping: The future of VT ablation? JACC Cardiovasc. Imaging 2012, 5, 211–213. [Google Scholar] [CrossRef]
- Mamalakis, M.; Garg, P.; Nelson, T.; Lee, J.; Wild, J.M.; Clayton, R.H. MA-SOCRATIS: An automatic pipeline for robust segmentation of the left ventricle and scar. Comput. Med. Imaging Graph. 2021, 93, 101982. [Google Scholar] [CrossRef] [PubMed]
- Gudbjartsson, H.; Patz, S. The Rician distribution of noisy MRI data. Magn. Reson. Med. 1996, 36, 332. [Google Scholar] [CrossRef]
- Zheng, Q.; Duchateau, N.; Ayache, N. 3D Consistent & Robust Segmentation of Cardiac Images by Deep Learning with Spatial Propagation. 2021; 1–12. [Google Scholar]
- Amado, L.C.; Gerber, B.L.; Gupta, S.N.; Rettmann, D.W.; Szarf, G.; Schock, R.; Nasir, K.; Kraitchman, D.L.; Lima, J.A. Accurate and objective infarct sizing by contrast-enhanced magnetic resonance imaging in a canine myocardial infarction model. J. Am. Coll. Cardiol. 2004, 44, 2383–2389. [Google Scholar] [CrossRef]
- Moccia, S.; Banali, R.; Martini, C.; Moscogiuri, G.; Pontone, G.; Pepi, M.; Caiani, E.G. Automated Scar Segmentation from CMR-LGE Images Using a Deep Learning Approach. In Proceedings of the 2018 Computing in Cardiology Conference (CinC), Maastricht, The Netherlands, 23–26 September 2018; pp. 1–4. [Google Scholar]
- Chen, C.; Qin, C.; Qiu, H.; Tarroni, G.; Duan, J.; Bai, W.; Rueckert, D. Deep Learning for Cardiac Image Segmentation: A Review. Front. Cardiovasc. Med. 2020, 7, 25. [Google Scholar] [CrossRef]
- Schuster, A.; Lange, T.; Backhaus, S.J.; Strohmeyer, C.; Boom, P.C.; Matz, J.; Kowallick, J.T.; Lotz, J.; Steinmetz, M.; Kutty, S.; et al. Fully Automated Cardiac Assessment for Diagnostic and Prognostic Stratification Following Myocardial Infarction. J. Am. Heart Assoc. 2020, 9, e016612. [Google Scholar] [CrossRef]
- Baessler, B.; Mannil, M.; Oebel, S.; Maintz, D.; Alkadhi, H.; Manka, R. Subacute and Chronic Left Ventricular Myocardial Scar: Accuracy of Texture Analysis on Nonenhanced Cine MR Images. Radiology 2018, 286, 103–112. [Google Scholar] [CrossRef]
- Wamil, M.; Borlotti, A.; Liu, D.; Briosa e Gala, A.; Bracco, A.; Alkhalil, M.; De Maria, G.L.; Piechnik, S.K.; Ferreira, V.M.; Banning, A.P.; et al. Combined T1-mapping and tissue tracking analysis predicts severity of ischemic injury following acute STEMI—An Oxford Acute Myocardial Infarction (OxAMI) study. Int. J. Cardiovasc. Imaging 2019, 35, 1297–1308. [Google Scholar] [CrossRef] [PubMed]
- Haaf, P.; Garg, P.; Messroghli, D.R.; Broadbent, D.A.; Greenwood, J.P.; Plein, S. Cardiac T1 Mapping and Extracellular Volume (ECV) in clinical practice: A comprehensive review. J. Cardiovasc. Magn. Reson. 2016, 18, 89. [Google Scholar] [CrossRef] [PubMed]
- Almutairi, H.M.; Boubertakh, R.; Miquel, M.E.; Petersen, S.E. Myocardial deformation assessment using cardiovascular magnetic resonance-feature tracking technique. Br. J. Radiol. 2017, 90, 20170072. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, A.S.; El-Rewaidy, H.; Nezafat, M.; Nakamori, S.; Nezafat, R. Automated analysis of cardiovascular magnetic resonance myocardial native T1 mapping images using fully convolutional neural networks. J. Cardiovasc. Magn. Reson. 2019, 21, 7. [Google Scholar] [CrossRef]
- Zhu, Y.; Fahmy, A.S.; Duan, C.; Nakamori, S.; Nezafat, R. Automated Myocardial T2 and Extracellular Volume Quantification in Cardiac MRI Using Transfer Learning-based Myocardium Segmentation. Radiol. Artif. Intell. 2020, 2, e190034. [Google Scholar] [CrossRef]
- Ferdian, E.; Suinesiaputra, A.; Fung, K.; Aung, N.; Lukaschuk, E.; Barutcu, A.; Maclean, E.; Paiva, J.; Piechnik, S.K.; Neubauer, S.; et al. Fully Automated Myocardial Strain Estimation from Cardiovascular MRI–tagged Images Using a Deep Learning Framework in the UK Biobank. Radiol. Cardiothorac. Imaging 2020, 2, e190032. [Google Scholar] [CrossRef]
- Swift, A.J.; Lu, H.; Uthoff, J.; Garg, P.; Cogliano, M.; Taylor, J.; Metherall, P.; Zhou, S.; Johns, C.S.; Alabed, S.; et al. A machine learning cardiac magnetic resonance approach to extract disease features and automate pulmonary arterial hypertension diagnosis. Eur. Heart J. Cardiovasc. Imaging 2020, 22, 236–245. [Google Scholar] [CrossRef]
- Dawes, T.J.W.; de Marvao, A.; Shi, W.; Fletcher, T.; Watson, G.M.J.; Wharton, J.; Rhodes, C.J.; Howard, L.S.G.E.; Gibbs, J.S.R.; Rueckert, D.; et al. Machine Learning of Three-dimensional Right Ventricular Motion Enables Outcome Prediction in Pulmonary Hypertension: A Cardiac MR Imaging Study. Radiology 2017, 283, 381–390. [Google Scholar] [CrossRef]
- Bello, G.A.; Dawes, T.J.W.; Duan, J.; Biffi, C.; de Marvao, A.; Howard, L.S.G.E.; Gibbs, J.S.R.; Wilkins, M.R.; Cook, S.A.; Rueckert, D.; et al. Deep learning cardiac motion analysis for human survival prediction. Nat. Mach. Intell. 2019, 1, 95–104. [Google Scholar] [CrossRef]
- Aung, N.; Vargas, J.D.; Yang, C.; Cabrera, C.P.; Warren, H.R.; Fung, K.; Tzanis, E.; Barnes, M.R.; Rotter, J.I.; Taylor, K.D.; et al. Genome-Wide Analysis of Left Ventricular Image-Derived Phenotypes Identifies Fourteen Loci Associated With Cardiac Morphogenesis and Heart Failure Development. Circulation 2019, 140, 1318–1330. [Google Scholar] [CrossRef]
- Trevethan, R. Sensitivity, Specificity, and Predictive Values: Foundations, Pliabilities, and Pitfalls in Research and Practice. Front. Public Health 2017, 5, 307. [Google Scholar] [CrossRef]
- Overmars, L.M.; van Es, B.; Groepenhoff, F.; De Groot, M.C.H.; Pasterkamp, G.; den Ruijter, H.M.; van Solinge, W.W.; Hoefer, I.E.; Haitjema, S. Preventing unnecessary imaging in patients suspect of coronary artery disease through machine learning of electronic health records. Eur. Heart J.-Digit. Health 2022, 3, 11–19. [Google Scholar] [CrossRef]
- De Fauw, J.; Ledsam, J.R.; Romera-Paredes, B.; Nikolov, S.; Tomasev, N.; Blackwell, S.; Askham, H.; Glorot, X.; O’Donoghue, B.; Visentin, D.; et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 2018, 24, 1342–1350. [Google Scholar] [CrossRef]
- Jeddi, Z.; Bohr, A. Remote Patient Monitoring Using Artificial Intelligence; Bohr, A., Memarzadeh, K.B.T.-A.I., Eds.; Academic Press: Cambridge, MA, USA, 2020; Chapter 9; pp. 203–234. [Google Scholar]
- Lanier, J. Ten Arguments for Deleting Your Social Media Accounts Right Now; Henry Holt and Co., Inc.: New York, NY, USA, 2018. [Google Scholar]
- McConnell, M.; Turakhia, M.; Harrington, R.; King, A.; Ashley, E. Mobile Health Advances in Physical Activity, Fitness, and Atrial Fibrillation. J. Am. Coll. Cardiol. 2018, 71, 2691–2701. [Google Scholar] [CrossRef]
- Sharma, A.; Harrington, R.A.; McClellan, M.B.; Turakhia, M.P.; Eapen, Z.J.; Steinhubl, S.; Mault, J.R.; Majmudar, M.D.; Roessig, L.; Chandross, K.J.; et al. Using Digital Health Technology to Better Generate Evidence and Deliver Evidence-Based Care. J. Am. Coll. Cardiol. 2018, 71, 2680–2690. [Google Scholar] [CrossRef]
- Lim, L.; Tison, G.; Delling, F.N. Artificial Intelligence in Cardiovascular Imaging. Methodist DeBakey Cardiovasc. J. 2020, 16, 138–145. [Google Scholar] [CrossRef]
- Antoniades, C.; Oikonomou, E.K. Artificial intelligence in cardiovascular imaging—Principles, expectations, and limitations. Eur. Heart J. 2021; ehab678, Online ahead of print. [Google Scholar] [CrossRef]
- Vasey, B.; Nagendran, M.; Campbell, B.; Clifton, D.A.; Collins, G.S.; Denaxas, S.; Denniston, A.K.; Faes, L.; Geerts, B.; Ibrahim, M.; et al. Reporting guideline for the early-stage clinical evaluation of decision support systems driven by artificial intelligence: DECIDE-AI. Nat. Med. 2022, 28, 924–933. [Google Scholar] [CrossRef]
- Vokinger, K.N.; Feuerriegel, S.; Kesselheim, A.S. Mitigating bias in machine learning for medicine. Commun. Med. 2021, 1, 25. [Google Scholar] [CrossRef]
- Hussain-Gambles, M. Ethnic minority under-representation in clinical trials. J. Health Organ. Manag. 2003, 17, 138–143. [Google Scholar] [CrossRef]
- Tanne, J.H. US must urgently correct ethnic and racial disparities in clinical trials, says report. BMJ 2022, 377, o1292. [Google Scholar] [CrossRef]
- Balla, S.; Gomez, S.E.; Rodriguez, F. Disparities in Cardiovascular Care and Outcomes for Women from Racial/Ethnic Minority Backgrounds. Curr. Treat. Options Cardiovasc. Med. 2020, 22, 75. [Google Scholar] [CrossRef]
- Davis, A.M.; Vinci, L.M.; Okwuosa, T.M.; Chase, A.R.; Huang, E.S. Cardiovascular Health Disparities. Med. Care Res. Rev. 2007, 64 (Suppl. 5), 29S–100S. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.E.; Steinberg, J.R.; Weeks, B.T.; Rodriguez, F.; Cullen, M.R. Race/ethnicity reporting and representation in US clinical trials: A cohort study. Lancet Reg. Health-Am. 2022, 11, 100252. [Google Scholar] [PubMed]
- Collins, G.S.; Dhiman, P.; Andaur Navarro, C.L.; Ma, J.; Hooft, L.; Reitsma, J.B.; Logullo, P.; Beam, A.L.; Peng, L.; Van Calster, B.; et al. Protocol for development of a reporting guideline (TRIPOD-AI) and risk of bias tool (PROBAST-AI) for diagnostic and prognostic prediction model studies based on artificial intelligence. BMJ Open 2021, 11, e048008. [Google Scholar]
- Lujic, S.; Watson, D.E.; Randall, D.A.; Simpson, J.M.; Jorm, L.R. Variation in the recording of common health conditions in routine hospital data: Study using linked survey and administrative data in New South Wales, Australia. BMJ Open 2014, 4, e005768. [Google Scholar] [CrossRef]
- Emmanuel, T.; Maupong, T.; Mpoeleng, D.; Semong, T.; Mphago, B.; Tabona, O. A survey on missing data in machine learning. J. Big Data 2021, 8, 140. [Google Scholar] [CrossRef]
- Donders, A.R.T.; van der Heijden, G.J.M.G.; Stijnen, T.; Moons, K.G.M. Review: A gentle introduction to imputation of missing values. J. Clin. Epidemiol. 2006, 59, 1087–1091. [Google Scholar] [CrossRef]
- Demšar, J.; Zupan, B. Hands-on training about overfitting. PLoS Comput. Biol. 2021, 17, e1008671. [Google Scholar] [CrossRef]
- Volovici, V.; Syn, N.L.; Ercole, A.; Zhao, J.J.; Liu, N. Steps to avoid overuse and misuse of machine learning in clinical research. Nat. Med. 2022, 28, 1996–1999. [Google Scholar] [CrossRef]
- Stanford Medicine. Harnessing the Power of Data in Health. Stanford Medicine 2017 Health Trends Report. 2017. Available online: https://med.stanford.edu/school/leadership/dean/updates/healthtrends2017.html (accessed on 14 December 2022).
- Petersen, C.; Subbian, V. Special Section on Ethics in Health Informatics. Yearb. Med. Inform. 2020, 29, 77–80. [Google Scholar] [CrossRef]
- Grutters, J.P.C.; Govers, T.; Nijboer, J.; Tummers, M.; van der Wilt, G.J.; Rovers, M.M. Problems and Promises of Health Technologies: The Role of Early Health Economic Modeling. Int. J. Health Policy Manag. 2019, 8, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Ranschaert, E. The Cost and Value of AI for Radiology—What is the Healthcare Tipping Point? OSIMIS. 2022. Available online: https://www.osimis.io/post/the-cost-and-value-of-ai-for-radiology-what-is-the-healthcare-tipping-point (accessed on 12 February 2023).
- Van Leeuwen, K.G.; Meijer, F.J.A.; Schalekamp, S.; Rutten, M.J.C.M.; van Dijk, E.J.; van Ginneken, B.; Govers, T.M.; de Rooij, M. Cost-effectiveness of artificial intelligence aided vessel occlusion detection in acute stroke: An early health technology assessment. Insights Imaging 2021, 12, 133. [Google Scholar] [CrossRef] [PubMed]
- Wouters, O.J.; Naci, H.; Samani, N.J. QALYs in cost-effectiveness analysis: An overview for cardiologists. Heart 2015, 101, 1868–1873. [Google Scholar] [CrossRef]
- Arora, A.; Wright, A.; Cheng, M.; Khwaja, Z.; Seah, M. Innovation Pathways in the NHS: An Introductory Review. Ther. Innov. Regul. Sci. 2021, 55, 1045–1058. [Google Scholar] [CrossRef] [PubMed]

| Study Author | Year | Modality | Population | Description | Main Findings |
|---|---|---|---|---|---|
| Arsanjani et al. [20] | 2013 | MPI | 1181 | This study aimed to improve the accuracy of myocardial perfusion SPECT (MPS). 1181 MPS studies were examined, including 713 cases with correlating invasive coronary angiography data. Clinical data and quantitative image features were integrated with ML algorithms. TPD and stress/rest perfusion change were obtained from automated perfusion quantification software and combined with variables such as age and sex by LogitBoost. | Computational integration of quantitative image measures and clinical data by ML improves the diagnostic performance of automatic MPI analysis to the level rivalling expert analysis. |
| Knackstedt et al. [15] | 2015 | Echo | 255 | ML analysis was used for fully automated left ventricular measurements including EF, as well as longitudinal strain. A reference centre re-examined all datasets by visual estimation, as well as manual tracking. | Automated left ventricular measurements were completed in 98% of studies, with good reproducibility and an average analysis time of 8 s. |
| Avendi et al. [35] | 2016 | CMR | 45 | This study utilised DL algorithms combined with deformable models in order to design a fully automated left ventricular segmentation model from short-axis CMR datasets. DL was used for automatic detection and inferring left ventricular shape. | Excellent agreement and high correlation with reference contours were reported. |
| Dawes et al. [54] | 2017 | CMR | 256 | This study investigated whether patient survival in pulmonary hypertension (PH) could be predicted using ML of 3-D patterns of cardiac motion on CMR. All patients with new diagnosis of PH underwent CMR, right heart catheterisation, and a 6-minute walk. | The ML survival model was found to predict outcome independent of traditional risk factors in patients with newly diagnosed PH. |
| Motwani et al. [16] | 2017 | CT | 10,030 | This was a registry analysis of 10,030 patients with suspected CAD. 25 clinical and 44 CCTA parameters were assessed, and ML involving automated feature selection and model building with a boosted ensemble algorith, was used to combine a clinical and CCTA, modified Duke index and Framingham risk score to predict all-cause mortality. | ML was found to predict 5-year mortality significantly better than existing clinical or CCTA metrics alone. |
| Madani et al. [14] | 2018 | Echo | 267 | CNN was trained to recognize 15 standard echocardiographic views, using a training set of 200,000 images based on still images and videos from 267 transthoracic echocardiograms. | DL achieved expert-level classification, with researchers demonstrating an accuracy of 91.7% compared to 79.4% for board certified echocardiographers classifying a subset of the same test images. |
| Nakashini et al. [17] | 2018 | CT | 6814 | This study included data from 6814 asymptomatic patients undergoing coronary artery calcium scanning who were followed up for coronary heart disease and atherosclerotic cardiovascular events over a decade. ML utilised all available clinical and CT data including the CAC score, CAC volume scores, as well as extracardiac CAC scores. | ML of all available clinical and non-contrast CT variables was superior to clinical risk factors and CAC score in predicting both coronary heart disease and cardiovascular disease events. |
| Betancur et al. [19] | 2018 | MPI | 1638 | This study compared the automated prediction of obstructive disease from MPI by DL with total perfusion deficit (TPD). Patients without known CAD underwent stress 99mTc-Sestamibi or tetrofosmin myocardial perfusion imaging (MPI). DL was trained using raw and quantitative polar maps and evaluated for prediction of clinically significant stenosis in a stratified 10-fold cross-validation procedure. | DL was shown to improve automatic prediction of obstructive CAD, as compared to the current method. AUC from the ROC curve for disease prediction by DL was higher than for TPD (per patient: 0.80 vs. 0.78; per vessel: 0.76 vs 0.73, p < 0.01). |
| Zabihollahy et al. [36] | 2018 | CMR | 34 | DL was used to design a semi-automated method for fully automated segmentation of a left ventricular scar from 3-D late gadolinium CMR images from patients with ischaemic cardiomyopathy, without any operator interaction. | The new method was found to outperform alternative techniques. |
| Zheng et al. [41] | 2018 | CMR | 3078 from UK Biobank(training), 756 (testing) | DL was used to carry out cardiac segmentation with spatial propagation on CMR image stacks. The method was trained on a large database of 3078 cases and then tested on 756 cases | This technique achieved comparable and even improved results in terms of distance measures when compared with state-of-the-art methods. |
| Baessler et al. [46] | 2018 | CMR | 120 | This was a proof-of-concept study assessing whether texture analysis allowed for the diagnosis of subacute and chronic MI on CMR images. 120 patients undergoing CMR showing large transmural infarcts or small chronic ischaemic scars were entered retrospectively. Regions of interest for texture analysis involving the left ventricle were contoured by 2 blinded readers on cine images by using a software package. Texture feature selection based on reproducibility, ML and correlation were carried out for selecting features, allowing the diagnosis of MI on non-enhanced CMR images by using LGE as standard of reference. | The authors concluded that texture analysis enabled the diagnosis of subacute and chronic MI with high accuracy. |
| Fahmy et al. [50] | 2019 | CMR | 210 (training and testing), 455 (validation) | In this study, the authors describe an automated technique (deep fully convolutional neural network, FCN) which was used for myocardial segmentation in T1 weighted CMR images. | FCN enabled fast segmentation (<0.3 s per image) with a high Dice similarity coefficient, thus allowing fast automatic analysis of myocardial native T1 mapping images on CMR. |
| Schuster et al. [45] | 2020 | CMR | 1017 | CMR data from 2 MI multicentre trials (n = 1017 patients) were included and analysis of parameters such as EF were manually and automatically assessed using conventional and AI-based software. Obtained measurements entered regression analysis for prediction of MACE. | Volumetric analysis carried out by AI software was feasible, with results being reported to be equally predictive of MACE compared with traditional methods. |
| Ferdian et al. [52] | 2020 | CMR | 4508 from Biobank (3244 for training, 812 for validation, 452 for testing) | This was a retrospective cross-sectional study whereby neural networks (including CNN) were used to perceive and track the myocardial landmarks through each slice, and strain measurements were made from the landmarks’ motion. | The automated technique allowed unbiased strain assessment, with a typical processing time of 260 frames (13 slices) per second, compared with 6–8 min per slice for manual methods. |
| Swift et al. [53] | 2021 | CMR | 220 | This study investigated the use of a tensor-based ML approach to highlight features of PAH using CMR. Untreated patients with PAH or no evidence of pulmonary hypertension (PH) who underwent CMR and right heart catheterisation studies within 48 h were selected from the ASPIRE registry. A tensor-based ML model was developed, and the accuracy of this tool was measured against standard CMR assessments. | The authors reported high diagnostic accuracy as assessed by AUC at receiver operating characteristic analysis (ROC), p < 0.001:0.92 for PAH, which is slightly higher than standard CMR assessments. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doolub, G.; Mamalakis, M.; Alabed, S.; Van der Geest, R.J.; Swift, A.J.; Rodrigues, J.C.L.; Garg, P.; Joshi, N.V.; Dastidar, A. Artificial Intelligence as a Diagnostic Tool in Non-Invasive Imaging in the Assessment of Coronary Artery Disease. Med. Sci. 2023, 11, 20. https://doi.org/10.3390/medsci11010020
Doolub G, Mamalakis M, Alabed S, Van der Geest RJ, Swift AJ, Rodrigues JCL, Garg P, Joshi NV, Dastidar A. Artificial Intelligence as a Diagnostic Tool in Non-Invasive Imaging in the Assessment of Coronary Artery Disease. Medical Sciences. 2023; 11(1):20. https://doi.org/10.3390/medsci11010020
Chicago/Turabian StyleDoolub, Gemina, Michail Mamalakis, Samer Alabed, Rob J. Van der Geest, Andrew J. Swift, Jonathan C. L. Rodrigues, Pankaj Garg, Nikhil V. Joshi, and Amardeep Dastidar. 2023. "Artificial Intelligence as a Diagnostic Tool in Non-Invasive Imaging in the Assessment of Coronary Artery Disease" Medical Sciences 11, no. 1: 20. https://doi.org/10.3390/medsci11010020
APA StyleDoolub, G., Mamalakis, M., Alabed, S., Van der Geest, R. J., Swift, A. J., Rodrigues, J. C. L., Garg, P., Joshi, N. V., & Dastidar, A. (2023). Artificial Intelligence as a Diagnostic Tool in Non-Invasive Imaging in the Assessment of Coronary Artery Disease. Medical Sciences, 11(1), 20. https://doi.org/10.3390/medsci11010020






