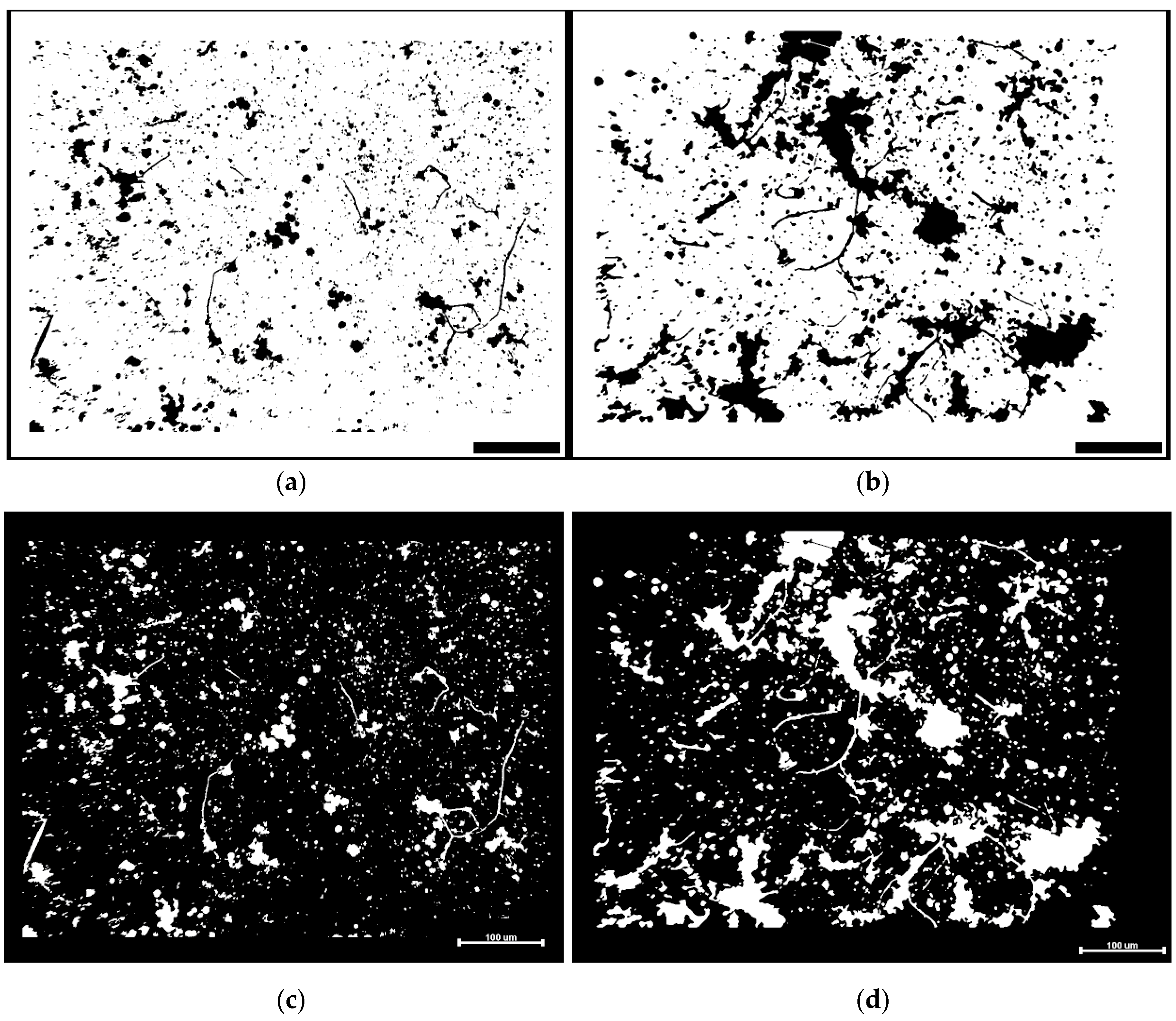
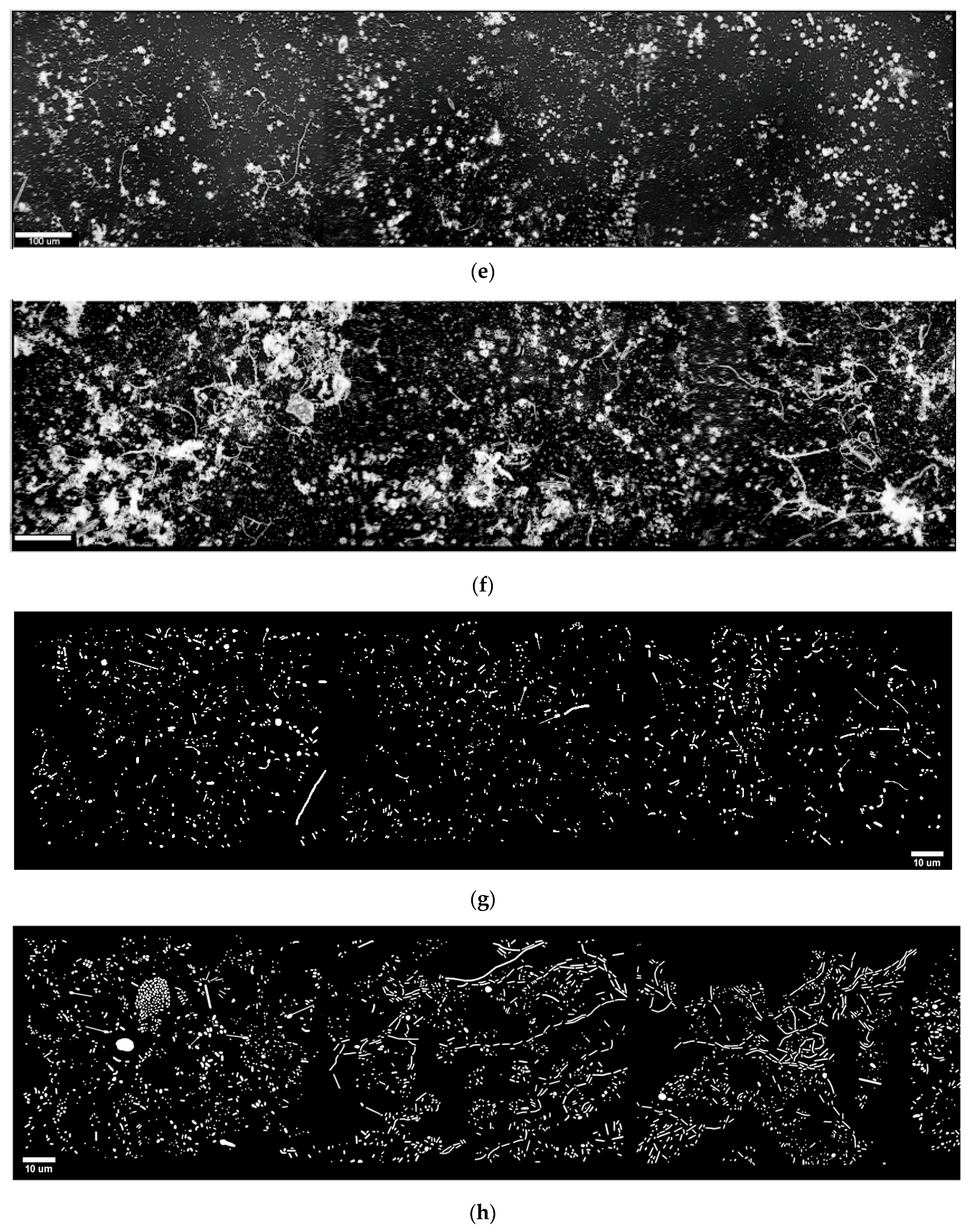
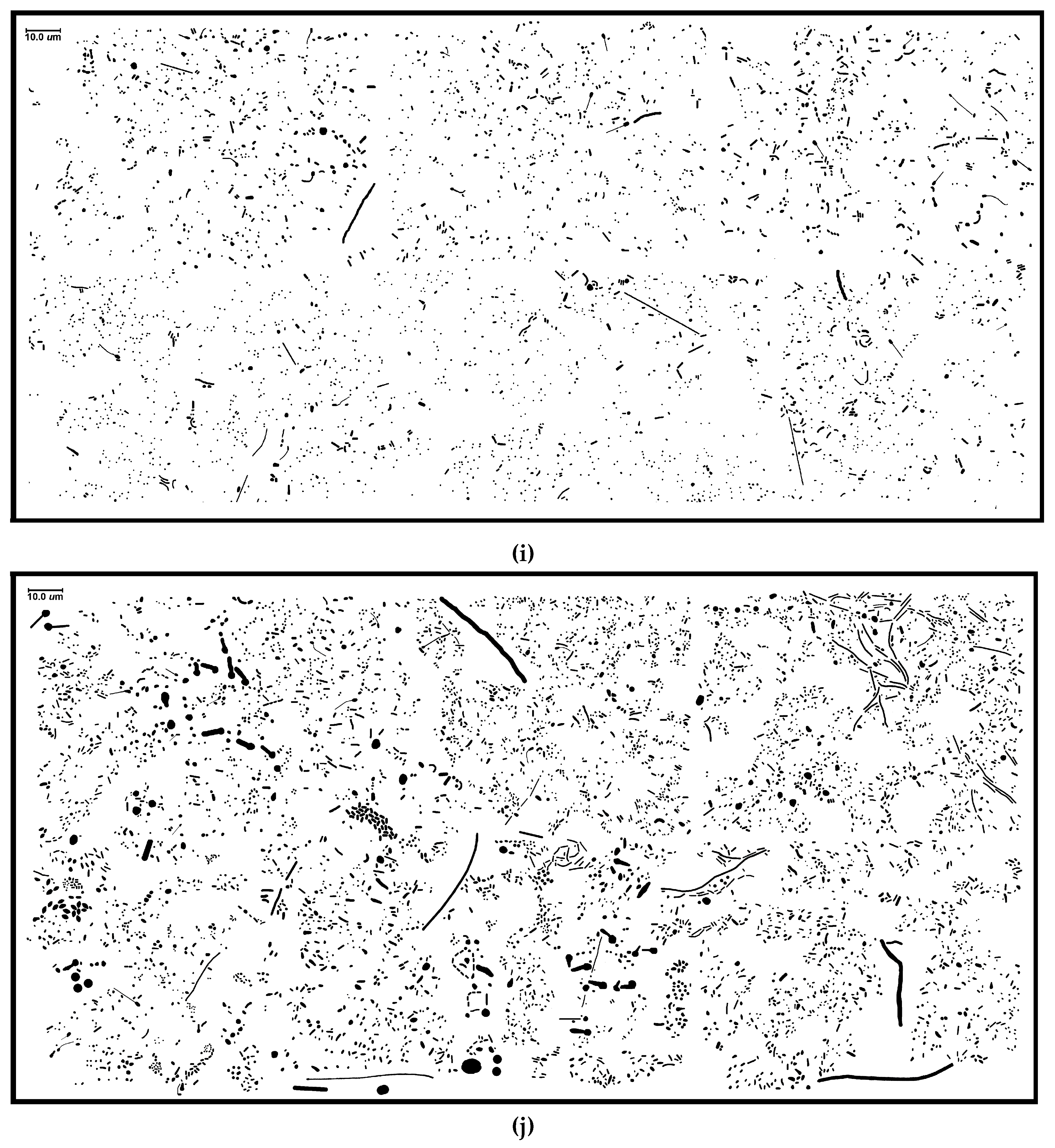
3.2. River Biofilm Architecture Analyzed at Microcolony Spatial Resolution
Five measurement attributes (area, perimeter, equivalent circular diameter, mean radius, and maximum radius) were used to compare the size distributions of microcolony biofilms developed on the plain glass (Community A) and polystyrene (Community B) substrata (
Table 2 and
Table 3). The percent proportional dissimilarities of their size distributions ranged between 5.4% and 7.7%. The
p-values (Ho of no difference) of multivariate parametric MANOVA and non-parametric Krustal–Wallis statistic tests for these five size metrics were 3.24 × 10
−34 and 0.000, respectively, indicating that the sizes of microcolony biofilms in communities A and B are not derived from the same distribution. Further analyses using the Student
t and Mann–Whitney two-tailed two-sample tests indicated that the means and medians for each of these size metrics are significantly different for the two biofilm communities (
Table 2). These results plus additional analyses in
Table 3 indicate that the microcolony biofilms are significantly larger in community B. This size differential reveals higher productivity of the community B developed on the polystyrene substratum in situ. Plausible (but not necessarily all inclusive) causes and consequences of this outcome [
5,
10,
20,
24,
25,
26,
27,
28,
29,
30,
31,
32,
38,
56,
57,
58,
59,
60,
61] include increased nutrient apportionment and utilization efficiency, positive cooperativity among neighbors, defense against protozoan bacteriovory, and longer distances (i.e., greater mean and maximum radii) that putative inhibitory metabolites must diffuse in order to reach all cell targets within microcolony biofilms.
A second architectural analysis of microcolony biofilms was implemented to assess their surface texture.
Figure 4a,b shows examples of heterogeneity in surface texture for biofilm communities A and B based on the varied distributions of their size and luminosity in pseudocolor rendered images prepared using the CMEIAS Color Segmentation tool [
14]. This result indicates a greater variation and intensity of surface texture for the biofilm community B developed on the polystyrene substratum (
Figure 4b). This brief test was followed by a grayscale brightness-based assessment of luminosity within images acquired using brightfield microscopy with transmitted light. In this case, microcolony biofilms display local heterogeneity in brightness intensity (on a scale of 0–255) due to variations in their height (third “
z” dimension) inversely proportional to the amount of transmitted light that has scattered (hence been subtracted) as it passes through them during microscopic examination and image acquisition. Analysis of inverted grayscale images (e.g.,
Figure 1e,f) then directly relates microcolony height to luminosity brightness.
Biofilm surface texture was analyzed in situ using the integrated density metric that combines both the size and luminosity of the individual microcolonies (computed as the product of the object’s pixel area times its mean gray level). For quantitative analysis of surface texture, the integrated densities of community A and B biofilms were compared in eighteen 8-bit inverted grayscale images following a brightness threshold of 85% to find and segment their individual microcolony biofilms. A pair-wise dissimilarity analysis indicated significant differences in their overall distributions, with distance coefficients of 12.18% proportional dissimilarity, average Euclidian distance of 23.02, and Canberra distance of 0.64. The mean values of cumulative integrated density per image were 41,597 and 84,003 for communities A and B, respectively. Two-tailed statistical tests rejected the Ho of no difference between sample means (p = 1.84 × 10−36), sample medians (p = 0.000) and variances (p = 0.000) of their integrated density, indicating that the surface texture of biofilm community B was more intensely heterogeneous with significantly more abundant, taller microcolony “mounds” in its three-dimensional (x, y, z) architecture on the polystyrene substratum, as further evidence of its greater productivity in this environment.
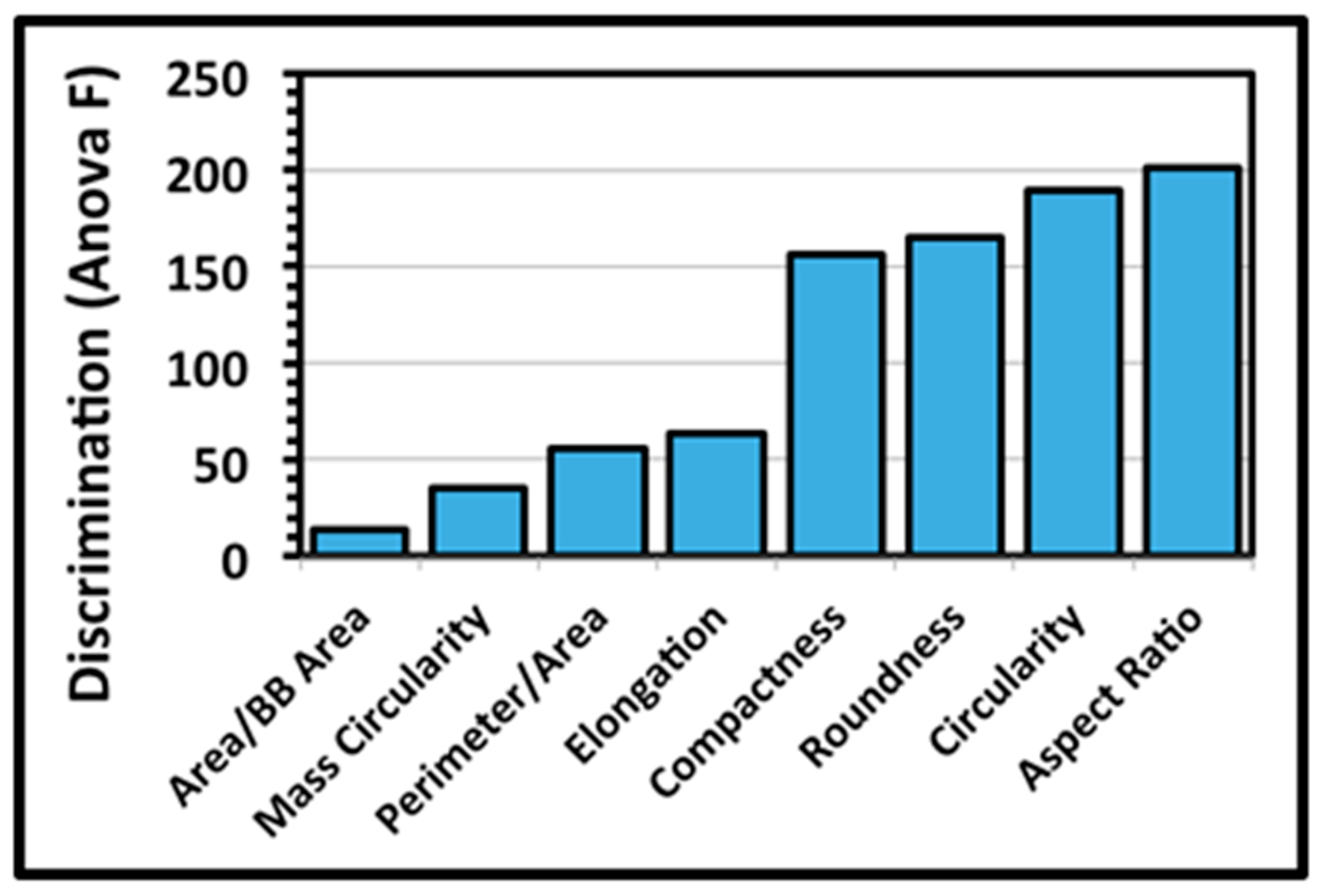
A third architectural analysis of the two communities of microcolony biofilms was implemented to assess differences in their two-dimensional shapes. The microcolonies >40 pixels in size within low-resolution binary images (25 per community) were analyzed by several metrics that evaluate the intensity at which their patch contour shapes deviate from a perfect circle of concentric radial growth. An ANOVA analysis indicated that the metrics of aspect ratio, circularity, roundness and compactness ranked highest in their ability to discriminate the microcolony shapes (
Figure 5).
The extent of differences in contour shapes of microcolony biofilms of communities A and B was then tested using these four top-ranked discriminating metrics. A two-tailed multivariate statistical T-test indicated that the values of this shape feature for the two communities were not derived from the same distribution (ANOVA F 60.933,
p (Ho of no difference) of 8.34 × 10
−51). A two-tailed Student
t-test of these four discriminating shape metrics analyzed separately indicated that the community A microcolonies on the plain glass substratum were significantly rounder and more compact (
Table 4), concurring with a visual inspection of their reduced intensity of radial dispersion (
Figure 1a,c and e).
3.3. Landscape Ecology of River Microbial Biofilms at Microcolony Spatial Resolution
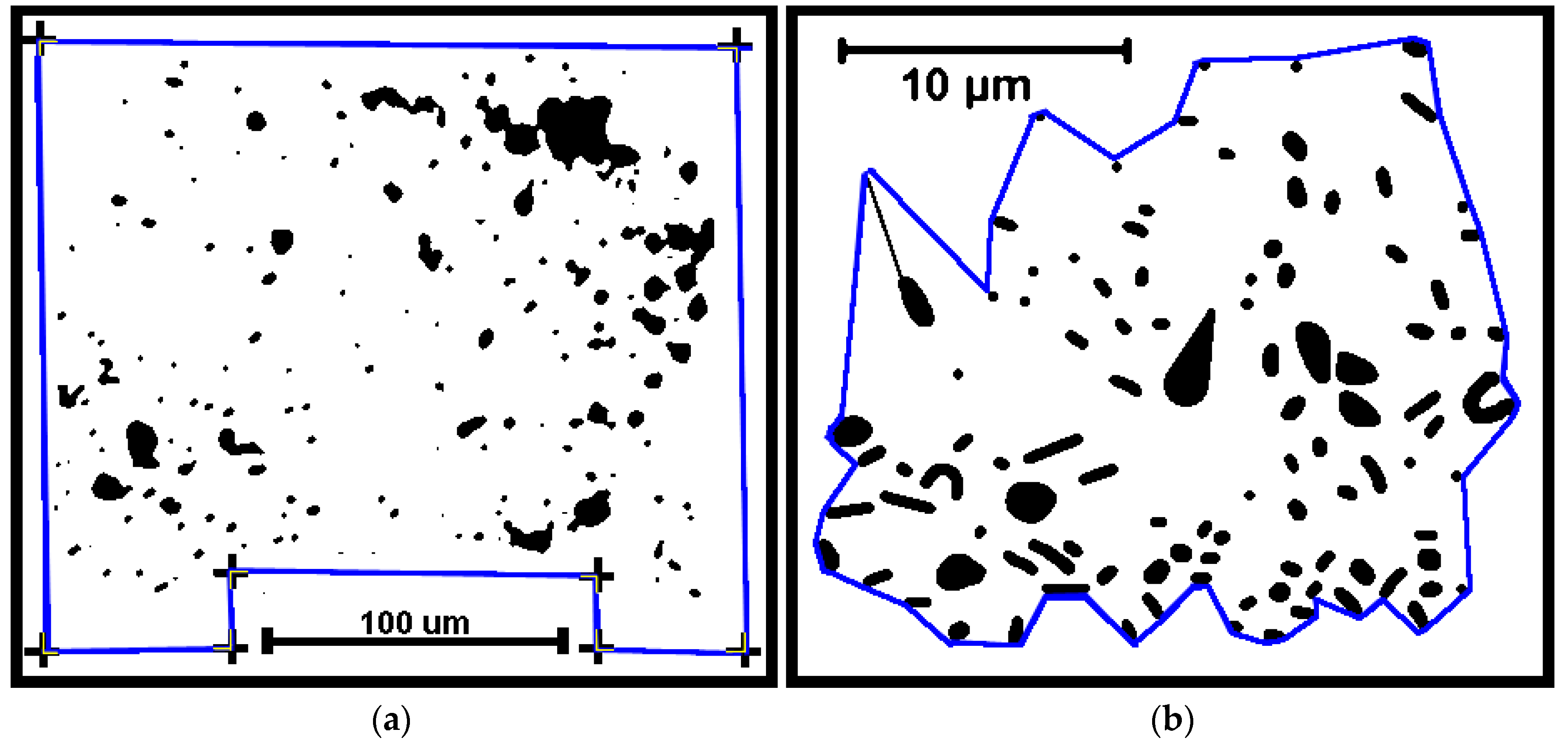
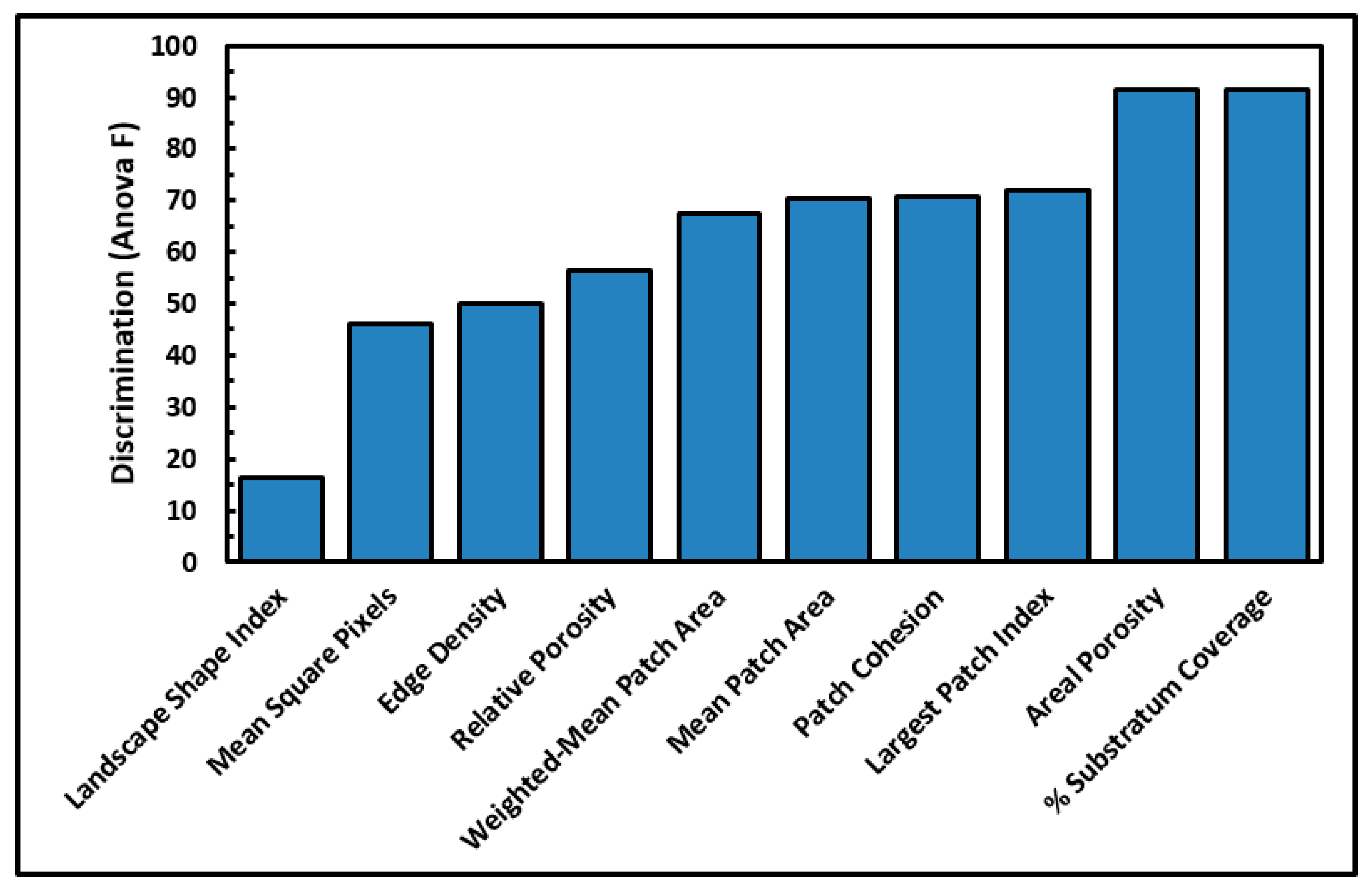
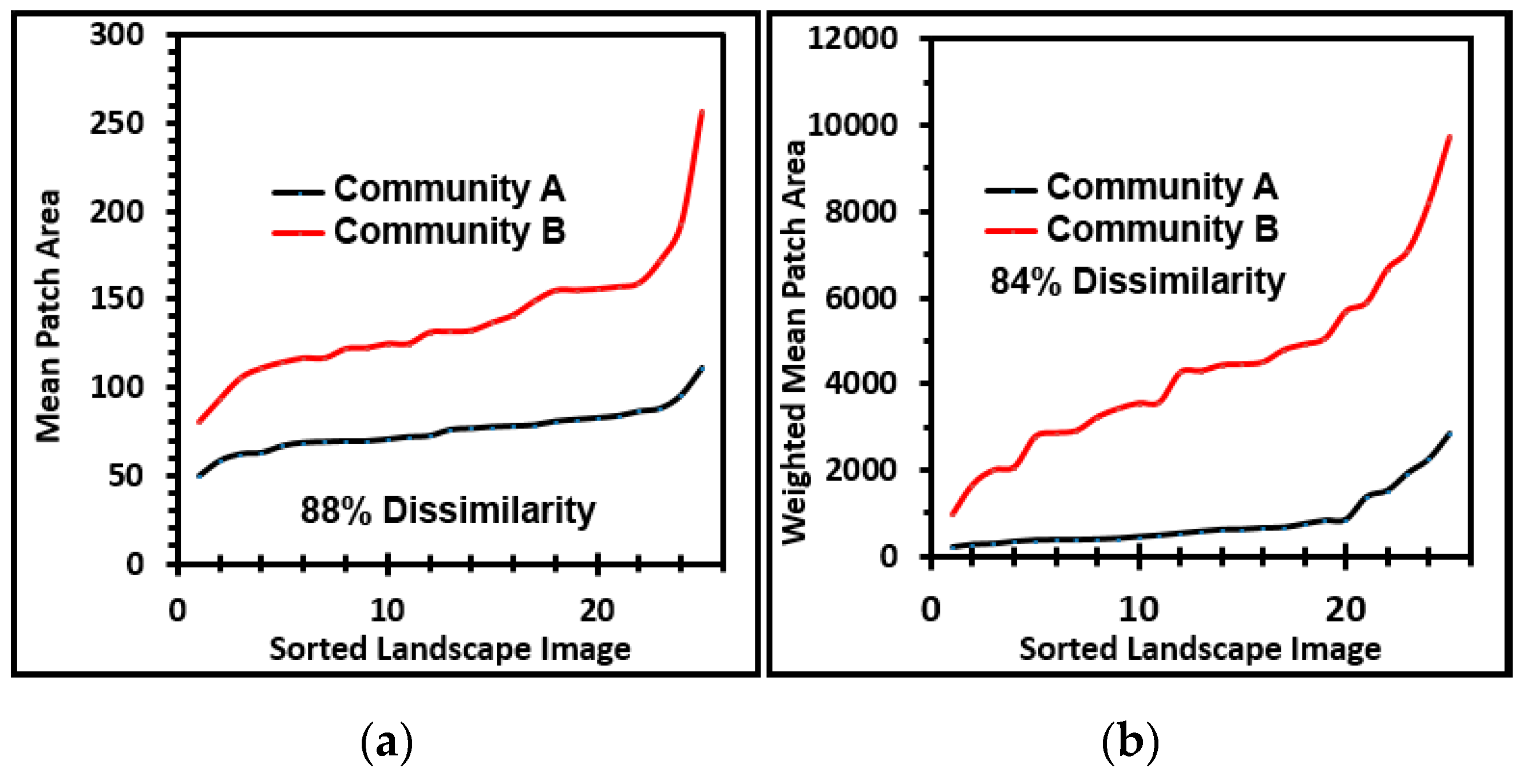
Several cumulative object analyses were done to characterize the mosaic of microcolony patches in the biofilm landscapes. They included categories of landscape ecology metrics that assess their patch area statistics (mean patch area, weighted-mean patch area, largest patch index), abundance and intensity of aggregated patches (percent substratum coverage), patch shape complexity (landscape shape index, mean square pixels), patch aggregation/dispersion/interspersion/fragmentation/connectivity (patch cohesion), patch edge intensity (edge density), and landscape porosity of fluid-filled channels between microcolony biofilm patches (areal porosity, relative porosity). Each metric was used to analyze microcolony biofilms >40 pixels in size in low-resolution binary images (25 per community), using the cross-hair method to define the area of interest polygon when that information was needed to compute metric intensities weighted by the substratum area (
Figure 2a).
The ranked ability of these landscape ecology metrics to discriminate biofilm architectures is shown in
Figure 6, revealing that percent substratum coverage and areal porosity had the greatest discriminating power among this group. The mean values for all 10 landscape ecology metrics extracted from images of the two biofilm communities were tested by the two-tailed Student
t-test, and their differences were all statistically very significant (
p << 0.01) (
Table 5).
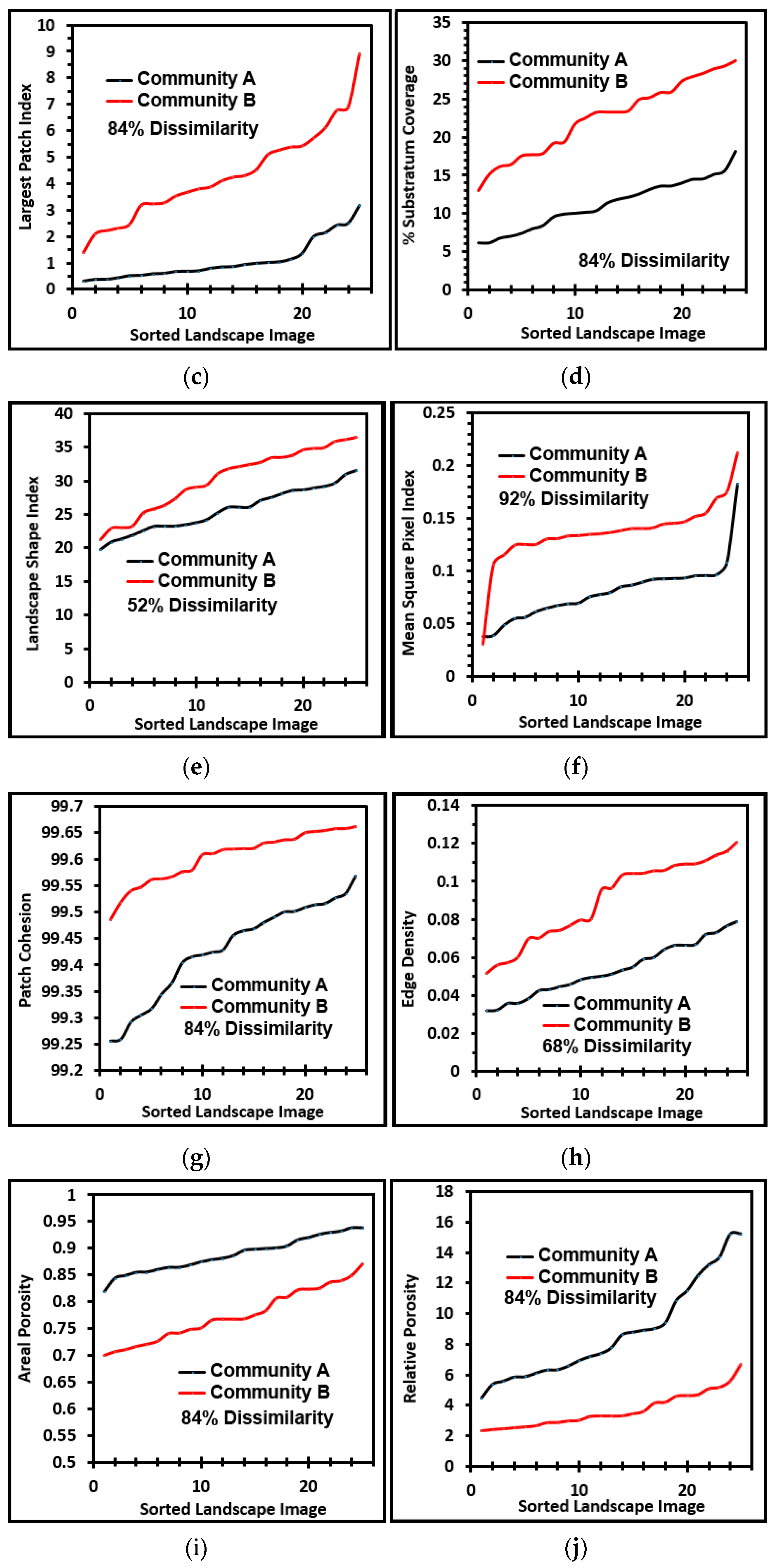
These metrics of landscape ecology were evaluated further by comparing paired ascending sort plots of their values extracted from images of the two biofilm communities. This analysis revealed clear separations in the distribution of ranked values for each metric of landscape ecology examined (
Figure 7a–j), confirming that all were able to discriminate the biofilm architectures of these two communities developed on contrasting substrata. Consistent with the visual evidence provided by representative biofilm images (
Figure 1a–f), these data further corroborate that the microcolony biofilms of community B developed a more dense and discrete landscape pattern, interpreted as having more productivity (more aggregated patches and larger sizes covering a significantly larger portion of the substratum area, hence less void spaces), greater edge intensities and complex shapes enabling their greater exploitation of external resources, significantly enhanced connectivity between neighboring microcolony patches (often via thin filamentous interconnections; see
Figure 1b,f) that would potentially extend their “calling distances” in cell–cell communication, and more access to additional opportunities for syntrophic-like cross-feeding and other positive interactions with community members, ultimately resulting in further growth expansion on the polystyrene substratum [
5,
18,
24,
29,
38,
53,
54,
55,
56,
61,
62].
These architectural differences between the two communities of microcolony biofilms prompted us to explore the fractal geometry component of their landscape architecture. We anticipated that the more productive microstructured biofilm of community B would exhibit a greater fractal dimension on the polystyrene substratum, reflecting its more intense colonization behavior of successful positioning to maximize nutrient resource acquisition and allocation within the hierarchical fractal-like nature of resource distribution networks within landscape microenvironments [
5,
7,
15,
25,
26,
27,
30,
31,
32,
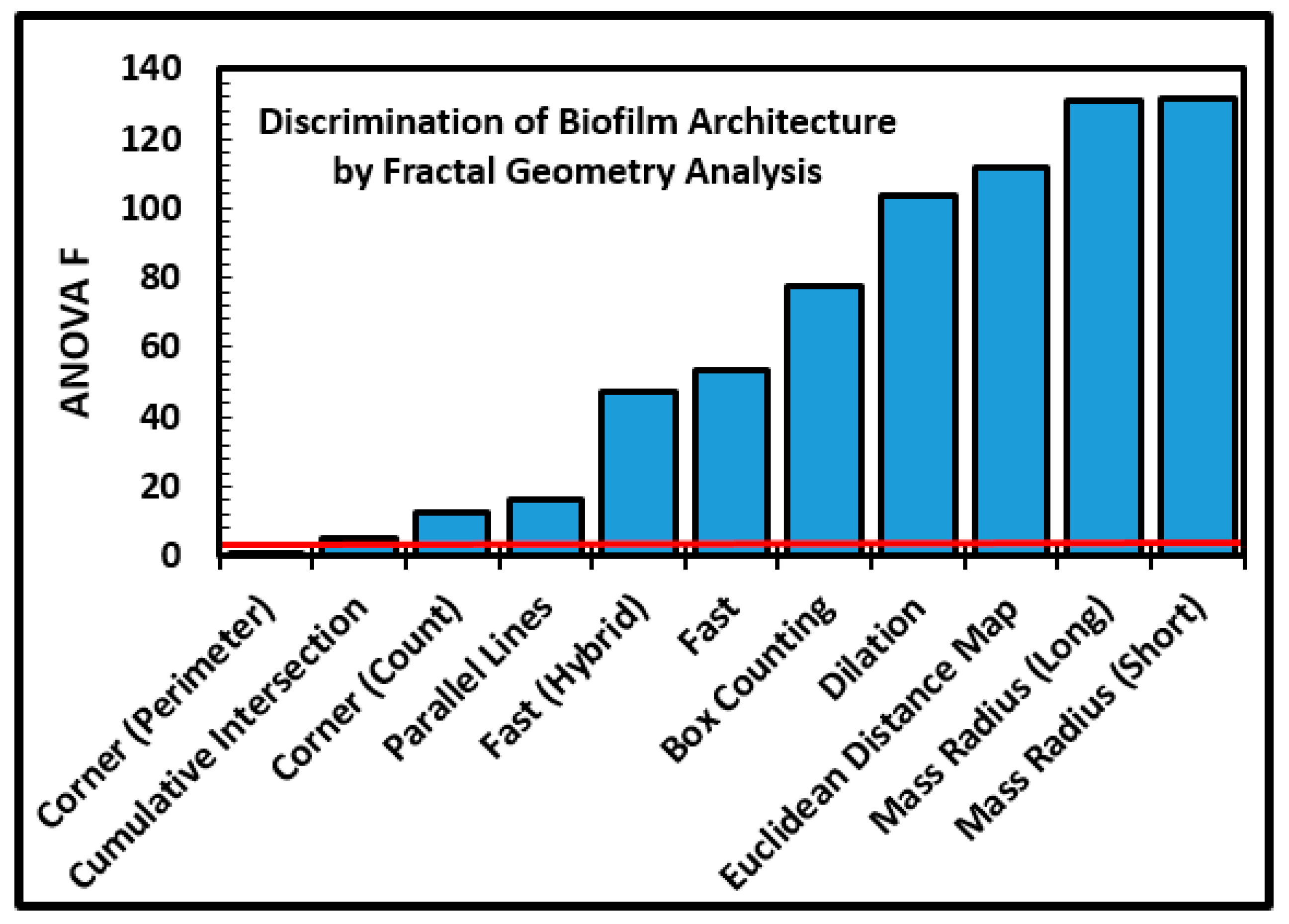
34]. The fractal dimensions of microcolony biofilm coastlines in the inverted grayscale images were analyzed using optimized segmentation settings (
Table 1), an 85% brightness threshold, and all 11 methods of fractal analysis using the automated batch process available in CMEIAS JFrad software [
15]. A univariate ANOVA analysis of the data indicated that nine of the 11 methods had sufficient discriminating power to discern substantially different intensities of fractal geometry between the two biofilm communities (
Figure 8). The variation in discriminating power is readily apparent among the 11 fractal methods in JFrad, illustrating the benefit of statistical data mining when the most discriminating methods of fractal analysis are not known beforehand.
A two-tailed two-sample multivariate Student
t test indicated that the nine discriminating methods combined together found very significant differences in fractal geometry of the microcolony biofilms in the two communities (
Table 6). In addition, the two-tailed two-sample statistical tests for each of these discriminating methods of fractal dimension analysis evaluated separately had very low
p values that rejected the null hypothesis of no difference between the two communities (
Table 6). The microcolony biofilm architecture was significantly more fractal when developed on the polystyrene substratum than on plain glass, predictably reflecting different fractal distributions of available growth-supporting resources and opportunities for species coexistence [
25,
26,
27,
31,
32].
3.4. Morphological Analysis of River Microbial Biofilms Analyzed at Single-Cell Resolution
Morphological analysis provides a strong complement to genotypic and other phenotypic methods of polyphasic taxonomy to deliver important insights on microbial community structure and function. The list of morphotype-weighted examples is long, including community productivity, biodiversity, dominance, conditional rarity, niche apportionments, food-web dynamics, ecological succession/resilience and other membership-environment relationships when competing for limiting resources, adaptations to various environmental stresses (e.g., starvation, predation, eutrophication, etc.) and spatio-temporal dispersal activities [
2,
3,
4,
5,
8,
16,
25,
28,
48,
49,
50,
51,
52,
57,
58,
59,
60,
61,
62,
63]. The unique, supervised, hierarchical morphotype classifier featured in CMEIAS uses mathematical rules of pattern recognition algorithms operating in 14-dimensional feature space to classify all major and several minor microbial morphotypes of individual cells, and performs with an overall 96% accuracy on properly edited images [
2,
13]. This classifier also automatically produces a rendered image containing each cell differentially pseudocolored in situ to indicate its assigned morphotype class [
2]. This latter software feature was used to produce the
Figure 9 composite image illustrating the diversity of microbial morphotypes present in the river biofilm assemblages.
High-resolution images of individual microbial cells spatially distributed in situ within each biofilm community were combined into montages and analyzed. The distributions of cell abundance among the ranked morphotype classes are presented in
Table 7. This equivalent sampling effort indicated that the biofilm community B had a 56.2% greater cell abundance. Both community assemblages had an equal richness of the same nine morphotypes, a dominance of cocci (77% of community A and 71% of community B), and a rare singleton of one branched filament.
The diversity of these class distributions was examined by several methods of community analysis [
5,
28,
42,
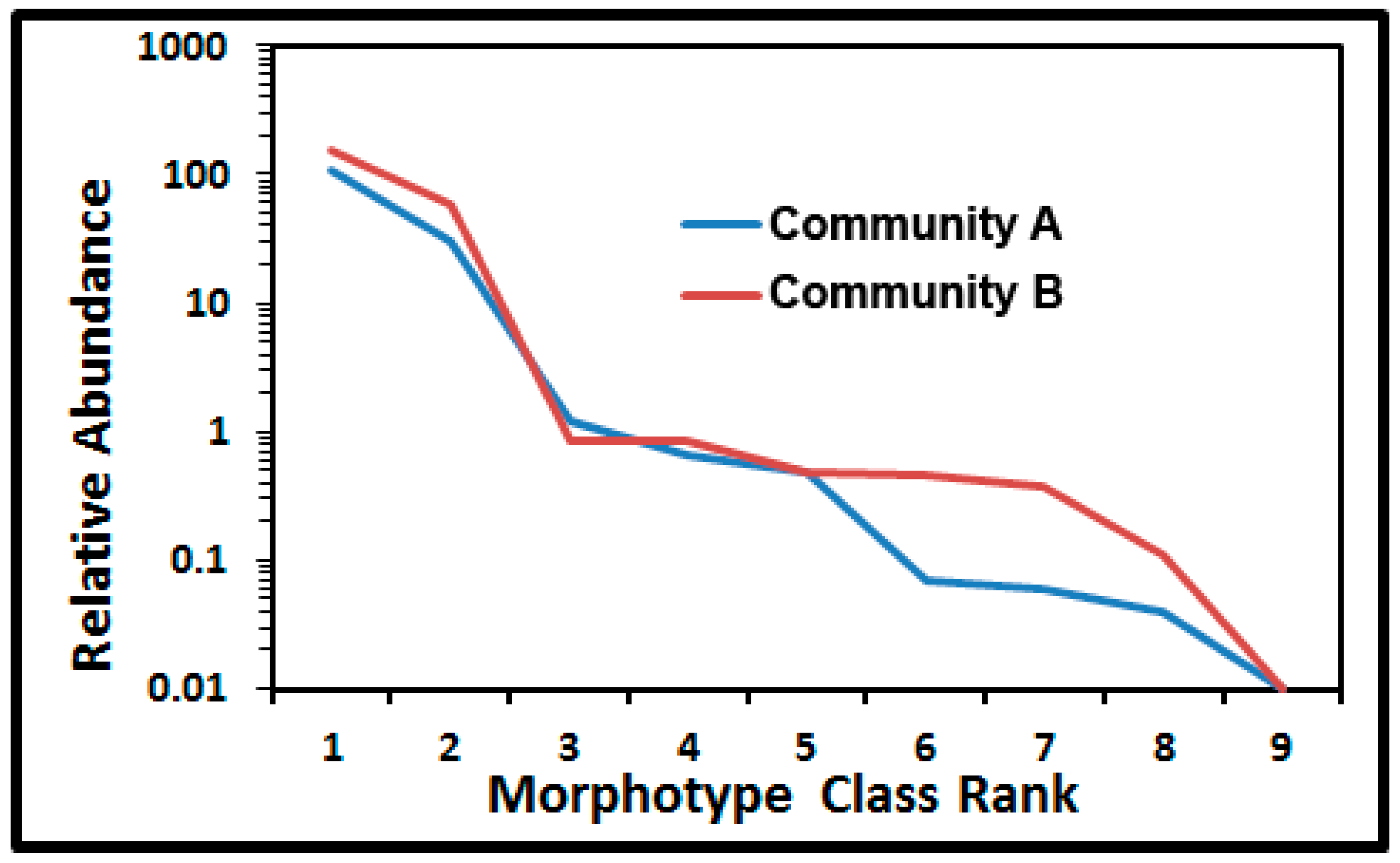
43]. The shape of the Whittaker ranked abundance plot [
28,
43] showcases differences in relative numerical abundances, dominance and evenness of each morphotype class in communities A and B (
Figure 10). The major separation of relative abundance in the curves occurred with rare morphotypes ranked as No. 6, 7, and 8 (clubs, ellipsoids and U-shaped rods). The abundance in these numerical ranked distributions was greater for six of the nine morphotypes colonizing the polystyrene surface. Based on the least difference between observed and expected values, the truncated logarithmic series model made the best fit to both curves because the abundance of intermediate classes was more common than predicted by the geometric model series, and their curves were steeper than the broken stick model or the sigmoid curve of the log normal model [
28,
43]. The singleton morphotype caused the slight truncation in the models for both communities.
Table 8 indicates various indices of community α-diversity, evenness and dominance computed from the data of raw numerical abundance (individual counts) (
Table 7), and after a relative normalized transformation (% ×100) of those same data to equalize community sample sizes. Data normalization only marginally affected the computed indices (likely because both communities had an equal richness of the same nine morphotype classes) without affect their ranking. The robust 10,000-iterated Solow statistic test [
43,
64] indicated that the diversity and evenness indices were significantly higher (
p ≤ 0.05) for the river biofilm community B on the polystyrene substratum. Correspondingly, the dominance index was significantly higher for the community A developed on plain glass. These results agree with other studies indicating that community diversity strongly correlates with larger sizes and complex structures of landscape patches [
24].
The Renyi ordering analysis provides a robust, entropy-based test of whether the trends of α-diversity that differ between communities change with the diversity index used, and allows the relative magnitude of α-diversity across a range of indices to be compared directly [
43,
65]. This analysis of the ranked abundance of morphotype classes for the two communities (
Figure 11) showed that their ranges of diversity indices do not cross one another indicating that they are validly comparable, and that community B developed on the polystyrene substratum had a higher Renyi index at each point of the scaled indices, validating its greater morphological diversity.
β-Diversity indications of the degree of differences in distribution of ranked abundance among morphotype classes of communities A and B are provided in computations of various dissimilarity (distance) coefficients (
Table 9) [
42] and in plots that examine the communal relationships of their ranked dominance and rarity (
Figure 12a,b) [
5,
25,
28,
66,
67].
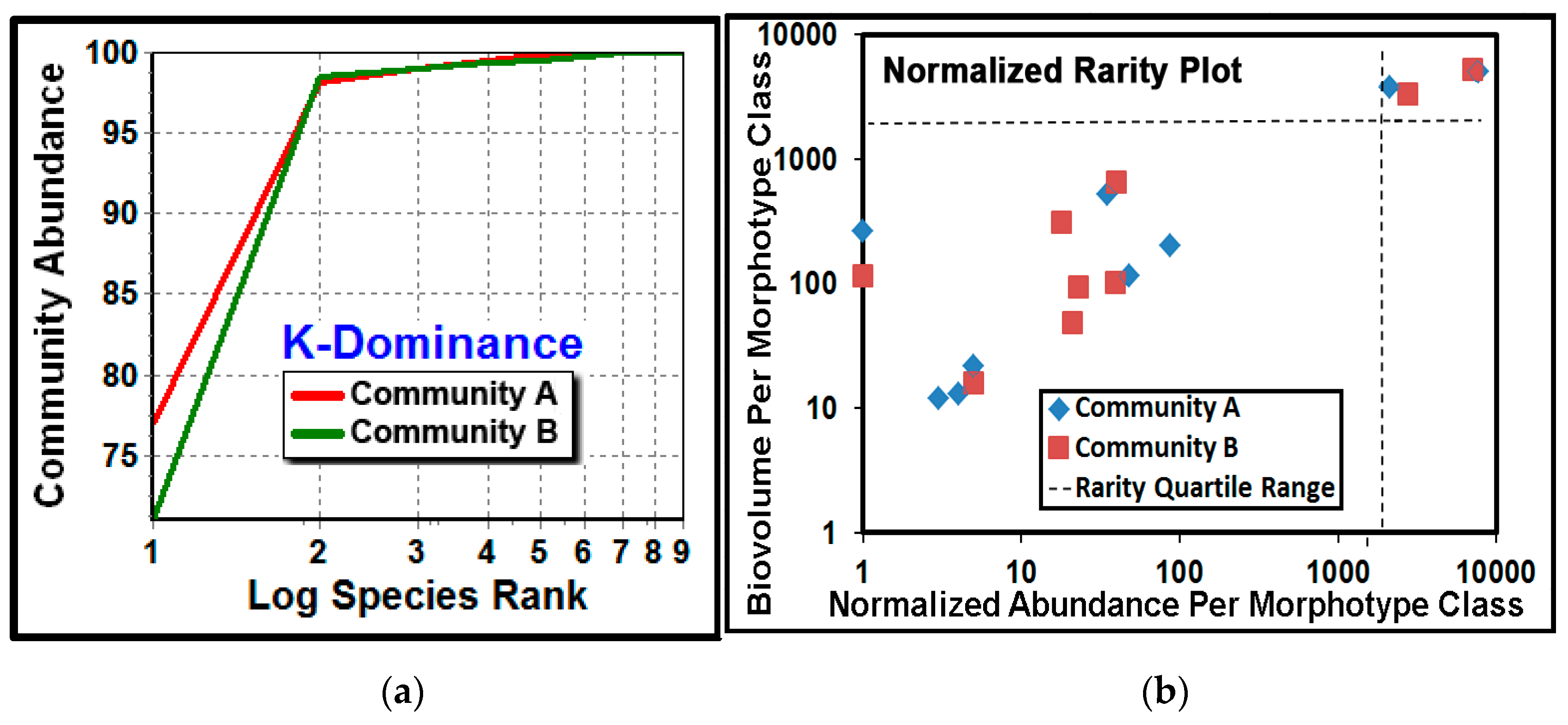
The
K-dominance analysis compares the cumulative abundance of classes as a percentage against their log class rank in the community [
5,
28,
43,
66]. The result showed that community A had a higher dominance of its most abundant cocci morphotype (
Figure 12a). The normalized rarity plot [
5,
28,
67] compares the relative abundance and cumulative biovolume for each morphotype class in the community, and identifies community classes that are considered “rare” when they locate within the lower left quadrant (25th percentile) of the plot range. This analysis indicated that most of the class richness was represented by the seven rare morphotypes (comprising ≤25% of the class abundances), and the relative abundances of their cumulative biomass were similar for some morphotype classes but different for others (
Figure 12b). This latter result also suggests that rarity for some morphotype classes may be “conditional” because their relative abundances were affected by the substratum environment upon which they had colonized [
68]. This finding has potential importance because conditionally rare classes can contribute significantly to community stability and resilience during the ecological succession that follows environmental perturbation [
2,
3,
4].
3.5. In Situ Ecophysiology of River Microbial Biofilm Communities Analyzed at Single-Cell Resolution
CMEIAS bioimage informatics were used to analyze traits of community ecophysiology in situ, including their intensity and productivity of biofilm colonization, allometric metabolic rate, and indicators of adaptations to starvation and predatory stresses [
5,
10,
13,
33,
34,
35,
36,
37,
48,
49,
50,
51,
52,
57,
58,
59,
60]. The strengths of these vital activities were compared in images of the two river biofilm communities using data of each cell’s biovolume, surface area/biovolume ratio, and relative lengths of elongated cells.
Table 10 presents data on the substratum area-weighted intensity of productive colonization and cell size-weighted allometric metabolic rates with equal sampling efforts for both communities. The total cell counts, spatial density, substratum coverage and cumulative biovolume intensities were 1.56–2.03-fold higher for community B. This greater intensity of biomass is consistent with earlier results (
Table 2 and
Table 3) indicating a larger, more abundant/widespread/highly structured architecture of microcolony biofilms for the corresponding community. Two-sample two-tailed statistical tests indicated that the differences between means were highly significant for spatial density (
p = 0.0001), and significant for biovolume intensity (
p = 0.02), mean cell biovolume (
p = 0.04) and median cell biovolume (
p = 3.81 × 10
−47). Thus, individual microbial cells were significantly bigger and more abundant when colonized on the polystyrene substratum. Since the metrics of biomass carbon and active allometric metabolic rates are derived from cell biovolume [
33,
34,
35,
36,
37,
46,
47,
48,
49], they had the same trend of significantly higher substratum area-weighted intensities and metabolic rate per individual cell in community B. Considered collectively, these results provide evidence to indicate that biofilm community B was more metabolically active and better able to convert resources into biomass resulting in its greater overall productivity on the polystyrene substratum in the river ecosystem.
The relative abundance of populations within a community assemblage to some extent reflects their success at competing for limited resources [
5,
28], and therefore the metric used to measure abundance in community membership can significantly influence how variations in that relationship are interpreted. This issue applies to all metrics used to measure class abundances in community analysis [
5]. Use of the CMEIAS morphotype classifier made it possible to examine the distribution of cell biovolumes and their size-scaled allometric metabolic rates among individual morphotype classes (singleton excluded). This analysis indicated a higher productivity for the biofilm community B (
Table 10,
Table 11 and
Table 12). Over 90% of the total biovolume was distributed among the cocci, unbranched filament, and regular rod morphotaxa classes. The difference between means for the biovolume intensity was statistically significant (
p < 0.05) for the cocci and marginally significant (
p = 0.054) for the regular rods. Although the cumulative biovolume and biovolume intensity were also greater for the U-shape rod, unbranched filament, ellipsoid and club morphotypes in community B (
Table 11), the range of their individual cell size was substantial, resulting in mean differences that were not statistically significant. The cocci, curved rods, U-shaped rods, regular rods, unbranched filaments, ellipsoids and clubs each had higher cumulative metabolic rates and metabolic rate intensities in community B (
Table 12).
Further morphotype-weighted analyses of biofilm communities at single-cell resolution provided additional insights on their productivity and adaptive responses to environmental stresses. For instance, sizing down to increase the cell’s surface area/biovolume ratio is one of several self-induced responses used particularly by
K strategists to adapt to starvation stress, and this morphological change is often accompanied by: (i) expression of transport systems with higher affinity and others with broader specificity that improve their ability to acquire essential nutrients when their local apportionment is low; (ii) enhanced distribution of those resources within the cell; and (iii) turnover of excess ribosomes and internal reserves of storage polymers [
5,
50,
51,
52]. An analysis of all cells in the two communities indicated that their surface area/biovolume ratios had dissimilar distributions (40.32% proportional dissimilarity, 495.65 average Eucledian distance, 0.515 Bray–Curtis distance, and 0.651 Canberra distance). Most of this pair-wise dissimilarity was attributed to the coccus morphotype, which differed between the two communities by distance coefficients of 51.34% proportional dissimilarity, 540.56 average Eucledian distance, 0.589 Bray–Curtis distance, and 0.662 Canberra distance. The mean and median values of the surface area/volume ratio were significantly higher for all cells and for the coccus morphotype in community A developed on the plain glass substratum, and the probability (
p) that these values differed by chance was extremely low (
Table 13).
Bacteriovory grazing activities by heterotrophic protozoan nanoflagellates and metazoan predators are important forces that shape the structure and composition of bacterial communities in aquatic ecosystems, largely because resistance to and refuge from selective bacteriovory are favored by large cell aggregates (e.g., microcolony biofilms) and elongated filamentous morphologies (e.g., unbranched filament) that exceed the oral diameter of the cytosome or lorica mouth opening of the predator, thereby increasing the predator’s difficulty to consume the microbial prey [
5,
48,
57,
58,
59,
60,
63]. Thus, bacteriovory predation is both size-selective and morphology-selective, and the relative abundance and length of the unbranched filament morphotype can provide insights on the intensity of the selective pressure of phagotrophic predatory stress that contributes to shaping the aquatic microbial community, in line with the evolutionary pressures to maximize resource intake [
5,
25]. That indicator morphotype had a 79.2% greater abundance (
Table 7), 21.1% greater cumulative length and 26.4% greater length intensity in the biofilm community B (
Table 14). The two-sample two-tailed Student
t tests indicated that these differences in communities A and B were statistically significant (
p = 0.04). These results plus the larger sizes of microcolony biofilms indicated earlier (
Table 2 and
Table 3) predict that bacteriovory grazing activities and adaptations to resist them were more intense in the biofilm assemblage of community B, and the increased fitness of the larger microcolonies and longer elongated unbranched filaments amidst the selective predatory stress likely contributed to their increased relative abundance and productivity in these river biofilms [
5,
57,
58,
59,
60,
61,
63]. These data-supported predictions also help to explain how the presence of predator bacteriovory tends to increase individual bacterial biomass under ambient nutrient conditions in aquatic ecosystems [
48].
3.6. Spatial Ecology of River Microbial Biofilm Communities Analyzed at Single-Cell Resolution
The dependence of spatially structured heterogeneity on ecosystem function provides the impetus to include analyses of spatial ecology in studies of microbial biofilm communities [
5,
6,
7,
9,
11,
12,
15,
25,
27,
30,
31,
32,
38,
45,
62,
69,
70,
71,
72,
73,
74]. Analysis of the in situ spatial patterns of microbes within biofilms reveals statistically defendable data that support ecological theories of biogeography indicating that their colonization behavior involves a spatially explicit process rather than occurs independent of their location in this microenvironment [
5,
6,
7,
25,
31,
38,
69,
70,
71,
72,
73,
74]. Spatial dependence is considered positive when neighboring organisms aggregate due to cooperative interactions that promote their localized productive growth, and is considered negative when conflicting/inhibitory interactions result in their uniform, self-avoiding colonization behavior [
5,
6,
7,
9,
27,
30,
38,
61,
69,
70,
71,
72,
73,
74]. This balance between positive cooperation (aggregation) vs. conflicting competition (over-dispersion) behaviors is crucial in biofilm ecology [
5,
38]. For instance, microbial cells exert stronger intensities of quorum-sensing communication when closely aggregated within biofilms [
9,
62]. The analysis protocol to measure these distinctions of spatial patterns typically involves initial statistical tests of the null hypothesis of complete spatial randomness, followed by additional quantitative measures of spatial dispersion/aggregation, and finally by geostatistical analyses that test for the spatial autocorrelation, variation and connectivity in continuously distributed “
z-variate” attributes of selected features (e.g., cluster index) at georeferenced
X,
Y coordinate locations of sampling points within the landscape domain [
5,
6,
7,
12,
27,
41,
69,
70,
71,
72,
73,
74].
Several features were extracted from each individual cell for this spatial analysis [
5,
6,
7,
9,
10,
11,
12,
13,
15,
41,
42,
44,
45,
71,
72], using optimized CMEIAS settings applied to high-resolution, fully segmented, spatially calibrated montage images of each community assemblage (
Table 1; examples are
Figure 1i,j). For instance, the proximity of neighboring cells impacts significantly on the intensity of successful cell–cell interactions in biofilms, e.g., in situ “calling distance” of quorum sensing-mediated communication [
5,
9,
62]. Statistical analyses of the 1st nearest neighbor distance indicated that neighboring cells were positioned further apart in the river biofilm community A developed on plain glass, indicating a greater proximity of cells in community B developed on the polystyrene substratum (
Table 15).
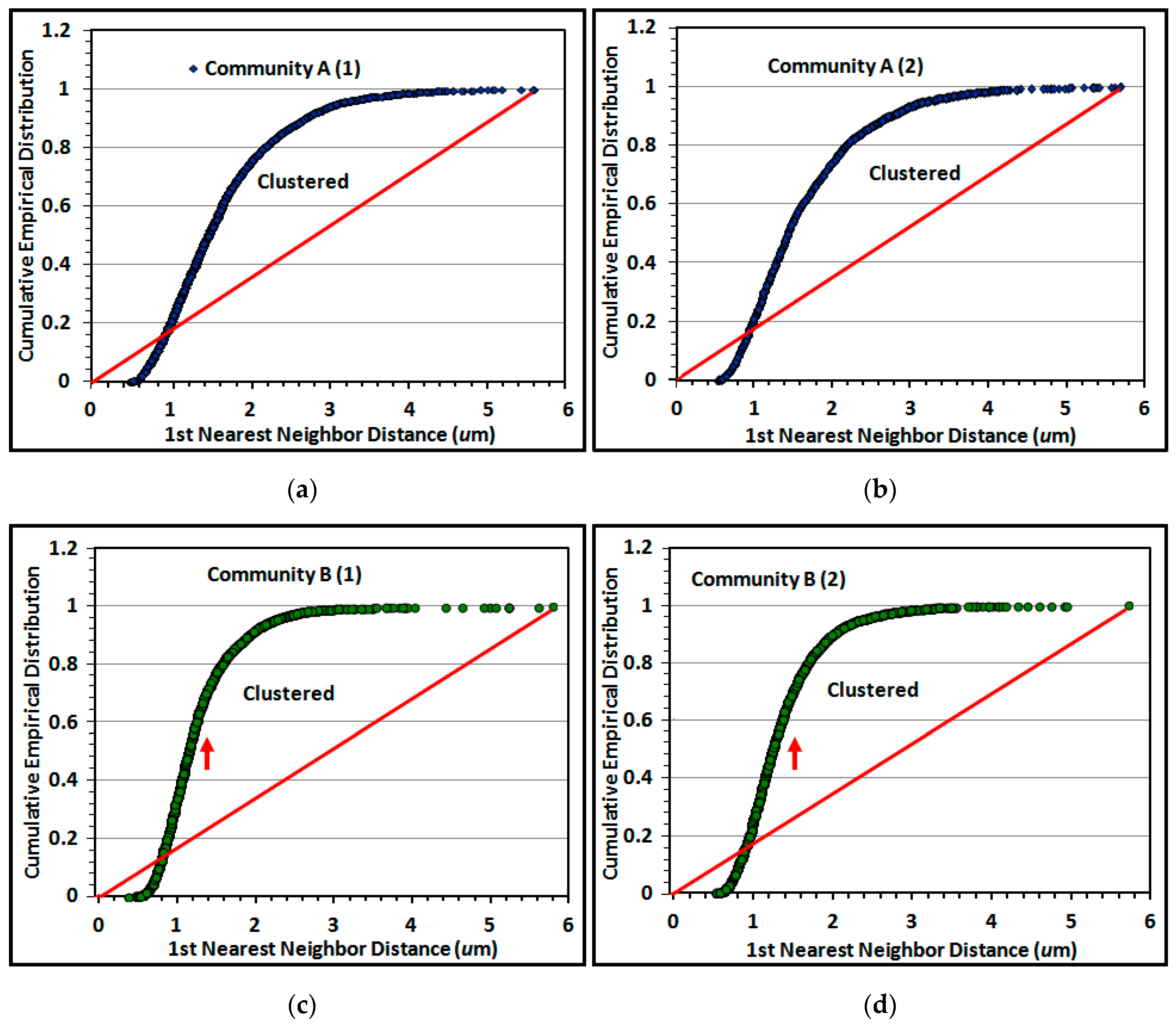
The Empirical Distribution Function (EDF) of 1st nearest neighbor distances between individual cells is a useful second test of spatial randomness. Its plot compares the cumulative rank of 1st nearest neighbor distances between individual cells in the biofilm community to the theoretical distribution that would occur if their pattern had complete spatial randomness [
5,
6,
7]. Replicate empirical distribution plots for cells in the two river biofilms are shown in
Figure 13a–d.
Random distributions in the EDF plot are defined by a diagonal line connecting the
XY intercept to the maximum 1st nearest neighbor distance calculated by analysis. Data points on EDF plots of spatially structured communities are commonly characterized by a sigmoidal curve, with positions representing uniform spatial patterns when located below the theoretical random trendline, and aggregated (clustered) patterns when they rise above the diagonal trendline to the 1.00 EDF asymptote [
5,
6,
7]. A random distribution is indicated if the EDF curve increases with a shallow slope close to the diagonal trendline. The results indicate that the proximity of cells in montage images of both communities has significant spatial structure, with a minority arranged in a uniformly equidistant spatial pattern and a significant majority that are spatially aggregated. Aggregated cells in community B on the polystyrene substratum display a steeper incline of their EDF curve (red arrows pointing upward) that reaches its asymptote at shorter (closer) distances between nearest neighbors (
Figure 13c–d). These results show similarities in EDF of spatially aggregated cells in replicated montages of the same biofilm community (
Figure 13a–d), and are consistent with visual inspection of local aggregate intensities within representative high-resolution images (
Figure 1i,j).
The next tests of spatial point-patterns for these two biofilm communities evaluated the Holgate and Clark and Evans indices of cell aggregation based on each cell’s 1st and 2nd nearest neighbor distances and centroid
X,Y coordinates, respectively [
41,
42,
75,
76]. These tests rejected the null hypothesis of complete spatial randomness for both communities (
p < 0.05), and indicated significant aggregation in their overall spatial patterns of distribution (
Table 16), consistent with the other spatial analyses of these two communities.
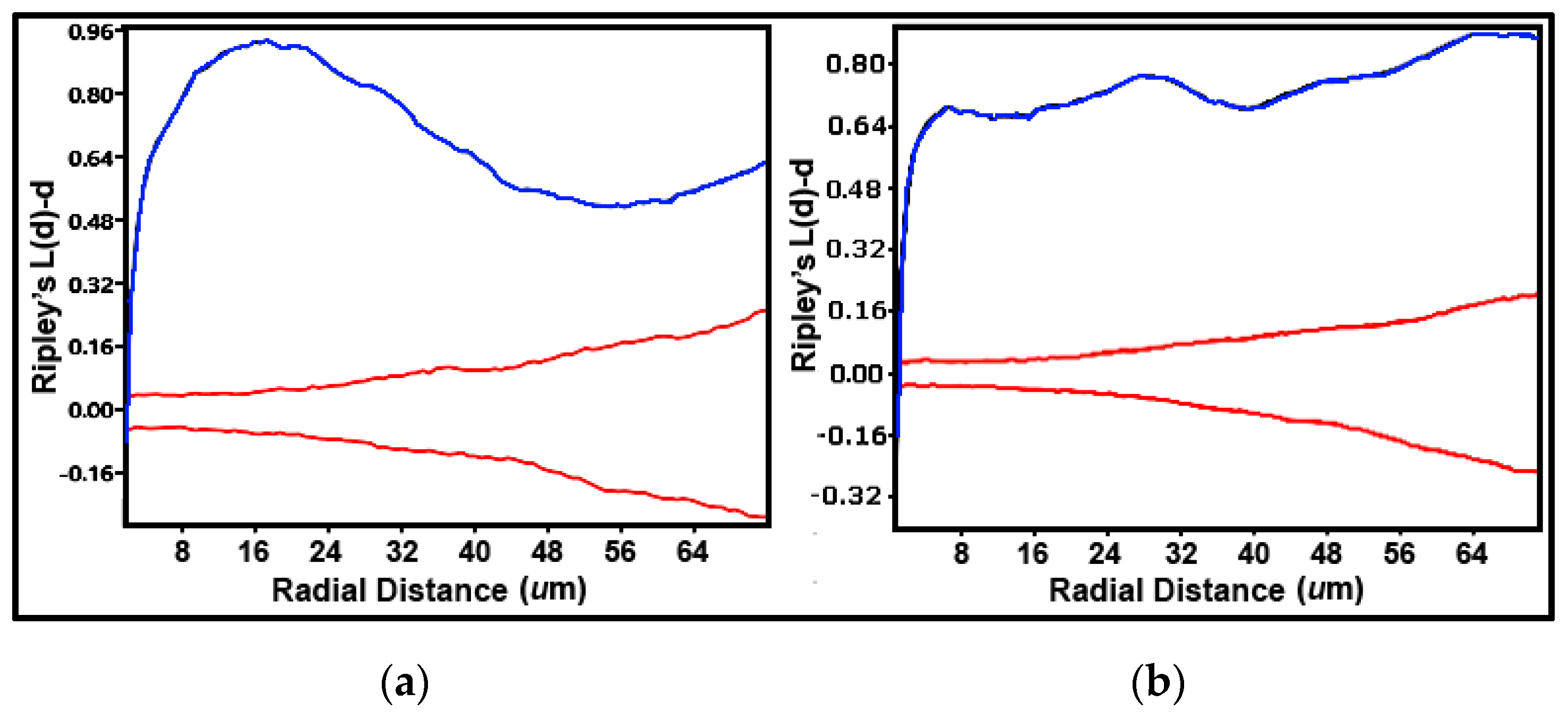
A useful counterpart to these point-pattern analyses is the Ripley’s K multi-distance clustering analysis [
5,
42,
71]. This second-order, point distribution statistic interprets multiple separation distances between objects to determine point pattern changes over a wide spatial scale [
5,
71]. K(
d)-
d measures average object counts within circles with a distinct radius
d centered on every object point in the landscape divided by the mean spatial density of all objects present [
42,
71]. A plot of K(
d)-
d vs. all radial separation distances in the landscape indicates uniform, random or clustered dispersion patterns determined by a Monte Carlo simulation of the 95% confidence interval representing the critical limits of complete randomness [
42]. K(
d)-
d values indicate overdispersed, uniform distribution patterns when located below the confidence envelope, and clustered distribution patterns when located above the envelope [
42,
71]. Peaks of K(
d)-
d values exhibiting the most intense aggregation can also be scrutinized at definable radial separation distances [
5]. The Ripley K plots over the same sampling interval range for both landscapes showed strong spatial structures with a uniform pattern at only one short radial distance, and clustered patterns above the 95% confidence envelope for the remaining 99 greater radial distances examined (
Figure 14a,b). Spatially aggregated patterns for cells in biofilm community A on plain glass had one peak of K(
d)-
d at a radial distance of ~16 μm (
Figure 14a), whereas cells in biofilm community B on polystyrene exhibited multimodal peaks of K(
d)-
d at radial distances of ~7, 30, and 66 μm (
Figure 14b).
Five additional methods of single-cell analysis were performed to capture the spatial relationships between neighboring cells and gain further insight on the predicted intensities of their in situ cell–cell interactions and colonization behaviors on different substrata. These analyses examined their fractal dimension, point kernel density, minimal spanning tree, linear point alignments, and geostatistical autocorrelation of pertinent z-variates.
Fractal analysis of structured biofilms can discriminate self-similar spatial patterns of biomass and deliver insights on intensity of cooperative microbial interactions, including their efficiency in positioning for optimal utilization of fractal-like apportionments of resource distributions and coexistence of multiple species among community members [
5,
7,
15,
25,
26,
27,
30,
31,
32]. A box counting analysis [
15] of inverted binary montage images (e.g.,
Figure 1g,h) indicated that the spatial pattern of individual cells in biofilm community B had a greater fractal dimension (mean ± std. dev. of 1.115 ± 0.046 compared to 1.019 ± 0.058 for community A) that was statistically significantly (Student
t of 2.562,
p same mean of 0.04). This greater fractal dimension of individual cell distributions in the biofilm community B indicates that they have higher spatial complexity, are responding to significantly different ecological processes that control their spatial structure on the polystyrene substratum, and predictably reflect an increased, fractal-like nutrient apportionment in that landscape [
15,
25,
26,
27].
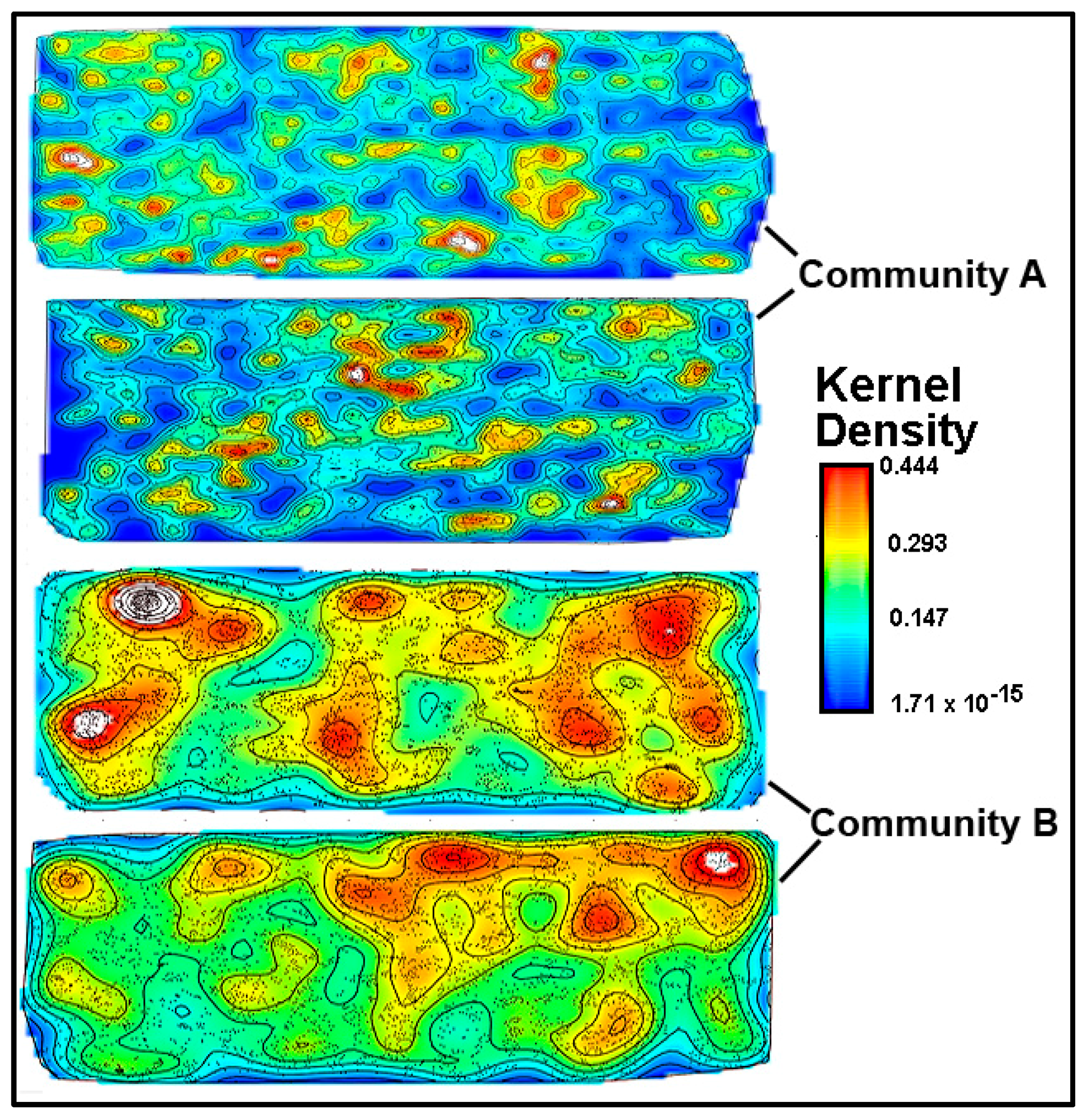
A kernel density analysis [
42] was performed on the data of spatial point coordinates to examine the in situ density of cells in both community landscapes. This spatial mapping tool uses a Gaussian smoothness method to estimate the probability of (dis)continuity in gradients of local cell density interpolated over the landscape area [
42].
Figure 15 shows equivalent pseudocolored scalings of spatial point kernel densities for cells in high-resolution montage images of the two river biofilm communities. A comparison of the landscape domains clearly reveals differences in the heterogeneity and discontinuity of georeferenced spatial intensity of the clustered cells in situ. Cells in community B congregated into several foci with greater kernel point densities that were spread over larger regions of the biofilm landscape, and had higher gradient connectivity with less discontinuity of kernel densities compared to cell locations in the biofilm of community A. Kernel densities of cells in community A had more discontinuity, as indicated by numerous internal gradients of aggregation that diminished to the minimum (blue) density within the full range present.
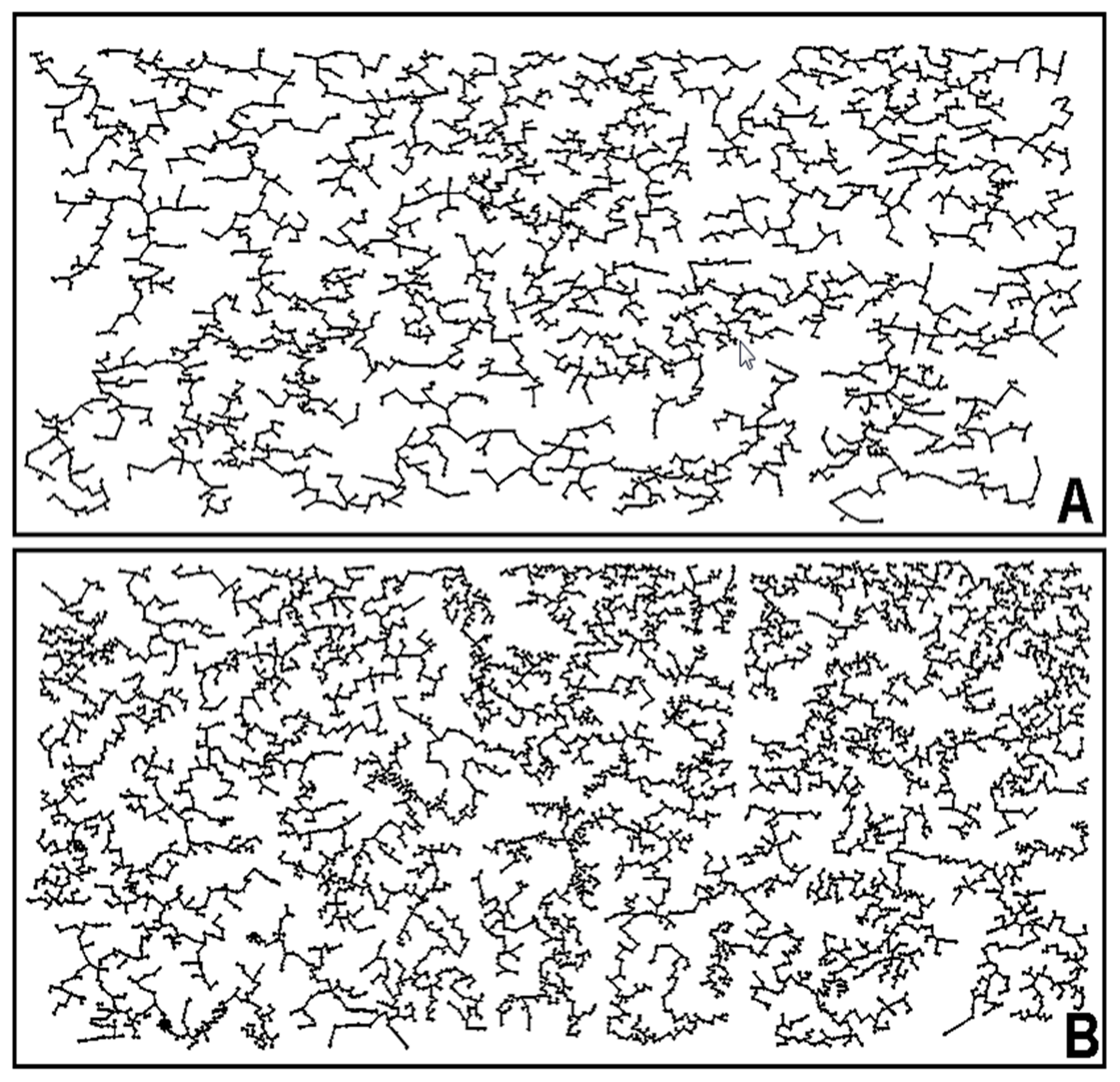
Analysis of the minimal spanning tree is another powerful guide to envision predicted opportunities of cell–cell interactions based on statistical analyses of the spatial connectivity between individual community members within the landscape [
77]. This method of spatial analysis creates a subgraph image of the original landscape with each cell point linked by the shortest linear vertex to its closest neighbor, ultimately producing a nearest-neighbor network of vertices inevitably connecting all cells to each other in a single tree with a multi-branched architecture having minimum total length and containing no closed loops [
27,
42]. Visual inspection of the tree reveals local aggregated patches where increased densities of vertices with short lengths predict high probability of intense cell–cell interactions. Minimal spanning trees derived from nearest neighbor point analysis of representative montage images of biofilm communities A and B (
Figure 1i–j) are presented in
Figure 16A,B. The minimal spanning tree of community B provides a vivid representation of greater connectivity among many more patch areas with branched vertices of shorter length.
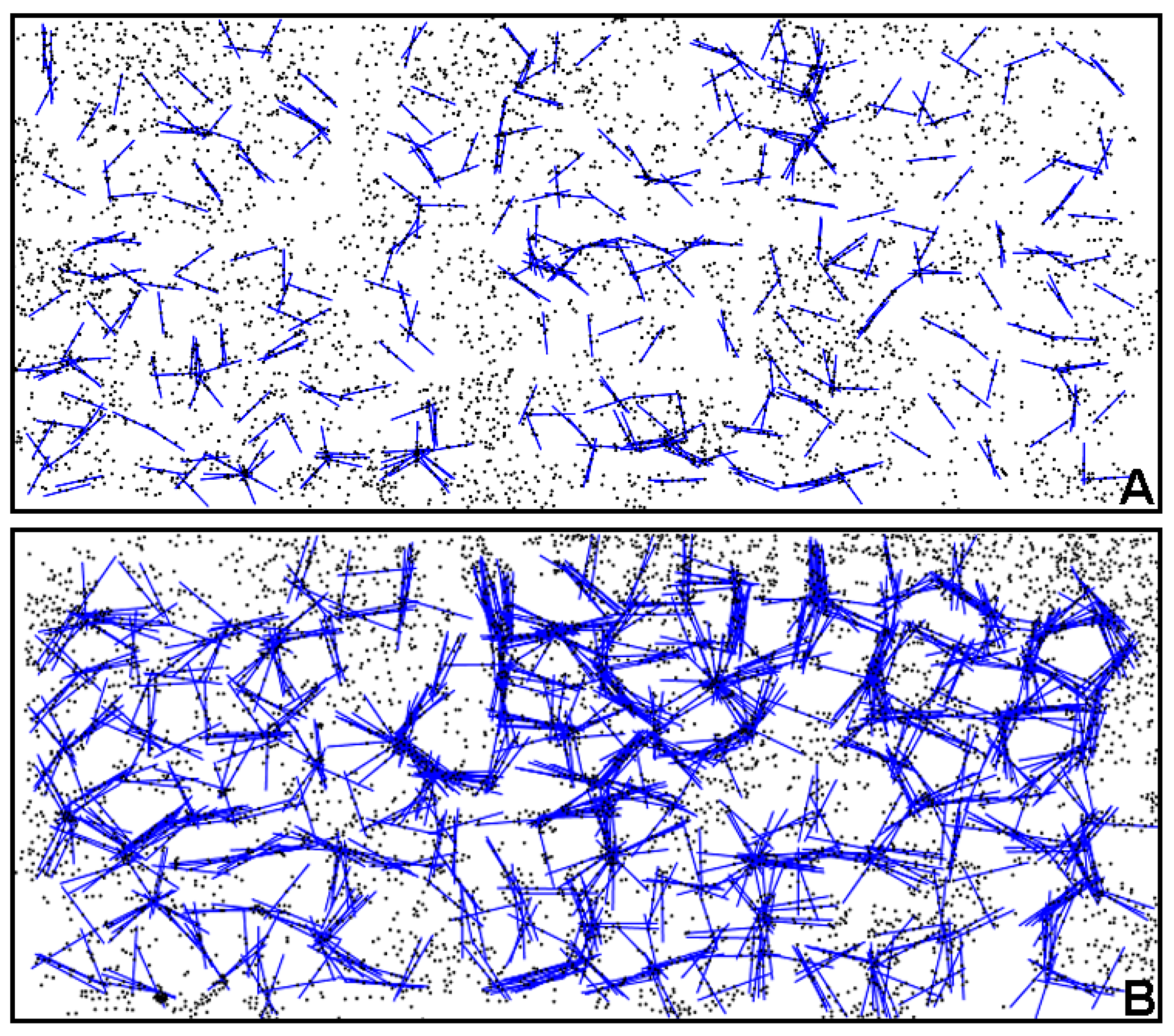
Further indications of the spatial location of intense cell–cell interactions are provided in two-dimensional directional plots that use a continuous sector method to transform individual object point positions noted by their Cartesian coordinates into another domain of statistically significant, linear alignments within the landscape [
5,
42,
78]. Plots of linear point alignments computed at equivalent sampling intervals for river biofilm communities A and B are presented in
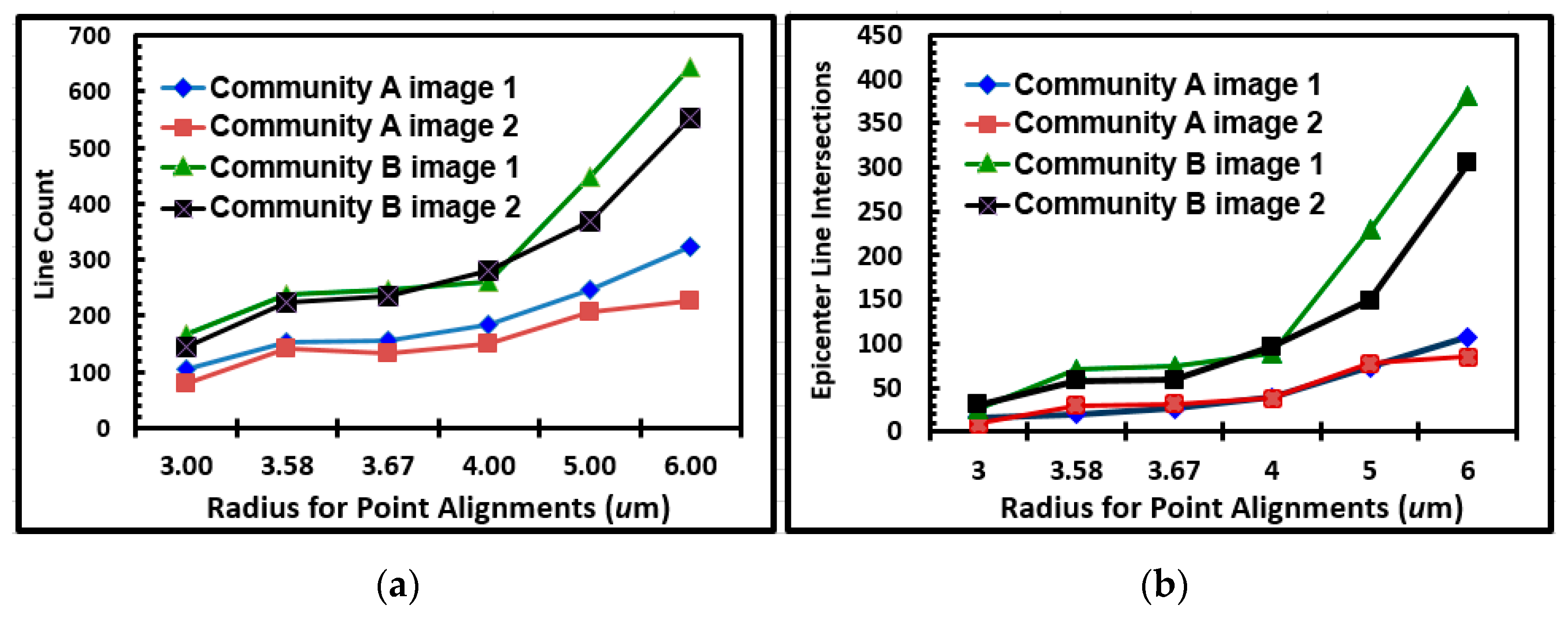
Figure 17A,B, respectively. These results indicated many linear alignments whose multi-directional angular orientations identified more “hot-spot epicenters” of interpoint intersections created by intense clusters of closely neighboring bacteria in the community B developed on the polystyrene substratum. Quantitative assessments of the number of linear alignments and their epicenters of clustered intersections confirmed their increased intensities for community B over a range of increasing radial distances of sampling (
Figure 18a,b). These results provide additional evidence supporting the hypothesis that cell–cell interactions are predictably more abundant and intense within the spatially clustered patterns of cells in the community B biofilm developed on the polystyrene substratum.
The final method of spatial ecology analysis for this study involved a geostatistical approach that measures the dependency among
z variate observations in georeferenced space in order to evaluate the continuity or continuous variation of spatial patterns over that entire landscape domain [
5,
7,
44,
72]. It does so by quantifying the resemblance between
z variate values at neighboring points as a function of their spatial separation distance [
5,
7,
44,
72]. The data indicate positive autocorrelation if the
z variate values of neighboring pairs are more similar when located nearby rather than far apart [
5,
72], as occurs when communities are spatially clustered to facilitate cell–cell communication and cross-feeding [
9,
38,
62]. When found, autocorrelated
z variates are then mathematically modeled using regionalized variable theory to connect various spatially dependent relationships of their ecology, including the range of real-world radial distances at which they occur in situ [
44,
68,
72].
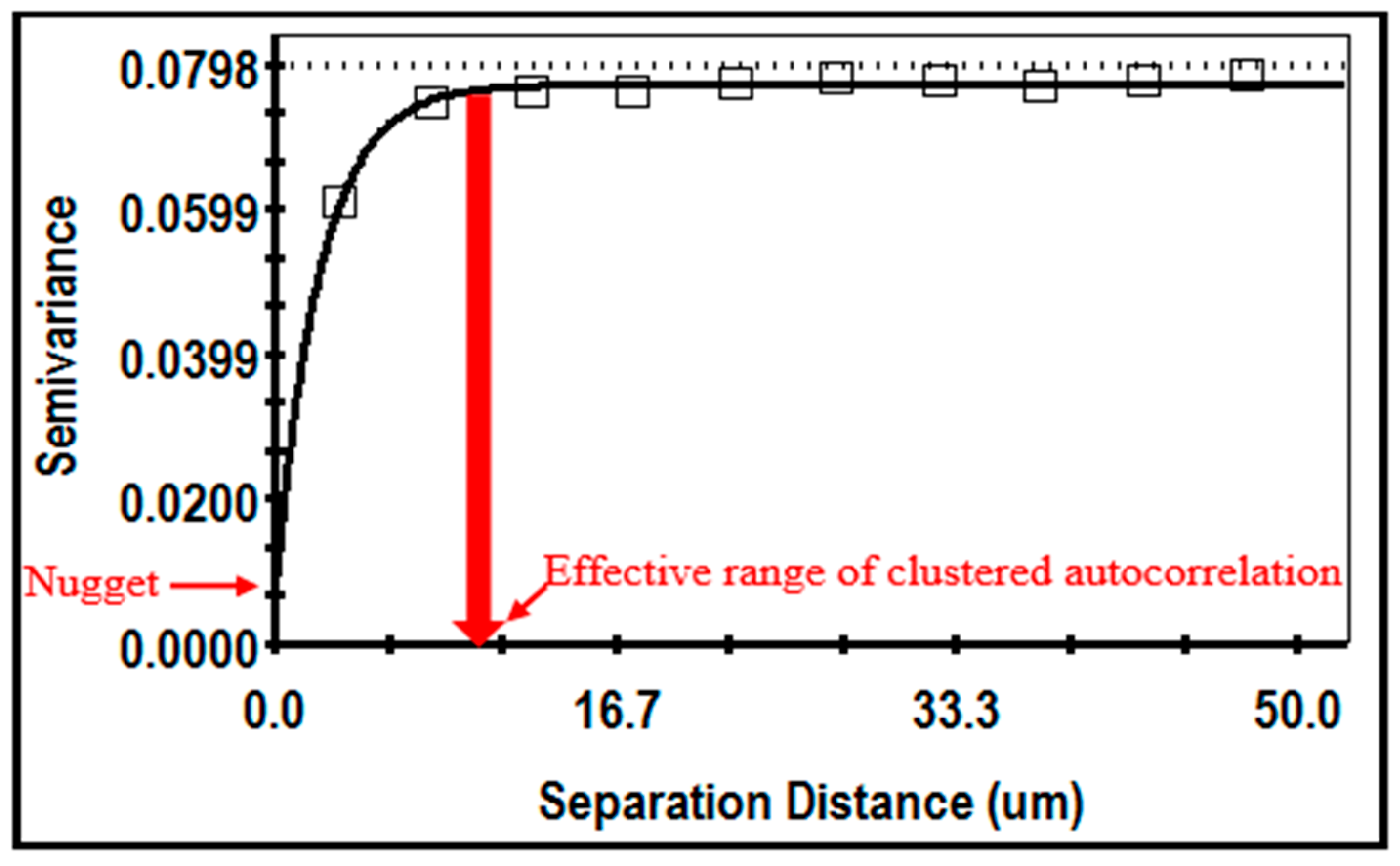
Geostatistical analysis produces a semivariogram (
Figure 19) describing the extent that the measured
z variate exhibits autocorrelated spatial dependence between all cell pairs at multiple sampled locations [
5,
7,
68,
72]. Spatial autocorrelation of two
z variates were evaluated in this study: a CMEIAS cluster index indicating the intensity of aggregated colonization behavior between nearest cell neighbors [
5,
45], and the cell biovolume to test for cell–cell interactions among neighbors affecting their allometric metabolism and growth ecophysiology [
5,
10,
37]. For microbial biofilm analyses, these two
z variates typically have units of μm
−1 and μm
3, respectively.
Figure 19 shows an example of the isotropic semivariogram for the cluster index of cells in a montage image of the biofilm community B. Important discriminating features include the nugget at the Y-axis intercept denoting the amount of measured microstructure that is not spatially dependent, and the effective separation range indicating the
X-axis value at 95% of the model’s asymptote height, representing the maximal separation distance between sampling points at which the
z variate is still autocorrelated [
5,
7,
68,
72]. This example indicates a strong spatial autocorrelation of the cluster index, with a very small nugget (sufficient points have been adequately sampled) and the autocorrelated effective range that defines the maximal radial distance between cells that still influences their neighbor’s ability to congregate locally in situ within the defined spatial domain. Geostatistical tests for geometric
anisotropy in the semivariograms at 0, 45, 90, and 135 compass degrees did not indicate a preferential bias in directionality of cells in the biofilms, suggesting no major directional influence of hydrodynamic forces exerted on their cell positioning during development of the biofilms within the gently flowing river.
Table 17 summarizes the geostatistical analyses of the cluster index and biovolume
z variates for equal sampling efforts of individual cells in the two biofilm community landscapes. Both biofilms exhibited spatially-dependent isotropic autocorrelation for both
z variates. Their nugget variances were small, indicating that the analyses were adequately sampled with little discontinuity of small-scale variation, and the majority of their measured microstructure was spatially autocorrelated. The exponential model made an acceptable fit (low residual sum of squares) to the semivariogram data for both community
z variates. Two-tailed Student
t and Mann–Whitney tests indicated that the cluster index for cells in community B had significantly greater mean and median values (
p of 1.52 × 10
−141 and 0.000, respectively). In addition, the effective separation ranges for both autocorrelated
z variates were somewhat longer (hence stronger) with the biofilm community B developed on the polystyrene substratum (statistically significant for biovolume). Most cells in both communities had nearest neighbor distances that positioned them well within the corresponding specific effective ranges of influence, and thus their “socially-adapted” proximity to each other was sufficient to enable spatially autocorrelated cell–cell interactions affecting these ecophysiologically relevant metrics. These geospatial analyses provide statistical proof of spatially structured biofilm communities with predominantly aggregated distribution patterns that exhibit positive cooperative interactions between proximal cells benefitting their colonization and productivity, and also reveal the real-world spatial dimensions at which these cell–cell interactions extend in situ.