Abstract
In 2008, as part of a feasibility study for radioactive waste disposal in deep geological formations, the French National Radioactive Waste Management Agency (ANDRA) drilled several boreholes in the transposition zone in order to define the potential variations in the properties of the Callovo–Oxfordian claystone formation. This consisted of a rare opportunity to investigate the deep continental biosphere that is still poorly known. Four rock cores, from 1709, 1804, 1865, and 1935 m below land surface, were collected from Lower and Middle Triassic formations in the Paris Basin (France) to investigate their microbial and geochemical composition. Rock leachates showed high salinities ranging from 100 to 365 g·L−1 NaCl, current temperatures averaging 65 °C, no detectable organic matter, and very fine porosity. Microbial composition was studied using a dual cultural and molecular approach. While the broad-spectrum cultural media that was used to activate microbial communities was unsuccessful, the genetic investigation of the dominant 16S rRNA gene sequences revealed eight bacterial genera considered as truly indigenous to the Triassic cores. Retrieved taxa were affiliated to aerobic and facultative anaerobic taxon, mostly unknown to grow in very saline media, except for one taxon related to Halomonas. They included Firmicutes and α-, β-, and γ-Proteobacteria members that are known from many subsurface environments and deep terrestrial and marine ecosystems. As suggested by geochemical analyses of rocks and rock leachates, part of the indigenous bacterial community may originate from a cold paleo-recharge of the Trias aquifer with water originating from ice melting. Thus, retrieved DNA would be fossil DNA. As previously put forward to explain the lack of evidence of microbial life in deep sandstone, another hypothesis is a possible paleo-sterilisation that is based on the poly-extremophilic character of the confined Triassic sandstones, which present high salinity and temperature.
1. Introduction
Over the past decades, many fundamental discoveries, especially the hyperthermophily concept, have enabled us to expand life boundaries, with prokaryotic life occurring up to 121 °C in deep-sea hydrothermal vents [1] and evidence of deep-sea chemolithoautotrophic methanogenic cell proliferation and activity at 122 °C when grown at high hydrostatic pressure [2]. Microorganisms are also well known to inhabit the deep terrestrial subsurface, at depths where life meets harsh physico-chemical constraints in terms of temperature and/or salinity, availability of nutrients, and limited rock porosity [3,4,5]. The subsurface, including the continental one, represents one of the most important microbial habitats on Earth and stores an important part of the world’s prokaryotic community [6,7]. Nevertheless, our knowledge about the composition, distribution, and origin of the deep biosphere is rather scarce, mainly because of limited abilities to access and acquire uncontaminated samples from the ultra-deep subsurface.
In deep environments, microorganisms are exposed to extreme physico-chemical conditions. The combination of hydrostatic pressure, temperature, salinity, and anoxia are major subsurface parameters that drive life boundaries, but hydrogeological and geochemical aspects should also be considered. They include rock porosity and permeability [8], water activity [9], electron acceptors, energy and carbon source availability [3,10], or pH values, which are strongly influenced by dissolved gas in the deep subsurface [11]. Subsurface microorganisms are present in rocks and consolidated sediment pores where microbial movements and activities are limited, leading to slow microbial growth [8]. Moreover, the liquid/solid ratio decreases with depth, thus granting only little space for life in ultra-deep environments [12]. In this respect, a decrease of the microbial biomass has been observed with depth in numerous studies [13,14]. Despite extreme conditions, biomass in the continental subsurface, estimated at 2 to 6 × 1029 cells, account for a significant fraction of the total prokaryote biomass on Earth [7].
Deep continental samples have mainly been acquired through open windows into the deep subsurface, such as mining, oil drilling, geothermal, and water wells [4,15,16,17,18]. Few microbiological studies have been conducted on the ultra-deep geosphere and especially on cores below 1500 m depth in the continental crust where the estimated biomass is low [17,19,20,21]. In the deep continental subsurface, studies report on various microbial community sizes. A prokaryotic PFLA (Phospholipid Fatty Acid Analysis) concentration of 16 pmol·g−1, equivalent to 4 × 105 prokaryotic cells·g−1, was measured in a shale of a natural gas-bearing formation (Virginia, USA) at 2700 mbsl (meters below surface level) and a current temperature of 76 °C [19]. Around 104 cells·g−1 were found in an ultra-high-pressure rock in the Chinese Dabie-Sulu metamorphic belt [20]. In comparison, 103 and less than 102 cells·g−1 were reported in the quartzite and the Carbon Leader, respectively, extracted at 3200 mbsl from a functional gold mine with a formation temperature of approximately 48 °C, in the South African Witwatersrand basin [17]. Moreover, a decrease of biomass with depth was measured in cores at 860 mbsl (10 pmol PLFA g−1), 1996 mbsl (about 0.4 pmol PLFA g−1), and 2090 mbsl (no PFLA) from a sedimentary Cretaceous sandstone in the Piceance Basin of western Colorado (USA) [21].
A rare opportunity to investigate the deep continental biosphere was enabled through the recognition campaign of the “Transposition Zone” (TZ) of the Meuse/Haute-Marne Underground Research Laboratory (URL) in the east of the Paris Basin. As a part of the feasibility study of radioactive waste disposal in deep geological formations, the French National Radioactive Waste Management Agency (ANDRA) drilled, in 2008, several boreholes in the TZ in order to define the potential variations in properties of the Callovo–Oxfordian claystone formation, the targeted layer for radioactive waste disposal. This huge «multi-drilling» campaign enabled a direct access to different geological facies extracted along a depth range extended from 500 to 2000 mbsl and a multidisciplinary scientific collaboration was conducted in the TAPSS 2000 project (present and past transfer in an aquifer–aquitard sedimentary system) [22].
In this context, the main aim of our study was to investigate the indigenous microbial diversity of four Triassic sandstones from 1709, 1804, 1865, and 1935 mbsl by culture-dependent and independent complementary approaches. The origin of the detected microorganisms is discussed based on the geological history, the hydrogeological functioning of the Paris Basin (France), and the physico-chemical data acquired from the targeted Triassic formations.
2. Materials and Methods
2.1. Site, Drilling and Sampling
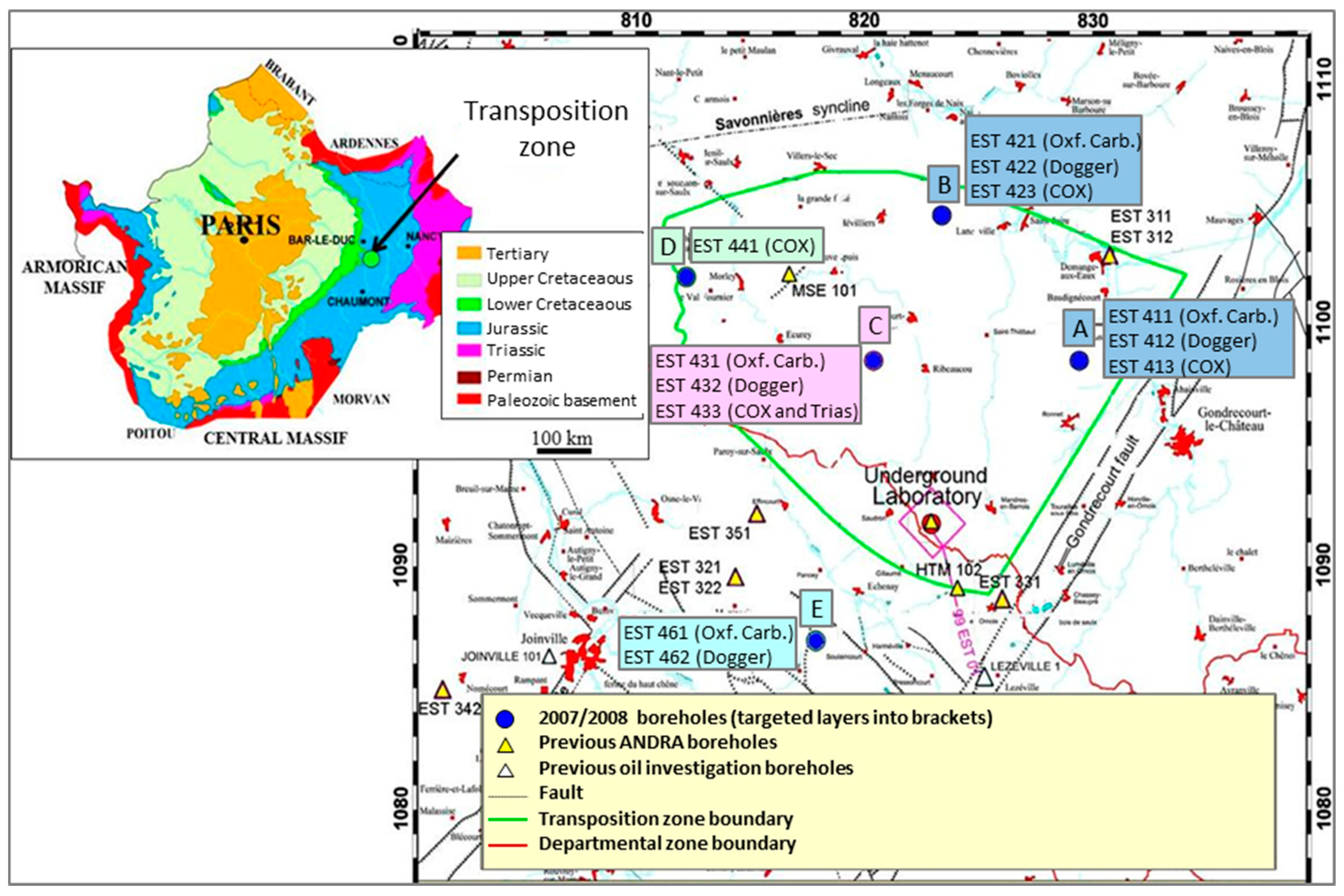
The drilling site (EST433 borehole on the C platform) is located close to Montiers-sur-Saulx (Meuse, France) within the TZ delimited by ANDRA in the Paris Basin. The Paris Basin is supported by the Hercynian basement and it contains a succession of sedimentary layers arranged in wide concentric rings; the latest outcrop is in the middle and the oldest on the periphery as a direct result of the Paris Basin subsidence in the Secondary and Tertiary era with the uplift of northern, eastern, and southern margins [23]. The TZ represents an area of approximately 250 km2 in the east of the Paris Basin where no fault was observed (Figure 1). The EST433 borehole was the only TZ borehole to reach Triassic layers. It represents one of the few deep continental holes drilled from the surface down to 1700 mbsl in undisturbed sediments. In situ temperature has been estimated at 60 °C at 1880 mbsl [22]. Drilling procedures, borehole investigations and geological description of the drilling cores were described by Landrein et al. [22]. Briefly, a destructive drilling with punctual core sampling was used for the deep Triassic formations between about 1700 and 2000 mbsl. Four 9 m long cores were obtained and named K2-17 to K2-20. These were then sub-divided into smaller pieces of rock (Figure 2).
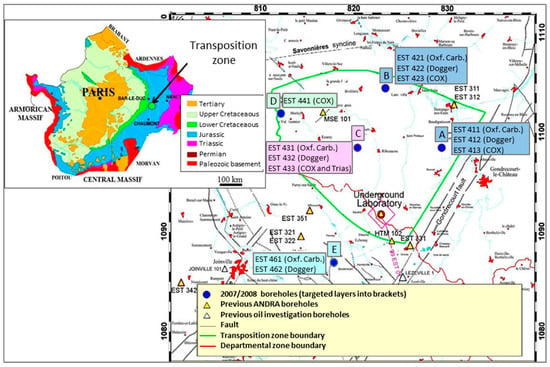
Figure 1.
Geological context of Paris Basin and map of the Transposition Zone with the location of the EST433 borehole (C platform).
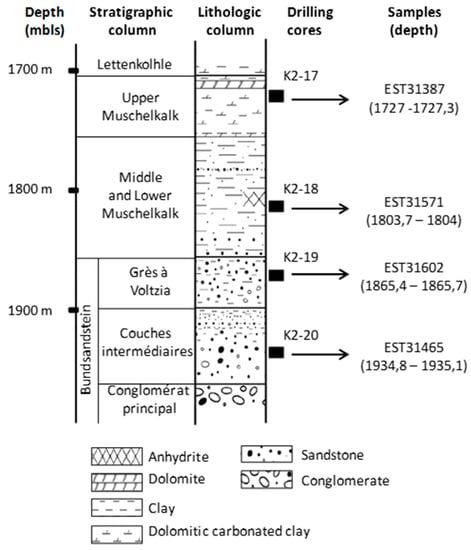
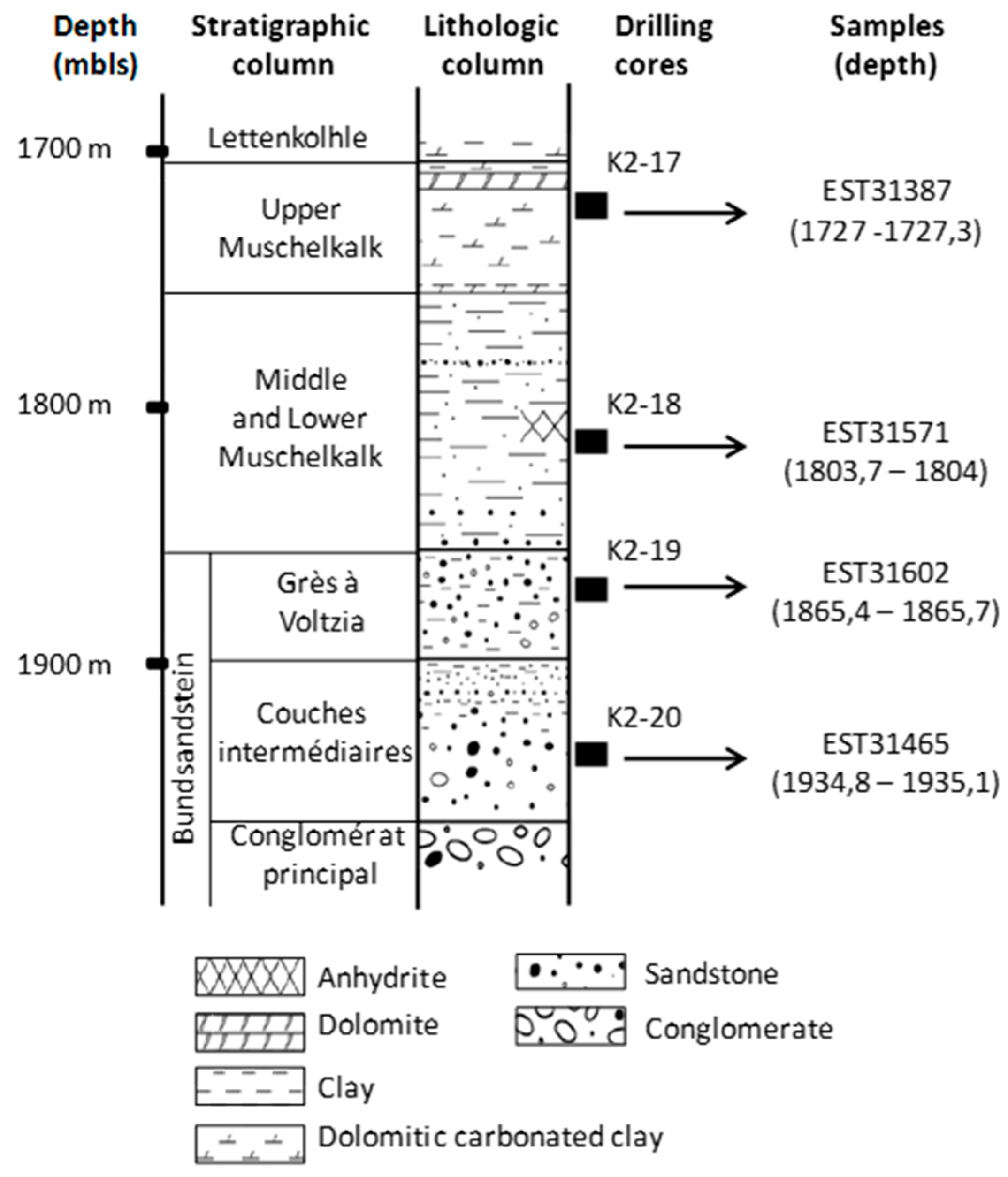
Figure 2.
Location of drilling cores and samples from the EST433 deep borehole (Paris Basin, France) into the lithostratigraphic column.
On site, four samples, named K2-20 (EST31467), K2-19 (EST31602), K2-18 (EST31571), and K2-17 (EST31387), from four distinct Triassic formations and drilling fluids from the same depths were collected using sterile materials. As it was assumed that no oxygen was present at these depths, the samples were prepared for storage in an anaerobic glove box under N2 by cleaning the core surfaces with ethanol and then placing the cores in sterile double plastic bags and closing the bags by thermal sealing. The sealed plastic bags were then placed successively in two aluminized envelopes, each sealed under flowing N2 to ensure anaerobiosis. Samples were kept at 4 °C until analysis. A piece of core from each Triassic formation (9 cm long) was re-pressurized (190 bar) in a high-pressure chamber under N2 to preserve the indigenous piezophilic microbial community.
Triassic cores were aseptically sub-sampled in an anaerobic chamber filled with filtered pure N2 according to the Poulain et al. protocol [24]. The peripheral two centimeters were removed by cutting them off using a sterile saw, to prevent and limit the presence of contamination by allochthonous microorganisms in the inner part of the core. To obtain a representative sample and to overcome the spatial heterogeneity of microorganisms sequestered in rocks, about 200 g of each inner Triassic core were manually crushed with a sterile mortar and hammer, homogenized, and packed into sterile bottles capped and hermetically sealed under N2. Rock powder for the cultural approaches and geochemical analysis was stored at 4 °C, while sub-samples used for the molecular analyses were frozen at −20 °C.
2.2. Geochemical Analyses and Mineralogical Characterisation of the Triassic Cores
pH values, ammonium, alkalinity, cations, and anions were determined on rock leachates. Only the rock fraction <80 µm was leached with deaerated ultrapure deionized water (liquid/solid ratio of 10) with stirring (150 rpm) for seven days in a CO2- and O2-free N2 atmosphere. pH values were measured by electrometry according to the French standard procedure NF T 90-008 [25]. Ammonium concentration was measured by flow analysis coupled with spectrometry based on the International standard procedure ISO 11732 [26]. Alkalinity was determined by potentiometric titration according to the ISO 9963-1 procedure [27]. Concentrations of nitrate, chloride, fluoride, sulfate, bromide, and phosphate were determined by ionic chromatography according to the ISO 10304-1 procedure [28]. Sodium measurements were performed after tri-acid digestion (HF-HNO3-HClO4) by flame atomic absorption spectrometry (Spectra AA-200, Varian, Victoria, Australia), according the NF T 01-041 French procedure [29]. An alkaline fusion (Na2O2) of 2 g of rock was carried out to determine the rock chlorine concentrations by potentiometric titration (Mettler DL25, Mettler-Toledo, Columbus, OH, USA). The Total Organic Carbon (TOC, expressed in %) was determined by Rock Eval VI pyrolysis according to the protocol that was described by Copard et al. [30]. Finally, concentrations of Mg, K, and Ca were measured by ICP-AES (Ultima 2, Horiba-Jobin Yvon, Kyoto, Japan) according to the NF EN ISO 11885 [31].
Crystallized phase identification of rock cores was carried out by X-ray diffraction patterns (D5000 diffractometer, Siemens AG, Karlsruhe, Germany) on rock powder that was deposited on a slide. Powder X-ray diffractograms were recorded between 4 and 84°2θ with CoKα cross (α1 = 1.789 Å). Minerals were identified using the search-match software DIFFRACplus. X-ray diffraction provided qualitative and semi quantitative data. Well-polished thin sections of each core were prepared to perform optical microscopic observations in order to determine the textural relationships of minerals and to observe faultindicating microstructures. These data compiled with the Thermoddem database [32] were used to model the composition of Triassic formation pore waters in PHREEQC [33]. Rock porosity was determined using mercury injection porosimetry (Autopore IV 9500 V1.09, Micromeritics, Norcross, GA, US) on dry pieces of inner cores.
2.3. Microbial Enrichments
The cultivation approaches were based on three anaerobic metabolic guilds: methanogenic, sulfate-reducing, and fermentative prokaryotes. The presence of organotrophic aerobes, nitrate-, thiosulfate-reducing, and chemolithotrophic microorganisms that were able to reduce various inorganic electron acceptors was also investigated (Table 1). Each enrichment medium was tested under different salinities varying from 0 to 200 g·L−1 of NaCl, except for the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH) 372 culture medium, specific for extreme halophilic aerobic microorganisms, tested only at 250 g·L−1 of NaCl. pH was adjusted with KOH 10M to obtain media close to neutral (pH 7 ± 0.5) or slightly alkaline (pH 8).

Table 1.
Composition of enrichment culture media. The resazurin color indicator was added into anaerobic media (1 mg·L−1).
Regarding anaerobic cultures, anaerobic methods that were introduced by Hungate were used [34]. Oxygen was removed by boiling and then cooling the media to room temperature under a sterile stream of O2-free N2. Each anaerobic medium was dispensed into Hungate tubes (10 mL) or serum bottles (20 mL), closed and hermetically sealed under a stream of O2-free N2. Resazurin, an oxidation-reduction indicator that is colorless when reduced, was used to confirm anaerobic conditions in the culture media. The vessels were autoclaved at 121 °C for 20 min. Prior to inoculation, substrate, electron acceptors, Na2S·9H2O, NaHCO3, and vitamins, when necessary, were added to media from sterile anaerobic stock solutions. Media were inoculated in an anaerobic chamber by adding 1 or 2 g rock powder to 10 or 20 mL media. Reconstituted pore waters (obtained by PhreeqC modelling) and saline waters were inoculated with 8.5 g rock powder for 17 mL of water. Two inocula were used for each core: crushed rock preserved either at atmospheric pressure or under pressure (190 bars). Five temperatures, i.e., 30, 37, 50, 55, and 70 °C were tested for incubations at atmospheric pressure. Incubations were also carried out under pressure (150 bars) at 55 °C, on media F2, B, and C, as well as on reconstituted pore waters amended with yeast extract (Table 1). Incubations under pressure were performed in Hungate tubes that were completely filled with media to avoid bubble formation and hermetically sealed with rubber stoppers, as described by Kallmeyer et al. [35]. Tubes were placed into a thermo-regulated high-pressure vessel that was previously heated to the incubation temperature and then pressurized using a hydrostatic pump. The pressure and the temperature inside the vessel were checked after equilibration and again before depressurisation. Microbial growth in the enrichments was assessed by direct microscopic observation, after two to 12 months of incubation.
The lack of toxicity of each crushed rock on bacterial growth was tested with a selection of deep subsurface isolates, including Thermosipho japonicus (DSM 13481) isolated from a deep-sea hydrothermal vent, Petrotoga mexicana (DSM 14811) isolated from an oil-producing well, and Desulfovibrio profundus (DSM 11384) isolated from deep marine sediments. They were grown on recommended DSMZ media (respectively, 283, 718, and 383) amended with 1 g crushed rock in 10 mL media. Growth was determined by microscopic observations. Negative controls for each condition were performed to identify media modifications resulting from culture conditions that could be misinterpreted and lead to false-positive growth. These consisted of uninoculated media and media that were inoculated with rocks sterilised three times at 121 °C with 24 h interval.
Efforts to identify a potential contaminant microflora carried by drilling fluids were performed by plating fluids on media to detect aerobic (media AEM1, AAEM1) and anaerobic (media ANM1, AANM1) organotrophic, neutrophilic, and alkaliphilic microorganisms (Table 1). Alkaliphilic microorganisms were targeted since the pH values of drilling fluid ranged between 11 and 12. Contaminant microorganisms that are present in the anaerobic chamber were also targeted by exposing AEM1 and ANM1 media agar plates to the chamber atmosphere during the core conditioning. Incubations that were performed at 30 °C lasted seven days for aerobic plates and four weeks for anaerobic ones.
2.4. DNA Extraction and Optimisation
Genomic DNA was extracted from drilling fluids, microbial isolates, and inner cores. For drilling fluids and isolates, DNA extractions were performed on pellets with the FastDNA Spin Kit for Soil, as recommended by the manufacturer (MP Biomedicals, Illkirch, France). Before extraction, pellets from the drilling fluids were treated three times with phosphate buffer (pH 8) in order to decrease their initial pH from 11/12 to 8. For Triassic rock, five grams of crushed rocks were used and three different extraction protocols were applied in order to optimise DNA extraction.
Protocol 1 used the DNA extraction protocol of Lerner et al. [36] with slight modifications: before incubation at 37 °C, slurries were subjected to two sonications and one vigorous agitation. SDS at a concentration of 10% was added at the same time as Proteinase K (Sigma-Aldrich, Sigma-Aldrich, St. Louis, MO, US). The second incubation was carried out at 65 °C for 2 h. Subsequently, samples were cooled on ice before addition of Tris-HCl (pH 8)-saturated phenol.
Protocol 2 was based on the FastDNA Spin Kit for Soil (MP Biomedicals). Extractions were performed on five distinct subsamples (ca. 1 g) of crushed rock and replicate DNA fractions were pooled so that the DNA was recovered from 5 g of sample. A lysis at speed 5 in the FastPrep instrument (MP Biomedicals) for 30 s and an extended spin of 30 min were applied at step 1 of the manufacturer’s protocol. Optimisation tests that were performed on Protocol 1 (no initial wash, cell desorption in a sodium pyrophosphate buffer, or addition of poly dIdC, a synthetic oligonucleotide acting as DNA competitors for binding sites instead of washing) and Protocol 2 (with and without addition of polydIdC and silicate beads at Step 1) did not improve DNA extraction yield, PCR efficiency, or diversity patterns.
Protocol 3 used the UltraClean Mega Soil DNA kit, as recommended by the manufacturer (MoBio Laboratories, Carlsbad, CA, USA). All crude DNA extracts were divided into two aliquots, one being further purified by dialysis (Slide A Lyser Mini Dialysis Unit, 3.5 k MWCO) against sterile TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) at 4 °C for 24 h. The dialysed DNA extracts were concentrated by precipitation with isopropanol, washed in 70% ethanol, and air dried. The final DNA products were resuspended in 20 to 50 µL of DNA-free water or sterile TE buffer.
Negative controls were routinely performed in parallel without any sample in order to exclude a contamination by traces of DNA carried by tubes and chemicals. DNA extraction efficiency and quality of DNA extracts were evaluated by PCR amplification and diversity profiles of 16S rRNA genes.
2.5. PCR Amplification of 16S rRNA Gene Sequences
For DNA extracts of isolates and drilling fluids, 16S rRNA genes were amplified with primers 8F and 1492R [37]. For crude and dialysed DNA extracted from Triassic rocks, the V3 region of bacterial 16S rRNA genes was amplified by nested PCR for denaturing gradient gel electrophoresis (DGGE) analysis. The first PCR was performed on 1 µL of purified DNA with the primer set 8F/1492R, with 32 amplification cycles and primer binding at 55 °C. The nested PCR was performed on 1 µL of 10-fold diluted PCR product with the primer set 341F-GC/907R [38], 15 amplification cycles and primer binding at 55 °C. Amplification of archaeal 16S rRNA genes was performed using the three forward primers A2Fa, A344F, and A109F combined with the same reverse primer A934R [39], 40 amplification cycles, and primer binding at 65 °C. A nested PCR was also tried with Arch344F/w34FAM primers [40]. PCR mixes contained 1X GoTaq® Flexi buffer (Promega, Charbonnières-les-Bains, France), 2.5 mM of MgCl2, 25 μM of each dNTP, 500 nM of each primer, 1 to 2 μL of DNA extract, and 1 U of GoTaq polymerase (Promega), in a final volume of 20 µL in nuclease- and DNA-free water (Sigma).
Negative controls without DNA were included in all PCR reaction sets. Absence of nested PCR contamination was systematically checked by re-amplification of the negative control from the first PCR. Several precautions were taken to exclude contamination by external DNA during PCR setup: DNA-free plastics, U.V. irradiation of non-biologic products (water, buffer, tubes, tips…), and setup in a Biocap hood (Captair®bio, Erlab).
2.6. DGGE Analysis of Bacterial Diversity
DGGE analyses of the bacterial 16S rRNA gene V3 region were performed on 200 to 400 ng of nested PCR products, in a 6% w/v polyacrylamide gel with a denaturant (formamide and urea) gradient varying from 35 to 65%. Electrophoreses were performed in a D-Code system (BioRad, Marnes-la-Coquette, France) at 80 V and 60 °C during 17 h in 1X TAE buffer. Gels were stained in SYBR Gold (0.5X, Invitrogen, Cergy-Pontoise, France) for 20 min and then visualised on a Dark Reader (Clare Chemical Research). Bands of interest were punched and eluted in 20 µL of nuclease- and DNA-free water, and 1 µL was used for re-amplification by PCR with primer pair 341F-GC/907R. The purity of bands was checked by DGGE. Re-amplified products were sequenced by the GATC Company while using the Sanger method with primer 341F.
2.7. Phylogenetic Analysis
Presence of chimeric sequences was checked using BELLEROPHON [41]. Partial 16S rRNA gene sequences were compared to the Genbank reference sequences by Blastn (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Sequence manipulation and alignment were performed in the BioEdit software version 7.0.4.1 [42] All positions containing gaps and ambiguous data were eliminated. Distances were calculated on 390 unambiguous nucleotides using the [43] method. Taxa were defined as clusters of 16S rRNA gene sequences showing more than 97% similarity. Nucleotide sequences were deposited in Genbank database under accession numbers KC861992 to KC862021 for isolates and KC862022 to KC862057 for environmental clones. Phylogenetic trees, including closest Blast related sequences, were constructed using the neighbor-joining tree construction and bootstrap (1000 replicates) analysis [44,45], with all phylogenetic programs being implemented in the MEGA X package [46].
3. Results
3.1. Characterisation of Deep Triassic Samples
Deep Triassic formations are located between the Permian basement and the Keuper saline formation (Figure 1). The deeper samples K2-20 and K2-19 are two Lower Triassic sandstones. The granulometry of the K2-20 core is heterogeneous (>200 µm up to mm). Detrital fraction consisted of rounded grains of dominant monocrystalline quartz with minor polycrystalline quartz, K-feldspar, and lithic clasts. Detrital grains were cemented essentially by authigeneous quartz. Tourmaline was an accessory detrital phase (Figure 3A). The sample from the K2-19 showed a homogeneous granulometry (average ~100–200 µm). Detrital fraction consisted of rounded grains of dominant monocrystalline quartz with minor polycrystalline quartz and K-feldspar. Flakes of muscovite (white mica) were frequent. Detrital grains were cemented by scarce detrital illite, and authigeneous quartz (Qtza) and anhydrite (Figure 3B). These findings were corroborated by XRD patterns. The other samples were two fine-grained sandstones belonging to the Middle Triassic formations. The sample from the K2-18 core was heterogeneous sandstone alternated with a few thin clay-rich layers underlying the bedding, and with carbonate-rich levels. The detrital fraction consisted of dominant quartz with minor K-feldspar, plagioclase, chlorite, muscovite, tourmaline, and clay minerals locally cementing the detrital grains (Figure 3C). Diagenetic cement consisted of anhydrite and in some levels of fine-grained carbonates (Figure 3D). The K2-19 core sample was carbonate sandstone. The detrital fraction consisted of dominant quartz with minor K-feldspar, plagioclase, and white mica; illite was identified by X-ray diffraction. Detrital grains were cemented by abundant fine-grained dolomite with minor anhydrite (Figure 3E). The sandstone was crosscut by a coarse-grained vein of poecilitic quartz and fibrolitic anhydrite (Figure 3F).

Figure 3.
Optical micrograph showing various mineral in the Triasic samples. Photo (A,B) were taken from the K2-17 (EST31387) core, (C,D) from the K2-18 core (EST31571) and (E,F) from K2-19 (EST31602) and K2-20 (EST31467) cores, respectively. Dol, dolomite; Mus, muscovite; Anh, anhydrite; Anhf, fibrolitic anhydrite; Chl, chlorite; Tur, tourmaline; Carb, carbonate; Kfs, K-felspars; Qtza, Quartz authigeneous; Qtzm, Quartz monocrystalline; Qtzp, Quartz polycrystalline.
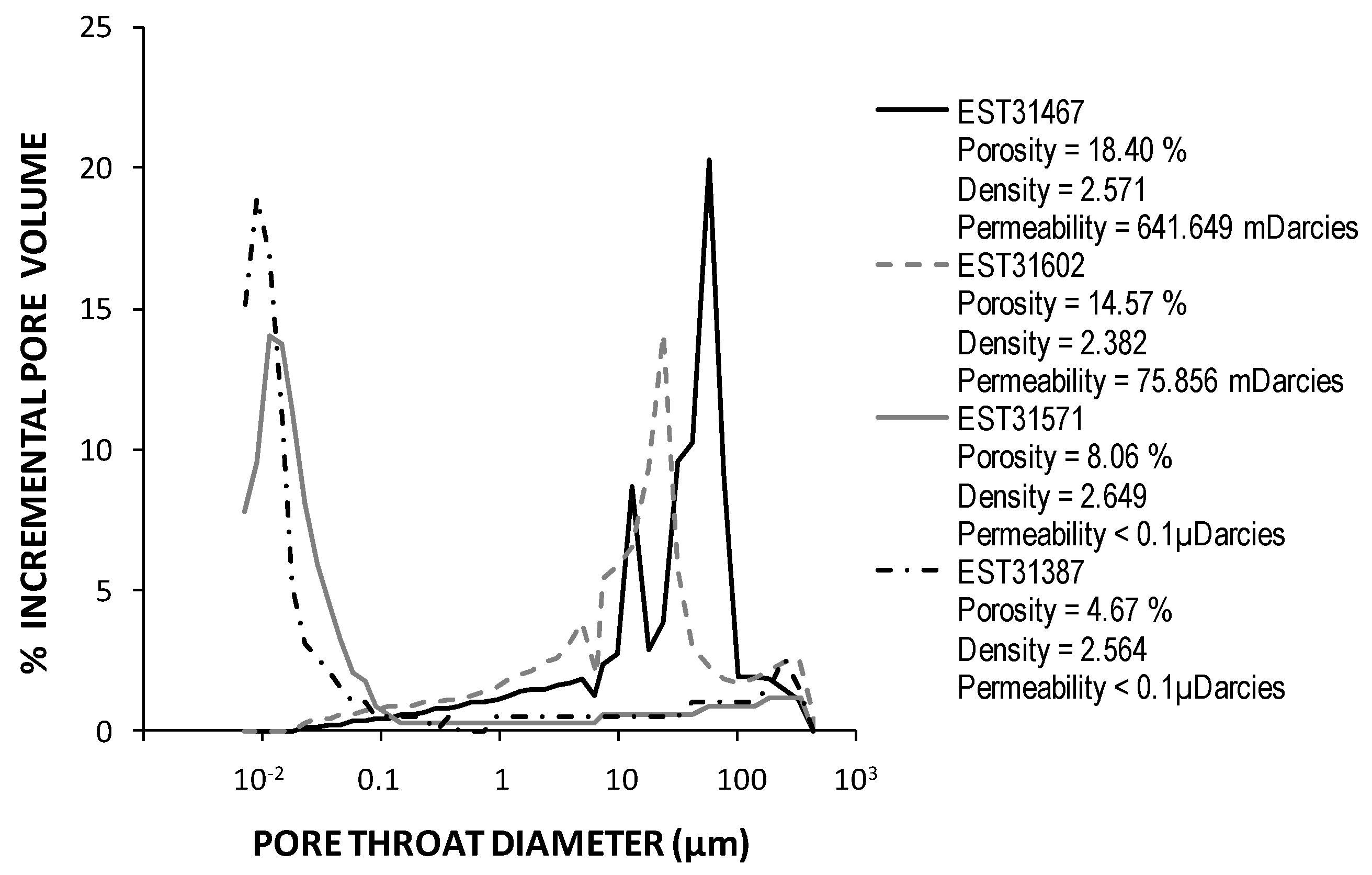
Rock porosities were estimated by mercury injection to be about 18, 15, 8, and 5%, for K2-20, K2-19, K2-18, and K2-17 cores, respectively. Triassic rock samples were classified into two groups: rocks with permeability lower than 0.1 µdarcy mainly formed by a pore system with diameters of less than 0.1 μm (K2-18 and K2-17 cores) and rocks with permeability higher than 50 mdarcy with pore diameters that are greater than 10 μm (K2-20 and K2-19 cores) (Figure 4).
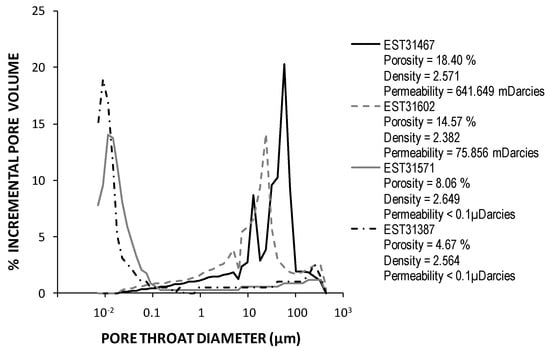
Figure 4.
Pore throat distribution of Triassic sandstones samples determined by Hg analyses. The permeability values were calculated from mercury porosimetry data according to the Katz and Thompson method [47].
No nitrate or nitrite was detected in Triassic rock leachates. As found in groundwater from Triassic formation EST03257 [48], sodium and chloride were amongst the main ionic compounds found in the four Triassic rock leachates with concentrations ranging from 104 to 191 mM and from 18 to 69 mM, respectively, together with sulfate (about 23–25 mM) and calcium K2-17 (respectively, 63 and 71 mM) and K2-18 rock leachates (Table 2). Groundwater from Triassic formation was more saline than leachates but higher amounts of sulfate were found in K2-18 and K2-17 rock leachates, about 23 and 25 mM, respectively, than in groundwater (12 mM). The groundwater salt content does not seem to be controlled by its host geological formation, except for the sulfate concentration exhibiting values of the same order of magnitude (9 for leachate and 12 for groundwater from Triassic formation). Sulfate concentrations could result from anhydrite dissolution, while other salts could be formed from the dissolution of various accumulations, such as over- and under-lying saline formations, for example. PHREEQC models estimated the pore water salinities of K2-18 and K2-17 samples to be, respectively, 365 and 100 g·L−1. The salinity of the groundwater from Triassic formation was 180 g·L−1 [20]. The amount of organic carbon (TOC) was below the quantification limit of the RE6 pyrolysis method (0.5%).

Table 2.
Geochemical analyses of pore water from Triassic sandstones and Triassic water (concentration in mM). Data of groundwater from Triassic formation were obtained from Rebeix et al. [48].
3.2. Exclusion of Potential Contamination Sources
The main source of potential contamination is the drilling fluids. Firstly, an estimation of the drilling fluid intrusion was performed for each core sample using nuclear magnetic resonance (NMR) and electrical resistivity patterns, which were acquired during the drilling procedure [22]. The low porosity and water mass content observed in NMR patterns indicated a very low drilling fluid intrusion in the K2-17 and K2-18 sample cores, estimated at about 1 cm maximum for the most porous K2-17 dolomitic parts. The mud-logging data (data not shown) of deeper geological formations K2-19 and K2-20 showed resistivity curves that were somewhat grouped, suggesting a minor drilling fluid intrusion of the order of one cm. In order to avoid potential microbial contamination from drilling fluid (limited to the first peripheral centimetre), the microbial diversity study was performed on inner cores only.
Secondly, one of the problems that was encountered in molecular analyses of low biomass environments is the misinterpretation of microbial community data by the detection of contaminant sequences [49,50]. To identify such sequences, a contaminants library was created by assembling the partial 16S rRNA gene sequences detected from drilling fluid, atmosphere of the anaerobic chamber, and all PCR negative controls (Table 3). It is of interest to stress that no amplification of the bacterial 16S rRNA gene was obtained directly from PCR negative controls; only weak amplifications were sometimes produced from nested PCR.

Table 3.
Distribution of contaminant phylotypes that were identified within control samples.
The contaminants library included 23 taxa that were affiliated to aerobic or facultative anaerobic bacterial species. Among them, 14 were often associated to Human beings, showing their allochthonous character. In contrast, the exogenous nature of some detected 16S rRNA gene sequences was more difficult to determine because of the metabolic properties of the associated bacteria. Indeed, some taxa were closely related to phylotypes adapted to oligotrophic environments; members of the genera Methylobacterium and Sphingomonas were able to survive in a nutrient limited clean room that was subjected to drastic cleaning procedures [51]. Some taxa were affiliated to members of the genus Bacillus, which have been retrieved from rocks or saline environments. This was particularly the case for taxa affiliated to B. halodurans and B. polygonii that were isolated from drilling fluids, previously isolated from soda lakes, hot springs, or sandstone ([52,53], GenBank EU676882). During the drilling procedure, drilling fluid were in direct contact with sedimentary formations in the borehole, thus they could remove microorganisms contained in host rocks or in pore water rocks. Thus, drilling fluid could reflect a mixture of indigenous and allochthonous microbial communities. In the present study, the drilling fluid 16S rRNA gene sequences were considered as contaminants. Although this library helped to discriminate between the indigenous or potentially indigenous bacterial communities of deep Triassic formations and allochthonous populations introduced during the drilling, handling, and analytical phases, it still remains difficult to validate the indigenous origin of the identified bacteria.
Thirdly, the absence from the inner Triassic cores of any viable and cultivable bacteria, including bacterial strains that could be recovered from drilling fluid, tends to show that the inner cores were uncontaminated by drilling fluid (see Section 3.3). Furthermore, low amounts of environmental DNA and limited biodiversity specifically adapted to extreme environments also suggest an absence of contamination within inner cores.
3.3. Microbial Enrichments from Triassic Sandstones
Despite more than 400 enrichments and several months of observation, no cultivable microorganisms either under aerobic or anaerobic conditions grew from any of the four inner Triassic core samples. Nevertheless, bacterial growth was observed in positive controls with known strains, native to deep environments, including Thermosipho japonicus (DSM 13481), Petrotoga mexicana (DSM 14811) and Desulfovibrio profundus (DSM 11384), and Triassic rocks, showing that inner cores were not toxic nor did they harbour substances that could inhibit microbial development.
The absence from the inner Triassic cores of any viable and cultivable bacteria support the hypothesis that there was no contamination from drilling fluids.
3.4. Molecular Biodiversity of Deep Triassic Formations
In deep consolidated environments, the low biomass and intrinsic adsorption properties of minerals forming geological formations limit the access to genetic material [49]. Among tests that were conducted to improve DNA quality, only a final dialysis step led to a better efficiency of 16S rRNA gene amplification, demonstrating a need for an additional purification step to obtain indigenous DNA suitable for further PCR-dependent studies on deep Triassic sandstone.
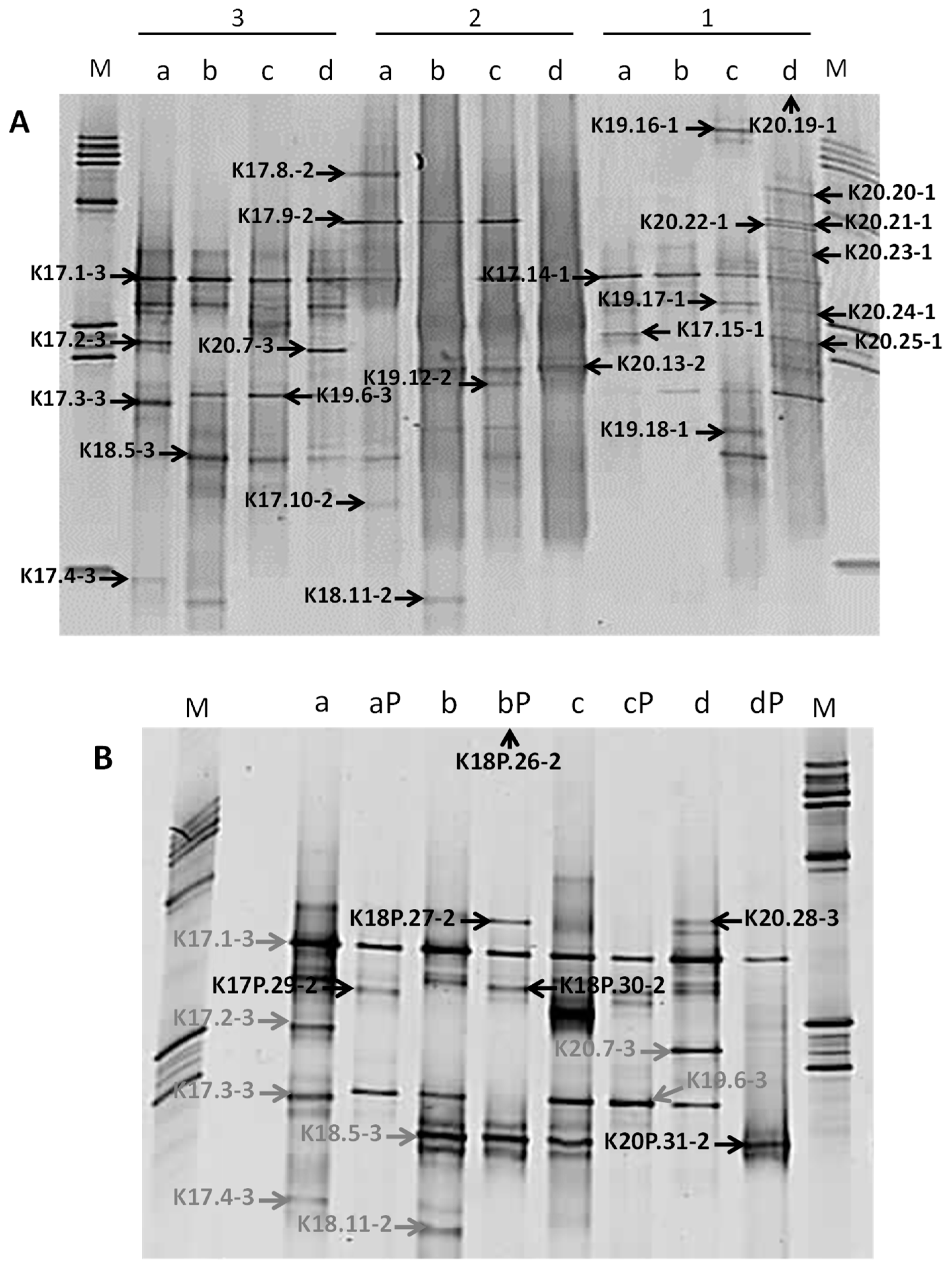
The molecular procedure was thus performed on dialysed DNA extracted from the inner cores, using the three DNA extraction protocols for samples that were stored at atmospheric pressure and only protocol 2 for samples stored under pressure because of the small amount of available sample. The diversity of the bacterial communities, as assessed by DGGE patterns of nested PCR products, was low. The most complex pattern showed 14 bands (Figure 5A). DNA extracted from samples preserved under pressure showed a least efficient 16S rRNA gene-nested-PCR amplification and the lowest bacterial diversity, suggesting that the repressurisation of samples may have decreased bacterial diversity (Figure 5B). Such a low degree of biodiversity was consistent with retrieved indigenous microbial communities from extreme environments, such as thermal springs [54], subsurface paleosols [55], or hypersaline environments and evaporites rocks [9]. Bacterial diversity also varied with samples and with DNA extraction protocols. Some bands were specific to a geological formation (DGGE band K18.11-2, Figure 5A) or to a DNA extraction procedure (DGGE band K20.7-3, Figure 5A). For the same Triassic rock sample, the parallel use of DNA extracted with three distinct protocols resulted in a better representation of the bacterial community. Several authors have previously shown a direct impact of DNA extraction methods on the estimation of microbial diversity targeted with 16S rRNA genes [56,57].
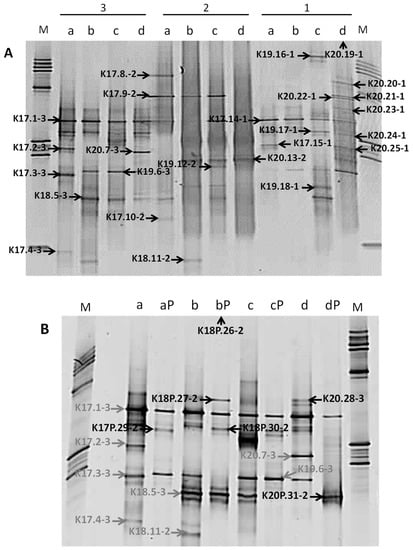
Figure 5.
DGGE profiles of bacterial 16S rRNA gene fragments from deep Triassic sandstones. (A) Bacterial diversity as assessed by three DNA extraction methods (1 to 3) from rocks stored under atmospheric pressure. (B) DGGE profiles of bacterial community from Triassic sandstones stored under high pressure. For samples stored under high pressure, environmental DNA was extracted with the method 2 only. Excised DGGE bands were identified with an arrow. M: ladder, a: K2-20 core (EST31467), b: K2-19 core (EST31602), c: K2-18 core (EST31571), d: K2-17 core (EST31387), P: stored under high pressure (190 bars).
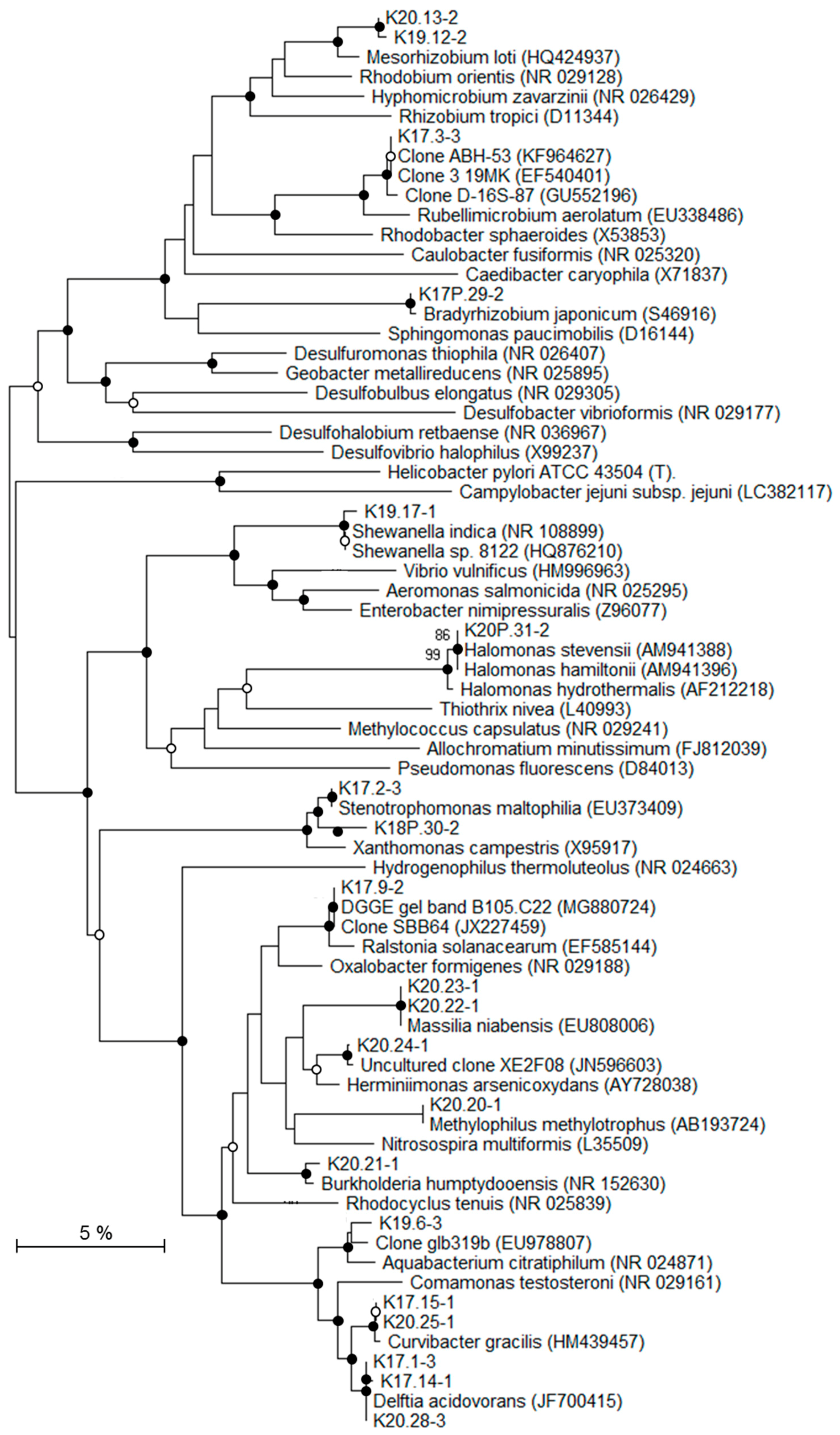
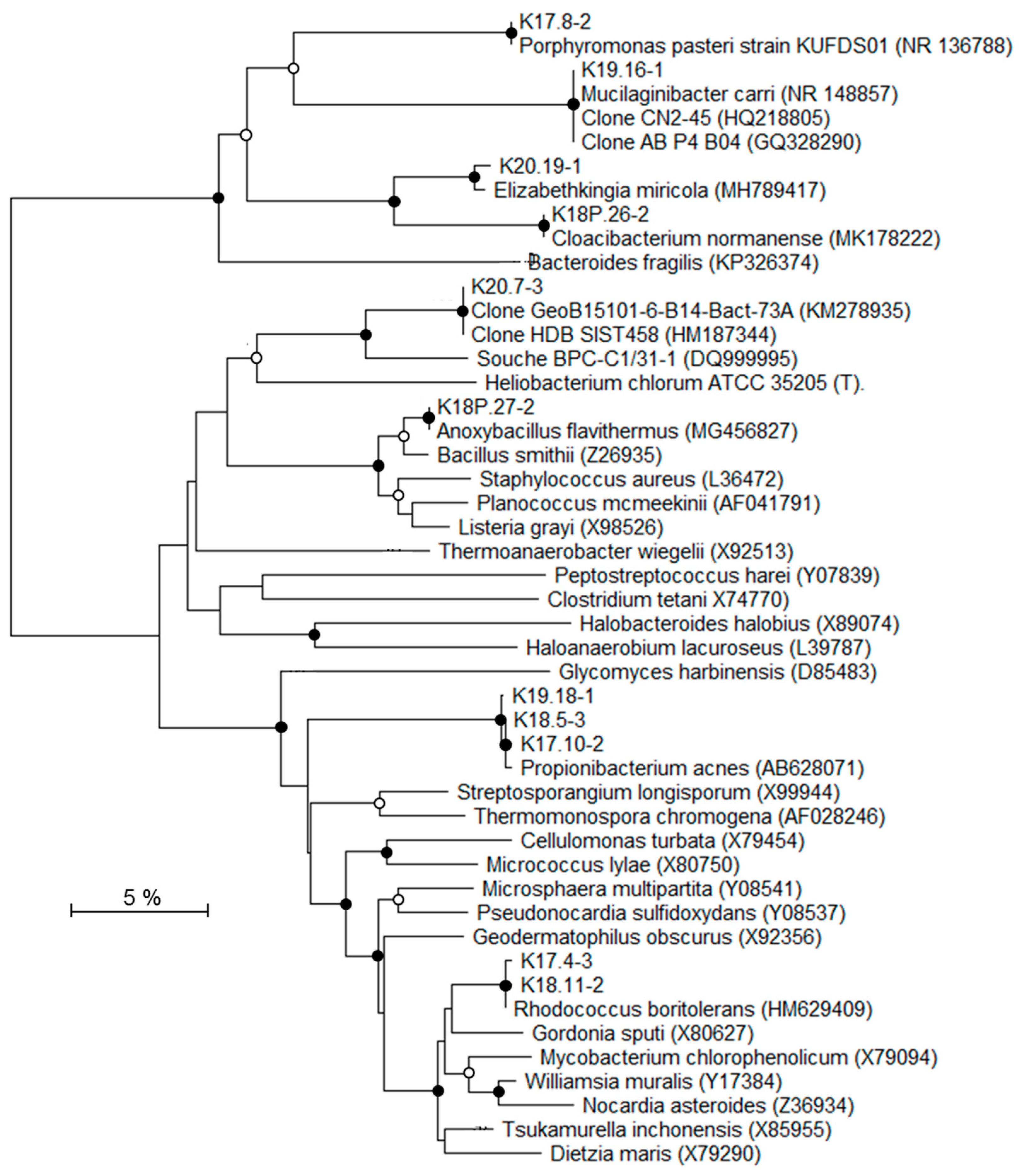
To identify the affiliations of the deep bacterial community, the most intense DGGE bands (n = 31) were considered and the derived 16S rRNA gene sequences were grouped into 22 distinct taxa (Table 4; Figure 6 and Figure 7). According to the classification of Gérard et al. [50], 16S rRNA gene sequences were considered either as potentially indigenous, if not found in the contaminant library of this study, but found once in previously published contaminant libraries, or as truly indigenous. Seven taxa (n = 9) fell into the potentially indigenous category. This was the case of the 16S rRNA gene sequence affiliated to Bradyrhizobium sp. S23321, an oligotrophic bacterium with an “ancestral-type genome” [58]. Sequences that are related to Bradyrhizobium were also retrieved in deep marine sediments [59]. Among the β–Proteobacteria, four taxa, related to Ralstonia, Aquabacterium, Curvibacter, and Methylophilus genera, were considered as potentially indigenous. Although, some members of Ralstonia, Aquabacterium, and Curvibacter were associated with laboratory contaminants in the literature [50], their distribution is widespread in many environments. Some Ralstonia spp. have specific metabolic and physiologic capabilities, such as hydrogenotrophy or heavy metal resistance, enabling them to live in harsh environmental conditions [60,61]. The Ralstonia-related sequence from K2-17 Trias core is highly related to clones retrieved from a hypersaline Tunisian Lake (Genbank MG880724) and from the deep-sea environment (Genbank JX227459). The ability of a Ralstonia to enter a dormant state may explain its survival and sustainability over geological times [62]. Members of Aquabacterium were often detected in low nutrient environments, including drinking, fresh water and glacier ice [63,64,65]. A Curvibacter sp. was recorded in the deep subsurface, at the interface between basaltic oceanic crust and deep seawater [66]. The taxon belonging to Methylophilus was closely related to sequences found in crude oil [67] and accretion ice samples from the subglacial Vostok Lake [68]. The two last taxa related to the potentially indigenous category were affiliated to Rhodococcus and Anoxybacillus. Marine strains of Rhodococcus have been isolated from deep-sea sediments [69] and they are well-known hydrocarbon-degrading bacteria, suggesting their capacity to survive on fossil fuel. Anoxybacillus spp. are aerobic spore-forming bacteria, often thermophilic, which have been isolated from thermal springs [70].

Table 4.
Phylogenetic analysis of bacterial 16S rRNA genes detected in Triassic sandstones by DGGE.
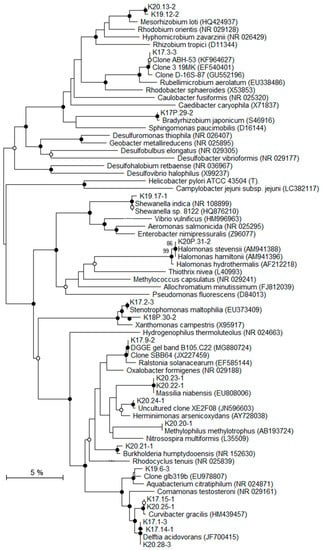
Figure 6.
Phylogenetic tree showing the relationships of recovered Proteobacteria-related 16S rRNA gene sequences with reference Genbank sequences. Black circles: bootstrap > 70%; white circles: 50% < bootstrap < 70% (1000 replicates).
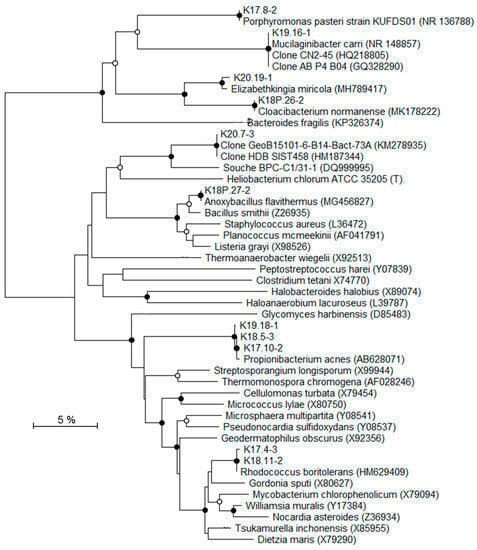
Figure 7.
Phylogenetic tree showing the relationships of recovered 16S rRNA gene sequences (excluding Proteobacteria-related sequences) with reference Genbank sequences. Black circles: bootstrap > 70%; white circles: 50% < bootstrap < 70% (1000 replicates).
Seven taxa (n = 8), affiliated to Firmicutes, α-, β, and γ-Proteobacteria, were detected exclusively in the inner Triassic cores and were thus regarded as truly indigenous in the deep bacterial community. These taxa were more or less distantly related to sequences retrieved from soils and deserts, but also from deep terrestrial environments, subsurface marine sediments, and hydrothermal fluids. The closest Firmicutes sequences were environmental clones found in oil reservoirs [71], subsurface terrestrial sediments [72], and subseafloor sediments (Genbank KM278935). The bacterial strain BPC-C1/31-1, isolated from a deep clay formation, was the closest known microorganism related to Firmicutes and it showed only 92% sequence similarity [24]. The α-Proteobacteria taxon affiliated to Mesorhizobium has been detected in seawater at 6000 m depth [73]. In a similar way, the Mesorhizobium-related taxon was mainly detected in saline aquifers from Triassic formations (K2-19 and K2-20 cores). Among β-Proteobacteria, organisms represented by the taxon that are related to Burkholderia seemed to be particularly well suited to live in subsurface sediments. Burkholderia spp. has been detected in Miocene terrestrial sediments [74]. Some species were able to use polycyclic aromatic hydrocarbons, such as naphthalene or phenanthrene, as a carbon source [75], suggesting, as for Rhodococcus-related taxon, their ability to survive using fossil fuel. Among the γ-Proteobacteria, two taxa were identified as indigenous to the drill cores. One of them was closely related to Shewanella indica, isolated from marine sediments [76] and a strain retrieved from a 2040 m deep marine sediment (GenBank HQ876210). One taxon that was retrieved from the K2-20 core is related to the halophilic Halomonas genus. Interestingly, several Halomonas strains have been isolated from deep-sea hydrothermal vents [77] and are able to grow in oligotrophic conditions and above seawater salinity, characteristics that are compatible with life in the studied core. Among the bacterial genera related to taxa from the Triassic sandstone, Halomonas is the only halophilic one that is known to grow at salt concentrations higher than seawater.
The eight remaining taxa (n = 14) were similar to taxa found in the contaminant library (this study) or had 16S rRNA gene sequences affiliated with bacteria found associated with human beings, and were considered as allochthonous in the deep geological formations.
Attempts to investigate the archaeal diversity were also made using both culture approaches and molecular ones, but no cultivable cells were detected and none or very weak 16S rRNA gene amplification were obtained (data not shown) and diversity studies were fruitless. Although we cannot totally exclude a technical reason, it appears that deep Triassic sandstones formations were mainly composed of Bacteria members, with Archaea constituting only a minority part in the deep Triassic microbial community.
Molecular fingerprinting by DGGE allowed for the comparison of the bacterial community structure of the four Trias cores, together with taxa identification, from band DNA recovery and sequencing, in five bacterial groups: α-, β-, and γ-Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes. If this fingerprinting approach has been widely used in diverse environments and until recently in the deep subsurface [78,79], it is, however, acknowledged that DGGE gives only a partial view of environmental diversity. Bomberg et al. (2014) [80] have compared molecular approaches to characterize deep terrestrial groundwater biosphere and highlighted the bias of DGGE, such as the detection of only dominant taxa and preferential recovery of certain taxa, such as α- and β-Proteobacteria. Extensive development of high-throughput gene sequencing during the past 10 years has led nowadays to its wide utilization in environmental microbial ecology. The low DNA recovery from the Trias cores was unfortunately not compatible with high-throughput sequencing, but efforts have now to be directed towards the application of such a sensitive and highly resolutive technique, to allow for deeper insights into the determination of native microbial diversity of the Trias cores, and more generally from the deep terrestrial subsurface.
4. Discussion
4.1. Life in Deep Geological Formations
From a microbiological point of view, deep Triassic formations could be seen as hot environments, highly salted, nutrient depleted, and subjected to important geostatic pressure. Deeper sandstones (KT-20 and KT-19 cores) present characteristics that are compatible to microbial colonisation. Pore size above 10 µm diameter, permeability above 50 mdarcy, and water contents of, respectively, 9.2% and 6.8% were considered to be sufficient to allow a flow of microorganisms [8,81] and an installation of microbial communities. On the contrary, clayey and carbonate sandstones (K2-18 and K2-17 cores) showed unfavourable properties for microbial growth. In addition to low water contents of close to 4% and less than 1%, respectively, as reported by Landrein et al. (2013) [22], the pore diameter of their network is so small that it prohibits the movement of microorganisms with diameters greater than 100 nm.
While the presence of microbial life in the depths of the Earth has been pointed out by many authors [5,10,76], a broad cultivation approach failed with deep Triassic sandstones in the present study. This lack of cultivable microorganisms could be explained by several hypotheses. First, the enrichment media used may have been inappropriate for obtaining growth and observing the activity of the resident microorganisms [50]. This possibility seemed very unlikely in view of the more than 400 enrichment media that were tested and their wide variety of possible metabolisms to be performed under anaerobic (various electron donors and electron acceptors tested) and aerobic conditions (various electron donors tested) as well. Second, the resident microbial communities may have grown slowly with low growth yields, making their detection impossible with current tools and methods [82]. Last, only very few or no viable prokaryotes were present in the deep Triassic sandstones. This absence of viability could be due to the combination of several stressful conditions [83], including in-situ temperature, salinity, and nutrient depletion, which may not be compatible for life. This is the case in oil reservoirs, considered to be a closed anaerobic system in which temperatures over 80 °C and salinities of 15–20% does not sustain life [11,84]. In this respect, our inability to recover native microorganisms from deep Triassic sandstones by enrichment cultures at high temperature and salinity is consistent with earlier failed attempts of recovery [84]. It is noteworthy that the maximal burial temperature of the Lower Triassic sandstones was estimated at 90 °C (±10 °C), while using a set of several independent paleothermometers [85], from about 65 million years ago. When considering such maximal paleo-temperatures, the deep Triassic formations may have been subjected to a paleo-sterilisation [86]. As indicated above, the paleo-sterilisation and the current extreme environmental conditions (salinity and temperature) could have finally prohibited microbial colonisation. This last hypothesis was previously put forward to explain the lack of prokaryotic biomarkers and cultivable microorganisms in 2091 mbsl sandstone [21].
4.2. Origins of the Indigenous Bacterial Community
In this study, most of the bacterial sequences showed more than 99% similarity to contemporary species, possibly because of their direct interaction with current ecosystems, as suggested by Gérard et al. in 2009 [50]. The “indigenous” and “potentially indigenous” 16S rRNA gene sequences belonged to Firmicutes and α-, β-, and γ-Proteobacteria. These phylogenetic groups are ubiquitous in surface environments but sequences that are related to Shewanella, Mezorhizobium, and Curvibacter have also been found in deep marine sediments [66,73,76] and those related to Methylophilus and Rhodococcus were found in oil fields and mines [67,69]. One of the Firmicute-related sequences, (K20.17-3), comes from bacteria that are cultivated from petroleum crude oil and subsurface sediments at the Hanford Site 300 Area [72,87] and seems to be restricted to the subsurface confined biosphere, arguing in favor of its indigenous origin. Some 16S rRNA sequences were also close to species with metabolic abilities that are necessary for sustaining life in deep confined compartments over geological times. This is the case for the sequences that are related to Ralstonia and Burkolderia [62,75].
Several hypotheses may explain the results that were obtained in this study. One is that the extracted DNA could have originated from viable and metabolically active but not cultivable bacteria, or dormant bacteria with low or no metabolic activity. A second hypothesis was put forward by Fish et al. in 2002 [88], when they found 16S rRNA sequences from ancient halite to be phylogenetically close to species that were detected in current environments. They explained these low divergences by a slow genetic evolution of some phylogenetic lineages and a preservation of biological material, such as microorganisms and DNA in saline rocks over geological times. Finally, it has also been reported that fossil DNA sequestered in intact cells or free in the environment could be a source of environmental DNA [88]. Indeed, several authors have suggested that at high salinities, such as deep Triassic sandstones, hypersaline oil reservoir core where aerobic hyperhalophilic Archaea were detected [89], or under desiccation conditions, DNA could be preserved for more than 10 million years [88,90].
However, there is also a controversial hypothesis with the theoretical prediction for which environmental DNA cannot persist for more than one million years [71]. Indeed, a potential contamination of Triassic formations by modern microorganisms could have occurred during geological events or during water flow through aquifer formations. Thus, the age of the environmental DNA may be more recent than that of the surrounding rocks and may have derived from dead bacteria that lived or circulated more recently through deep geological formations.
In this study, the clayed sandstone formations (cores K2-17, 1727 mbsl, and K2-18, 1804 mbsl) had very small pore sizes (<100 nm) (Figure 4). These two geological formations could be considered as natural microbial filters and exclude microbial community exchanges by passive diffusion phenomena between Triassic aquifers and upper geological formations. Despite their low porosity, the existence of micro-niches in clayey Triassic sandstone where microorganisms and various bio signatures could be sequestered and conserved is also possible. The 16S rRNA gene sequence related to the ancestral type genome of Bradyrhizobium sp. S23321, which is only found in the K2-17 core (1727 mbsl), could be evidence of this sequestration. These two clayey Triassic sandstones belong to the low permeability Triassic cap rocks that isolate Triassic aquifers from water leakage from the surface or from a more superficial productive aquifer, like the Dogger aquifer [63]. In the aquitard compartment, Marty et al. [91] supposed diffusive transfers to be negligible over a time scale greater than several million years. This hypothesis has been validated by Rebeix et al. [48] who have not detected any local connections between the different aquifers under the EST433 drilling. Furthermore, the groundwater recharge of Triassic aquifers of the Paris Basin may have occurred exclusively in faults and outcrop areas from Vosges and Morvan that are located in the eastern part of the basin (Lorraine region, France) [91,92]. According to Fontes and Matray [93], groundwater from Triassic formation could have been the outcome of highly evolved seawater diluted with a secondary brine derived from salt dissolution of upper saline geological formations or Permian basements by more recent continental or meteoritic water. Rebeix et al. [48] also suggested this possibility based on the high chloride and bromide content and the δ18O and δ2H values of the groundwater from Triassic formation collected from the EST433 borehole. Its Cl/Br molar ratio value was 167 and it corresponds to primary brine [93]. By contrast, values of δ18O and δ2H suggested a meteoritic water supply.
In the same way, the detection of non-halophilic and halophilic species from the same inner core samples suggested a fluid mixture with different salt contents (from fresh water to brine). The prokaryotic community from Triassic sandstones of the EST433 drilling could have come from upper geological formations leached by paleo-recharges and from recharge areas.
Interestingly, in this study we detected two sequences found in ice environments (Methylophilus, Aquabacterium). One explanation for this could be that the paleo-recharge partly resulted from melting ice that formed in the last Quaternary ice age, and especially during the period between the Holocene and the Pleistocene, less than one million years ago [92]. The possibility that a groundwater from the Triassic formation recharged under cold conditions has also been raised by Rebeix et al. [48]. Thus, some of the microorganisms that were detected in deep Triassic sandstones would then appear to be younger than surrounding rocks, but still far older than contemporary periods.
5. Conclusions
In the four Triassic rock cores that were harvested from down to 2000 m depth, we found a low biomass consistent with previous findings. Molecular approaches evidenced eight genera that were considered as truly indigenous and belonging to α, β, and γ-Proteobacteria and Firmicutes, all more or less distantly related to genera that are found in the deep subsurface. Despite extensive cultivation approaches, no cultivable bacteria were retrieved. This raises many questions about metabolic states, adaptive capacities, and origins of these resident microorganisms. Physicochemical constraints combining high temperature, salinity, high hydrostatic pressures, anoxia, and possibly geological events might have had a direct impact on deep indigenous microbial communities. A study of the microbial behavior in response to the change of environmental constraints induced by a geological event could help us to improve our knowledge on the potential metabolic and physiological abilities of deep microorganisms and help to establish the boundaries of life in the deep subsurface.
Author Contributions
Funding acquisition, C.S. and F.G.; Investigation, V.L., J.H., M.-L.F., S.K., B.O. and C.J.; Methodology, V.L., J.H., C.S. and C.J.; Project administration, F.G.; Supervision, C.S., F.G. and C.J.; Writing—original draft, V.L., J.H., B.O. and C.J.; Writing—review & editing, J.H., C.S., B.O. and C.J.
Funding
This research was funded by ANDRA, CNRS, and BRGM by GNR Forpro, and TAPSS 2000 research program «Present and past transfers in a sedimentary aquifer–aquitard system: a 2000 m deep drill-hole in the Mesozoic of the Paris Basin».
Acknowledgments
Authors would like to thank ANDRA for providing samples and informations. We thank C. Le Hénaff-Le Marrec of the Polytechnical Institute of Bordeaux for assistance and equipment loans during the project, V. Jean-Prost of the LAB Division of the BRGM for help with XRD analyses, C. Lerouge and P. Blanc of the SVP Unit of the BRGM for their help with petrography and modelling.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Kashefi, K.; Lovley, D.R. Extending the upper temperature limit for life. Science 2003, 301, 934. [Google Scholar] [CrossRef] [PubMed]
- Takai, K.; Nakamura, K.; Toki, T.; Tsunogai, U.; Miyazaki, M.; Miyazaki, J.; Hirayama, H.; Nakagawa, S.; Nunoura, T.; Horikoshi, K. Cell proliferation at 122 °C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc. Natl. Acad. Sci. USA 2008, 105, 10949–10954. [Google Scholar] [CrossRef] [PubMed]
- Amend, J.P.; Teske, A. Expanding frontiers in deep subsurface microbiology. Palaeogeogr. Palaeoclimatol. Palaeoecol. 2005, 219, 131–155. [Google Scholar] [CrossRef]
- Miranda-Tello, E.; Fardeau, M.-L.; Thomas, P.; Ramirez, F.; Casalot, L.; Cayol, J.-L.; Garcia, J.-L.; Ollivier, B. Petrotoga mexicana sp. nov., a novel thermophilic, anaerobic and xylanolytic bacterium isolated from an oil-producing well in the Gulf of Mexico. Int. J. Syst. Evol. Microbiol. 2004, 54, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Szewzyk, U.; Szewzyk, R.; Stenström, T.A. Thermophilic, anaerobic bacteria isolated from a deep borehole in granite in Sweden. Proc. Natl. Acad. Sci. USA 1994, 91, 1810–1813. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Phillips, R.; Milo, R. The biomass distribution on Earth. Proc. Natl. Acad. Sci. USA 2018. [Google Scholar] [CrossRef] [PubMed]
- Magnabosco, C.; Lin, L.-H.; Dong, H.; Bomberg, M.; Ghiorse, H.; Stan-Lotter, H.; Pedersen, K.; Kieft, T.L.; van Heerden, E.; Onstott, T.C. The biomass and biodiversity of the continental subsurface. Nat. Geosci. 2018, 11, 707–717. [Google Scholar] [CrossRef]
- Fredrickson, J.K.; Balkwill, D.L. Geomicrobial processes and biodiversity in the deep terrestrial subsurface. Geomicrobiol. J. 2006, 23, 345–356. [Google Scholar] [CrossRef]
- Oren, A. Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 1999, 63, 334–348. [Google Scholar]
- Krumholz, L.R.; McKinley, J.P.; Ulrich, G.A.; Suflita, J.M. Confined subsurface microbialcommunities in Cretaceous rock. Nature 1997, 386, 64–66. [Google Scholar] [CrossRef]
- Magot, M.; Ollivier, B.; Patel, B.K.C. Microbiology of petroleum reservoirs. Antonie Leeuwenhoek 2000, 77, 103–116. [Google Scholar] [CrossRef]
- Pedersen, K. Exploration of deep intraterrestrial microbial life: Current perspectives. FEMS Microbiol. Lett. 2000, 185, 9–16. [Google Scholar] [CrossRef] [PubMed]
- D’Hondt, S.; Jørgensen, B.B.; Miller, D.J.; Batzke, A.; Blake, R.; Cragg, B.A.; Cypionka, H.; Dickens, G.R.; Ferdelman, T.; Hinrichs, K.-U.; et al. Distributions of microbial activities in deep subseafloor sediments. Science 2004, 306, 2216–2221. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.F. Microbial life breathes deep. Science 2004, 306, 2198–2200. [Google Scholar] [CrossRef] [PubMed]
- Basso, O.; Lascourrèges, J.-F.; Jarry, M.; Magot, M. The effect of cleaning and disinfecting the sampling well on the microbial communities of deep subsurface water samples. Environ. Microbiol. 2005, 7, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Fardeau, M.-L.; Goulhen, F.; Bruschi, M.; Khelifi, N.; Cayol, J.-L.; Ignatiadis, I.; Guyot, F.; Ollivier, B. Archaeoglobus fulgidus and Thermotoga elfii, thermophilic isolates from deep geothermal water of the Paris Basin. Geomicrobiol. J. 2009, 26, 119–130. [Google Scholar] [CrossRef]
- Onstott, T.C.; Moser, D.P.; Pfiffner, S.M.; Fredrickson, J.K.; Brockman, F.J.; Phelps, T.J.; White, D.C.; Peacock, A.; Balkwill, D.; Hoover, R.; et al. Indigenous and contaminant microbes in ultradeep mines. Environ. Microbiol. 2003, 5, 1168–1191. [Google Scholar] [CrossRef] [PubMed]
- Purkamo, L.; Bomberg, M.; Nyyssönen, M.; Kukkonen, I.; Ahonen, L.; Itävaara, M. Heterotrophic communities supplied by ancient organic carbon predominate in deep Fennoscandian bedrock fluids. Microb. Ecol. 2015, 69, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Onstott, T.; Phelps, T.J.; Colwell, F.; Ringelberg, D.; White, D.; Boone, D.; Mckinley, J.P.; Stevens, T.O.; Long, P.E.; Balkwill, D. Observations pertaining to the origin and ecology of microorganisms recovered from the deep subsurface of Taylorsville Basin, Virginia. Geomicrobiol. J. 1998, 15, 353–385. [Google Scholar] [CrossRef]
- Zhang, G.; Dong, H.; Xu, Z.; Zhao, D.; Zhang, C. Microbial diversity in ultra-high-pressure rocks and fluids from the chinese continental scientific drilling project in China. Appl. Environ. Microbiol. 2005, 71, 3213–3227. [Google Scholar] [CrossRef]
- Colwell, F.S.; Onstott, T.C.; Delwiche, M.E.; Chandler, D.; Fredrickson, J.K.; Yao, Q.-J.; McKinley, J.P.; Boone, D.R.; Griffiths, R.; Phelps, T.J.; et al. Microorganisms from deep, high temperature sandstones: Constraints on microbial colonization. FEMS Microbiol. Rev. 1997, 20, 425–435. [Google Scholar] [CrossRef]
- Landrein, P.; Vigneron, G.; Delay, V.; Lebon, P.; Pagel, M. Lithologie, hydrodynamisme et thermicité dans le système sédimentaire multicouche recoupé par les forages Andra de Montiers-sur-Saulx (Meuse). Bull. de la Société Géologique de Fr. 2013, 184, 519–543. [Google Scholar] [CrossRef]
- Brunet, M.-F.; Le Pichon, V. Subsidence of the Paris Basin. J. Geophys. Res. 1982, 87, 8547–8560. [Google Scholar] [CrossRef]
- Poulain, S.; Sergeant, C.; Simonoff, M.; Le Marrec, C.; Altmann, S. Microbial investigations in Opalinus Clay, an argillaceous formation under evaluation as a potential host rock for a radioactive waste repository. Geomicrobiol. J. 2008, 25, 240–249. [Google Scholar] [CrossRef]
- NF T 90-008 2001. Water Quality—Determination of pH; AFNOR: Paris, France, 2001; ISBN 2-12-213141-1. (In French) [Google Scholar]
- ISO 11732 2005. Water Quality—Measurement of Ammonium—Method by Flow Analysis (CFA and FIA) and Spectrometrical Detection; AFNOR: Paris, France, 2005; ISBN 2-12-213141-1. (In French) [Google Scholar]
- ISO 9963-1 1994. Water Quality—Determination of Alkalinity—Part 1: Determination of Total and Composite Alkalinity; AFNOR: Paris, France, 1994; ISBN 2-12-213141-1. (In French) [Google Scholar]
- ISO 10304-1 2010. Water Quality—Measurement of Dissolved Anions by Ionic Chromatography in Liquid Phase—Part 1: Measurement of Bromide, Chloride, Fluoride, Nitrate, Nitrite, Phosphate and Sulphate; AFNOR: Paris, France, 2010; ISBN 2-12-213141-1. (In French) [Google Scholar]
- NF T 01-041 1975. Atomic Absorption Spectrometry—Use Introduction; AFNOR: Paris, France, 1975; ISBN 2-12-213141-1. (In French) [Google Scholar]
- Copard, Y.; Di-Giovanni, C.; Martaud, T.; Albéric, P.; Olivier, J.-E. Using Rock-Eval 6 pyrolysis for tracking fossil organic carbon in modern environments: Implications for the roles of erosion and weathering. Earth Surf. Process. Landf. 2006, 31, 135–153. [Google Scholar] [CrossRef]
- EN ISO 11885. Water Quality—Determination of Selected Elements by Inductively Coupled Plasma Optical Emission Spectrometry (ICP-OES); AFNOR: Paris, France, 2009. (In French) [Google Scholar]
- Blanc, P.; Lassin, A.; Piantone, P.; Azaroual, M.; Jacquemet, N.; Fabbri, A.; Gaucher, E.C. Thermoddem: A geochemical database focused on low temperature water/rock interactions and waste materials. Appl. Geochem. 2012, 27, 2107–2116. [Google Scholar] [CrossRef]
- Parkhurst, D.L.; Appelo, C.A.J. User’s Guide to Phreeqc (Version 2)—A Computer Program for Speciation, Batch-Reaction, One-Dimensional Transport, and Inverse Geochemical Calculations; Water-Resources Investigations Report; U.S. Department of the Interior, USGS: Denver, CO, USA, 1999; pp. 99–4259.
- Miller, T.L.; Wolin, M.J. A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl. Microbiol. 1974, 27, 985–987. [Google Scholar] [PubMed]
- Kallmeyer, J.; Ferdelman, T.G.; Jansen, K.H.; Jørgensen, B.B. A high-pressure thermal gradient block for investigating microbial activity in multiple deep-sea samples. J. Microbiol. Methods 2003, 55, 165–172. [Google Scholar] [CrossRef]
- Lerner, A.; Shor, Y.; Vinokurov, A.; Okon, Y.; Jurkevitch, E. Can denaturing gradient gel electrophoresis (DGGE) analysis of amplified 16s rDNA of soil bacterial populations be used in forensic investigations? Soil Biol. Biochem. 2006, 38, 1188–1192. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley Press: Chichester, UK, 1991; pp. 130–141. [Google Scholar]
- Schäfer, H.; Muyzer, G. Denaturing gradient gel electrophoresis in marine microbial ecology. Mar. Microbiol. 2001, 30, 425–468. [Google Scholar]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Zumstein, E.; Moletta, R.; Godon, J.-J. Examination of two years of community dynamics in an anaerobic bioreactor using fluorescence polymerase chain reaction (PCR) single-strand conformation polymorphism analysis. Environ. Microbiol. 2000, 2, 1462–2920. [Google Scholar] [CrossRef]
- Huber, T.; Faulkner, G.; Hugenholtz, P. Bellerophon: A program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 2004, 20, 2317–2319. [Google Scholar] [CrossRef] [PubMed]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Katz, A.J.; Thompson, A.H. Quantitative prediction of permeability in porous rock. Phys. Rev. Ser. B 1986, 34, 8179–8191. [Google Scholar] [CrossRef]
- Rebeix, R.; Le Gal La Salle, C.; Verdoux, P.; Gianesinni, S.; Lancelot, J.; Michelot, J.-L.; Noret, A.; Monvoisin, G.; Simler, R. Tracing the origin of water and solute transfers in deep groundwater from Oxfordian, Dogger and Trias formations in the east of the Paris Basin-France. Phys. Chem. Earth 2011, 36, 1496–1510. [Google Scholar] [CrossRef]
- Barton, H.A.; Taylor, N.M.; Lubbers, B.R.; Pemberton, A.C. DNA extraction from low-biomass carbonate rock: An improved method with reduced contamination and the low-biomass contaminant database. J. Microbiol. Methods 2006, 66, 21–31. [Google Scholar] [CrossRef]
- Gérard, E.; Moreira, D.; Philippot, P.; van Kranendonk, M.J.; López-García, P. Modern subsurface bacteria in pristine 2.7 Ga-old fossil xtromatolite drillcore samples from the fortescue group, Western Australia. PLoS ONE 2009, 4, e5298. [Google Scholar] [CrossRef]
- Moissl, C.; Osman, S.; La Duc, M.T.; Dekas, A.; Brodie, E.; DeSantis, T.; Venkateswaran, K. Molecular bacterial community analysis of clean rooms where spacecraft are assembled. FEMS Microbiol. Ecol. 2007, 61, 509–521. [Google Scholar] [CrossRef]
- Hashim, S.O.; Delgado, O.; Hatti-Kaul, R.; Mulaa, F.J.; Mattiasson, B. Starch hydrolysing Bacillus halodurans isolates from a Kenyan soda lake. Biotechnol. Lett. 2004, 26, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Ulfah, M.; Helianti, I.; Wahyuntari, B.; Nurhayati, N. Characterization of a new thermoalkalophilic xylanase-producing bacterial strain isolated from Cimanggu hot spring, west Java, Indonesia. Microbiol. Indones. 2012, 5, 139–143. [Google Scholar] [CrossRef]
- Reysenbach, A.-L.; Ehringer, M.; Hershberger, K. Microbial diversity at 83 °C in Calcite Springs, Yellowstone National Park: Another environment where the Aquificales. Extremophiles 2000, 4, 61–67. [Google Scholar] [PubMed]
- Chandler, D.P.; Brockman, F.J.; Fredrickson, J.K. Use of 16S rDNA clone libraries to study changes in a microbial community resulting from ex situ perturbation of a subsurface sediment. FEMS Microbiol. Rev. 1997, 20, 217–230. [Google Scholar] [CrossRef]
- Luna, G.M.; Dell’Anno, A.; Danovaro, R. DNA extraction procedure: A critical issue for bacterial diversity assessment in marine sediments. Environ. Microbiol. 2006, 8, 308–320. [Google Scholar] [CrossRef]
- Martin-Laurent, F.; Philippot, L.; Hallet, S.; Chaussod, R.; Germon, J.C.; Soulas, G.; Catroux, G. DNA extraction from soils: Old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 2001, 67, 2354–2359. [Google Scholar] [CrossRef]
- Okubo, T.; Tsukui, T.; Maita, H.; Okamoto, S.; Oshima, K.; Fujisawa, T.; Saito, A.; Futamata, H.; Hattori, R.; Shimomura, Y. Complete genome sequence of Bradyrhizobium sp. S23321: Insights into symbiosis evolution in soil oligotrophs. Microbes Environ. 2012, 27, 306–315. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Weightman, A.J.; Cragg, B.A.; Parkes, R.J.; Fry, J.C. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia Margin as revealed by 16S rRNA molecular analysis. FEMS Microbiol. Ecol. 2001, 34, 221–228. [Google Scholar] [CrossRef]
- Mergeay, M.; Monchy, S.; Vallaeys, T.; Auquier, V.; Benotmane, A.; Bertin, P.; Taghavi, S.; Dunn, J.; van der Lelie, D.; Wattiez, R. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: Towards a catalogue of metal-responsive genes. FEMS Microbiol. Rev. 2003, 27, 385–410. [Google Scholar] [CrossRef]
- Tiemeyer, A.; Link, H.; Weuster-Botz, D. Kinetic studies on autohydrogenotrophic growth of Ralstonia eutropha with nitrate as terminal electron acceptor. Appl. Microbiol. Biotechnol. 2007, 76, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Grey, B.E.; Steck, T.R. The viable but nonculturable state of Ralstonia solanacearum may be involved in long-term survival and plant infection. Appl. Environ. Microbiol. 2001, 67, 3866–3872. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.M.; Cho, N.-T.; Yang, S.-H.; Arun, A.; Young, C.-C.; Sheu, S.-Y. Aquabacterium limnoticum sp. nov., isolated from a freshwater spring. Int. J. Syst. Evol. Microbiol. 2012, 62, 698–704. [Google Scholar] [CrossRef] [PubMed]
- Kalmbach, S.; Manz, W.; Wecke, J.; Szewzyk, U. Aquabacterium gen. nov., with description of Aquabacterium citratiphilum sp. nov., Aquabacterium parvum sp. nov. and Aquabacterium commune sp. nov., three in situ dominant bacterial species from the Berlin drinking water system. Int. J. Syst. Bacteriol. 1999, 49, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Wiezer, A.; Strittmatter, A.W.; Daniel, R. Phylogenetic diversity and metabolic potential revealed in a glacier ice metagenome. Appl. Environ. Microbiol. 2009, 75, 7519–7526. [Google Scholar] [CrossRef] [PubMed]
- Santelli, C.M.; Orcutt, B.N.; Banning, E.; Bach, W.; Moyer, C.L.; Sogin, M.L.; Staudigel, H.; Edwards, K.J. Abundance and diversity of microbial life in ocean crust. Nature 2008, 453, 653–656. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.-C.; Liu, Z.-S.; Guo, P.; Chi, C.-Q.; Chen, J.; Wang, X.-B.; Tang, Y.-Q.; Wu, X.-L.; Liu, C.-Z. Bacteria in crude oil survived autoclaving and stimulated differentially by exogenous bacteria. PLoS ONE 2012, 7, e40842. [Google Scholar] [CrossRef] [PubMed]
- Christner, B.C.; Royston-Bishop, G.; Foreman, C.M.; Arnold, B.R.; Tranter, M.; Welch, K.A.; Lyons, W.B.; Tsapin, A.I.; Studinger, M.; Priscu, J.C. Limnological conditions in subglacial lake Vostok, Antarctica. Limnol. Oceanogr. 2006, 51, 2485–2501. [Google Scholar] [CrossRef]
- Colquhoun, J.A.; Heald, S.C.; Li, L.; Tamaoka, J.; Kato, C.; Horikoshi, K.; Bull, A.T. Taxonomy and biotransformation activities of some deep-sea actinomycetes. Extremophiles 1998, 2, 269–277. [Google Scholar] [CrossRef]
- Kevbrin, V.V.; Zengler, K.; Lysenko, A.M.; Wiegel, J. Anoxybacillus kamchatkensis sp. nov., a novel thermophilic facultative aerobic bacterium with a broad pH optimum from the Geyser valley, Kamchatka. Extremophiles 2005, 9, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Willerslev, E.; Cooper, A. Ancient DNA. Proc. R. Soc. B 2005, 272, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Kennedy, D.; Fredrickson, J.; Bjornstad, B.; Konopka, A. Vertical stratification of subsurface microbial community composition across geological formations at the Hanford Site. Environ. Microbiol. 2012, 14, 414–425. [Google Scholar] [CrossRef]
- Eloe, E.A.; Shulse, C.N.; Fadrosh, D.W.; Williamson, S.J.; Allen, E.E.; Bartlett, D.H. Compositional differences in particle-associated and free-living microbial assemblages from an extreme deep-ocean environment. Environ. Microbiol. Rep. 2011, 3, 449–458. [Google Scholar] [CrossRef]
- Elhottová, D.; Krištůfek, V.; Frouz, J.; Nováková, A.; Chroňáková, A. Screening for microbial markers in Miocene sediment exposed during open-cast brown coal mining. Antonie Leeuwenhoek 2006, 89, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.S.; Herrick, J.B.; Jeon, C.O.; Hinman, D.E.; Madsen, E.L. Horizontal transfer of phnAc dioxygenase genes within one of two phenotypically and genotypically distinctive naphthalene-degrading guilds from adjacent soil environments. Appl. Environ. Microbiol. 2003, 69, 2172–2181. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Pandey, P.K.; Gupta, A.K.; Kim, H.J.; Baik, K.S.; Seong, C.N.; Patole, M.S.; Shouche, Y.S. Shewanella indica sp. nov., isolated from sediment of the Arabian Sea. Int. J. Syst. Evol. Microbiol. 2011, 61, 2058–2064. [Google Scholar] [CrossRef] [PubMed]
- Kaye, J.Z.; Márquez, M.C.; Ventosa, A.; Baross, J.A. Halomonas neptunia sp. nov., Halomonas sulfidaeris sp. nov., Halomonas axialensis sp. nov. and Halomonas hydrothermalis sp. nov.: Halophilic bacteria isolated from deep-sea hydrothermal-vent environments. Int. J. Syst. Evol. Microbiol. 2004, 54, 499–511. [Google Scholar] [CrossRef]
- Lauer, A.; Sørensen, K.B.; Teske, A. Phylogenetic Characterization of Marine Benthic Archaea in Organic-Poor Sediments of the Eastern Equatorial Pacific Ocean (ODP Site 1225). Microorganisms 2016, 4, 32. [Google Scholar] [CrossRef]
- Frank, Y.A.; Kadnikov, V.V.; Gavrilov, S.N.; Banks, D.; Gerasimchuk, A.L.; Podosokorskaya, O.A.; Merkel, A.Y.; Chernyh, N.A.; Mardanov, A.V.; Ravin, N.V.; et al. Stable and Variable Parts of Microbial Community in Siberian Deep Subsurface Thermal Aquifer System Revealed in a Long-Term Monitoring Study. Front. Microbiol. 2016. [Google Scholar] [CrossRef]
- Bomberg, M.; Nyyssönen, M.; Nousiainen, A.; Hultman, J.; Paulin, L.; Auvinen, P.; Itävaara, M. Evaluation of Molecular Techniques in Characterization of Deep Terrestrial Biosphere. Open J. Ecol. 2014, 4, 468–487. [Google Scholar] [CrossRef]
- Jenneman, G.E.; McInerney, M.J.; Knapp, R.M. Microbial penetration through nutrient-saturated berea sandstone. Appl. Environ. Microbiol. 1985, 50, 383–391. [Google Scholar] [PubMed]
- Alain, K.; Querellou, J. Cultivating the uncultured: Limits, advances and future challenges. Extremophiles 2009, 13, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Ollivier, B.; Alazard, D. The oil reservoir ecosystem. In Handbook of Hydrocarbon Microbiology: Microbial Interactions with Hydrocarbons, Oils, Fats and Related Hydrophobic Substrates and Products; Timmis, K.N., Ed.; Springer: Berlin, Bermany, 2010; pp. 2262–2268. [Google Scholar]
- McGenity, T.J.; Gemmell, R.T.; Grant, W.D.; Stan-Lotter, H. Origins of halophilic microorganisms in ancient salt deposits. Environ. Microbiol. 2000, 2, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Blaise, T.; Barbarand, J.; Kars, M.; Ploquin, F.; Aubourg, C.; Bribaud, B.; Cathelineau, M.; El Albani, A.; Gautheron, C.; Izart, A.; et al. Reconstruction of low burial (<100 °C) in sedimentary basins: A comparison of geothermometer sensibility in the intracontinental Paris Basin. Marine Petroleum Geol. 2014, 53, 71–87. [Google Scholar] [CrossRef]
- Wilhelm, A.; Larter, S.R.; Head, I.; Farrimond, P.; di-Primio, R.; Zwach, C. Biodegradation of oil in uplifted basins prevented by deep-burial sterilization. Nature 2001, 411, 1034–1037. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Maki, H.; Nakayama, T.; Nakajima, T.; Nomura, N.; Uchiyama, H.; Kitaoka, M. Diversity and similarity of microbial communities in petroleum crude oils produced in Asia. Biosc. Biotechnol. Biochem. 2008, 72, 2831–2839. [Google Scholar] [CrossRef] [PubMed]
- Fish, S.A.; Shepherd, T.J.; McGenity, T.J.; Grant, W.D. Recovery of 16S ribosomal RNA gene fragments from ancient halite. Nature 2002, 417, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Gales, G.; Tsesmetzis, N.; Neria, I.; Alazard, D.; Coulon, S.; Lomans, B.P.; Morin, D.; Ollivier, B.; Borgomano, J.; Joulian, C. Preservation of ancestral Cretaceous microflora recovered from a hypersaline oil reservoir. Sci. Rep. 2016, 6, 22960. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, F.; Okada, H.; Tsapin, A.I.; Nealson, K.H. Microbial survival: The Paleome: A Sedimentary Genetic Record of Past Microbial Communities. Astrobiology 2005, 5, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Marty, B.; Dewonck, S.; France-Lanord, C. Geochemical evidence for efficient aquifer isolation over geological timeframes. Nature 2003, 425, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Celle-Jeanton, H.; Huneau, F.; Travi, Y.; Edmunds, W.M. Twenty years of groundwater evolution in the Triassic sandstone aquifer of Lorraine: Impacts on baseline water quality. Appl. Geochem. 2009, 24, 1198–1213. [Google Scholar] [CrossRef]
- Fontes, J.C.; Matray, J.M. Geochemistry and origin of formation brines from the Paris Basin, France: 1. Brines associated with Triassic salts. Chem. Geol. 1993, 109, 149–175. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).