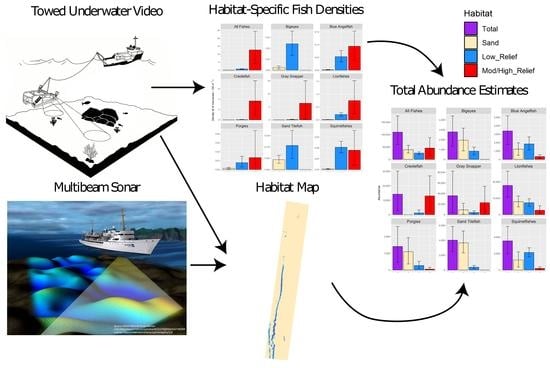
Integrating Towed Underwater Video and Multibeam Acoustics for Marine Benthic Habitat Mapping and Fish Population Estimation
Abstract
1. Introduction
2. Materials and Methods
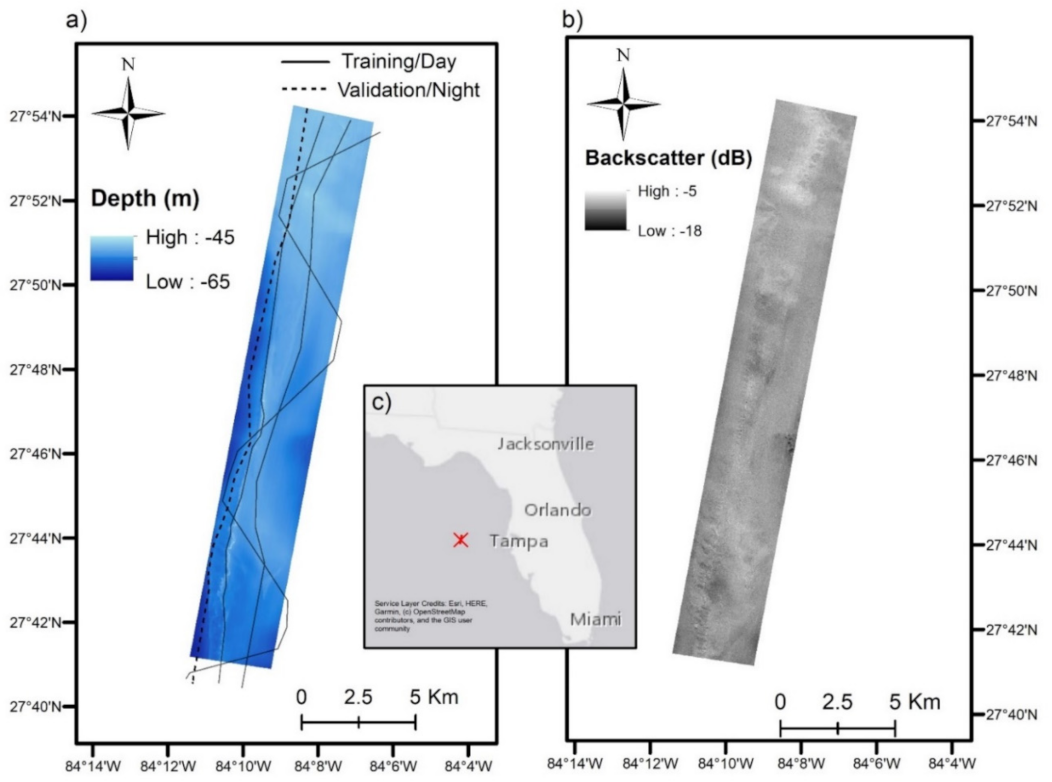
2.1. Study Area
2.2. Data Collection and Processing
2.2.1. Multibeam Echosounder Data
2.2.2. Towed Underwater Video
2.3. Predictive Habitat Mapping
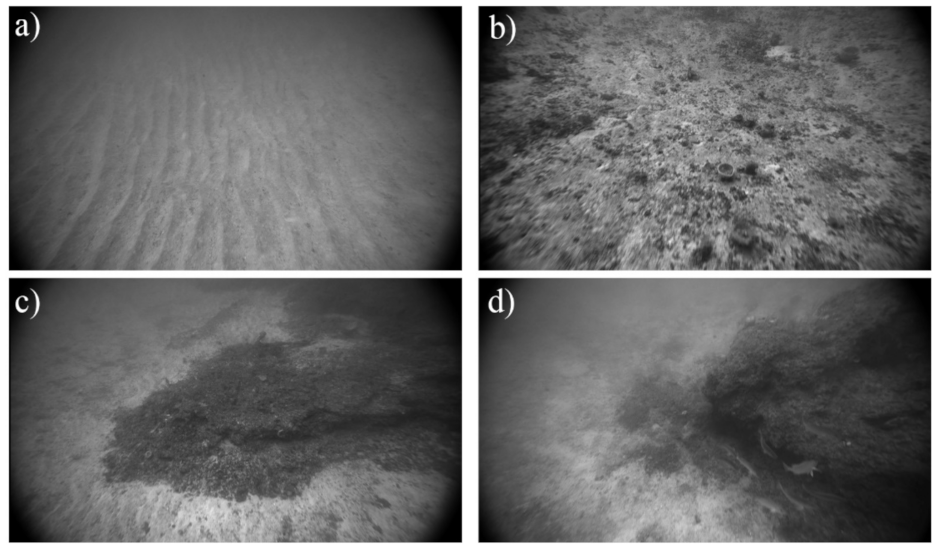
2.3.1. Response Variable: Ground-Truth Substrate Observations
2.3.2. Predictor Variables: MBES Data and Their Derivative Features
2.3.3. Estimating Towed Video Position

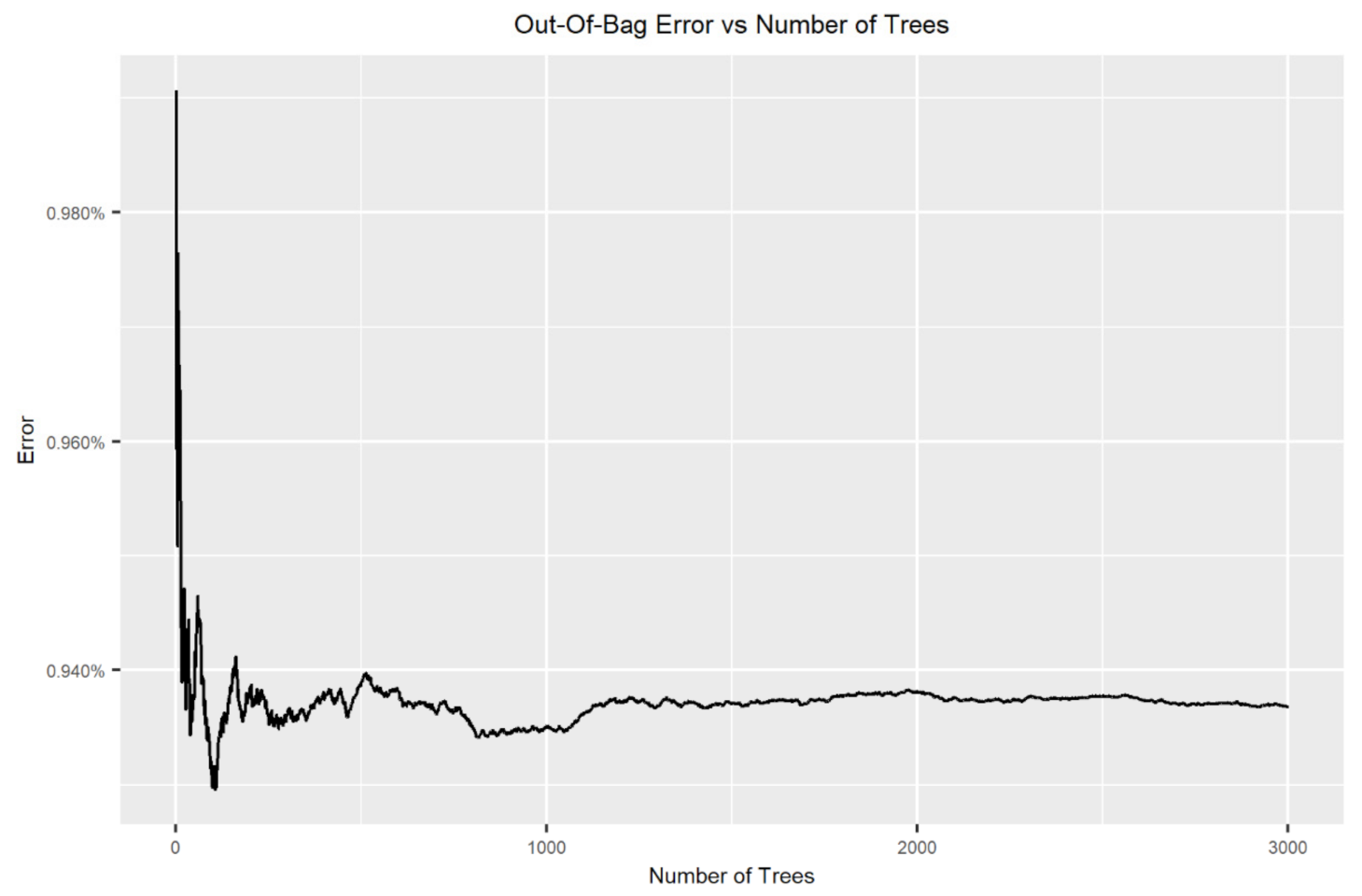
2.3.4. Classification Algorithms
2.3.5. Variable Selection and Dimensionality Reduction
2.3.6. Accuracy Assessment
2.3.7. Map Comparison
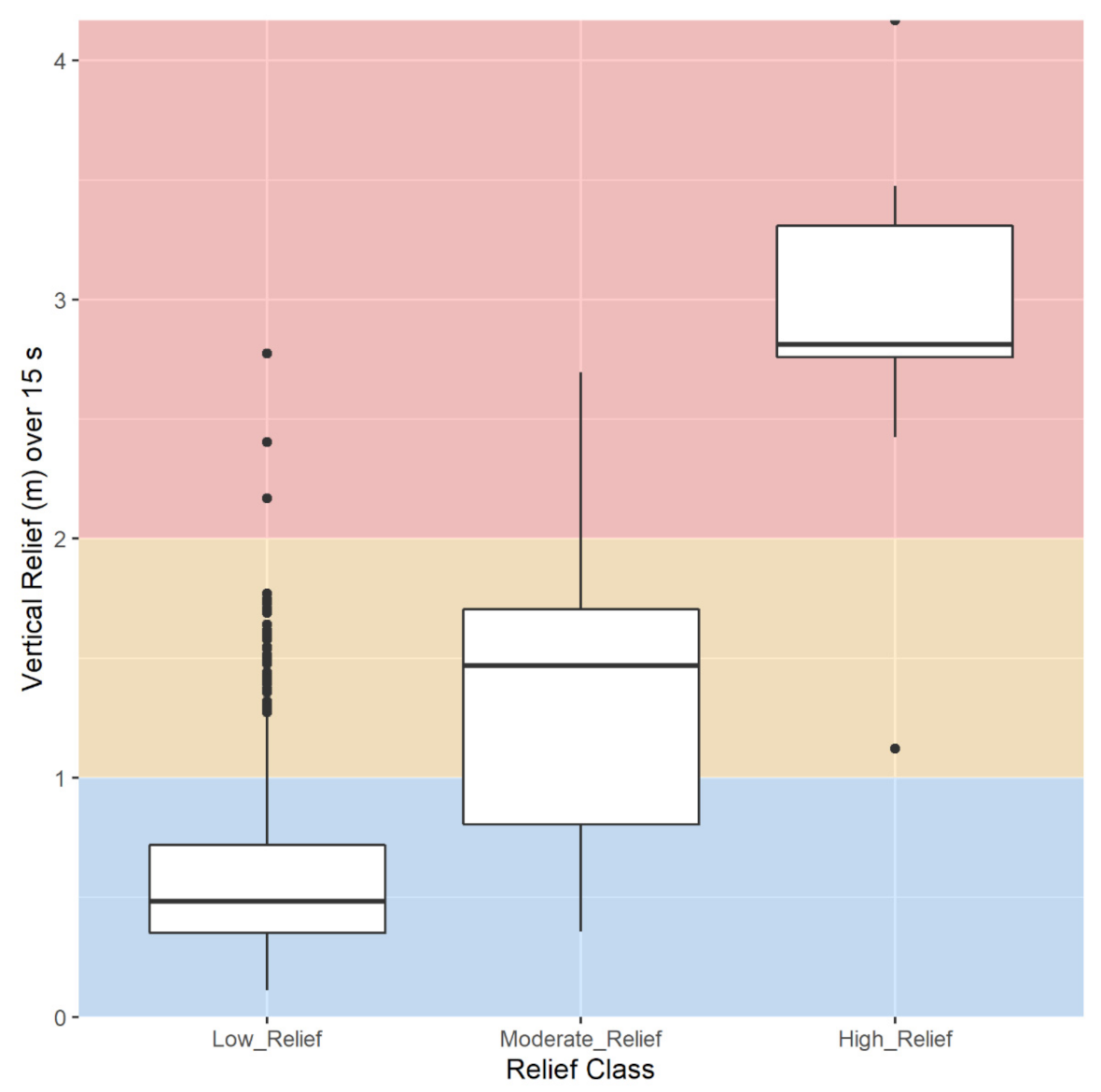
2.3.8. Vertical Relief
2.4. Fish Community Analyses and Abundance Estimates
2.4.1. Fish Densities
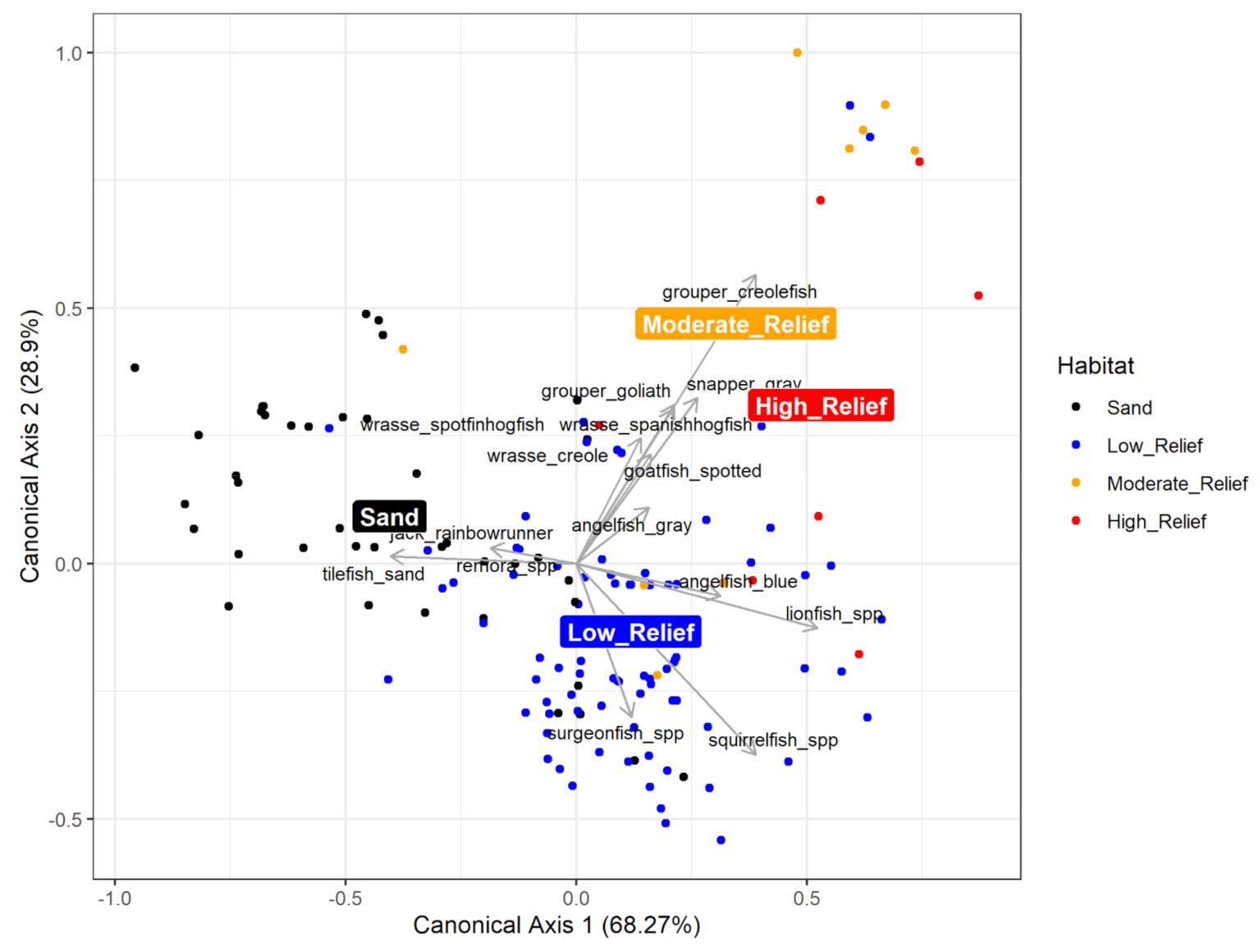
2.4.2. Multivariate Community Analysis
2.4.3. Fish Abundance Estimates
- Db: Average density of fish taxa for bin b;
- n: number of bins for a given habitat type;
- cb: number of fish of a given taxa in bin b;
- Ab: Aera viewed by the camera over bin b;
- : Average density of fish taxa for habitat type b;
- Abd: Abundance of fish taxa summed across all habitat types;
- nh: number of habitat classes;
- Ah: Area of habitat type h in the entire study area.
3. Results
3.1. Predictive Habitat Mapping
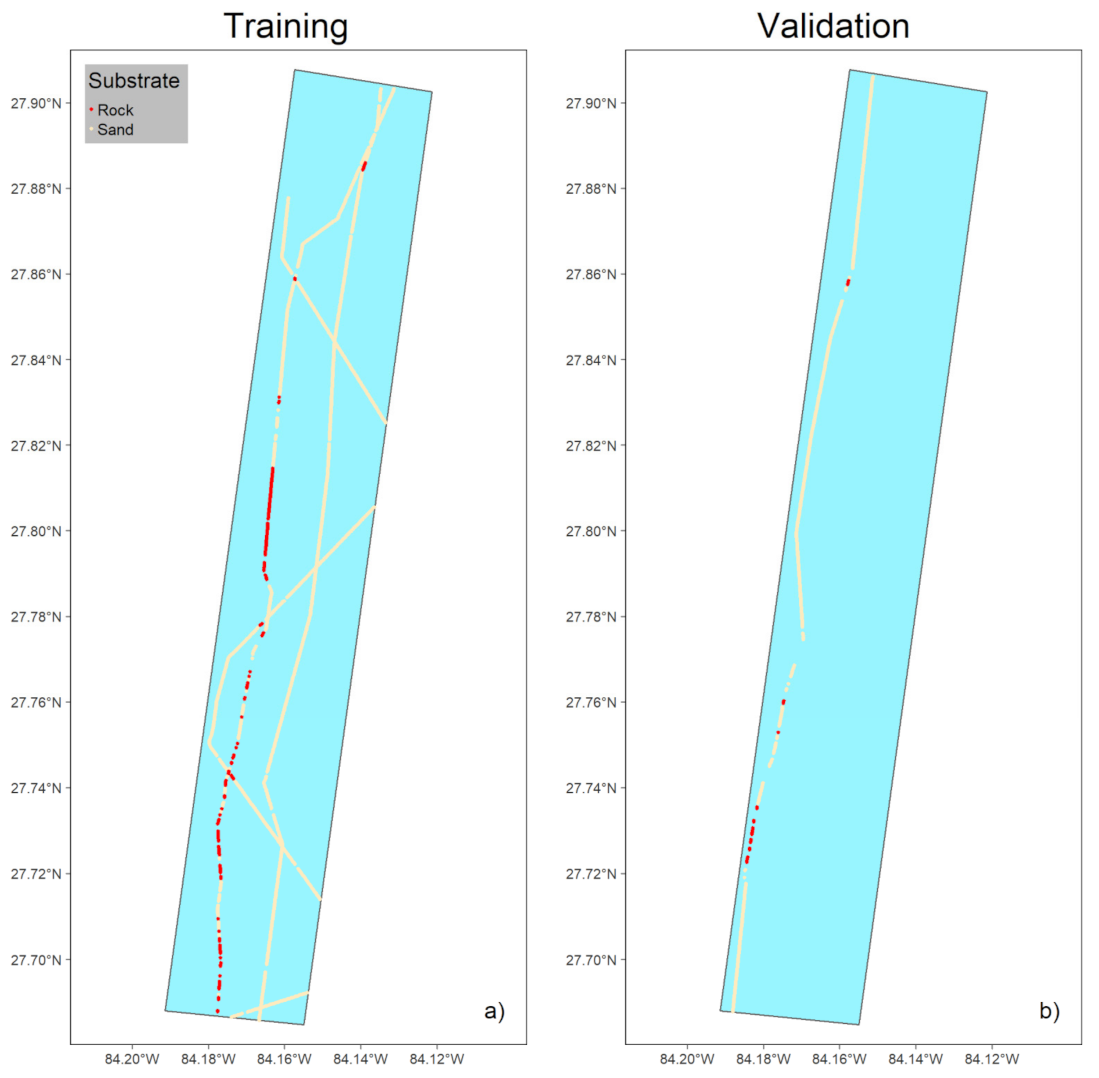
3.1.1. Preparing Ground-Truth Data
3.1.2. Supervised Classification
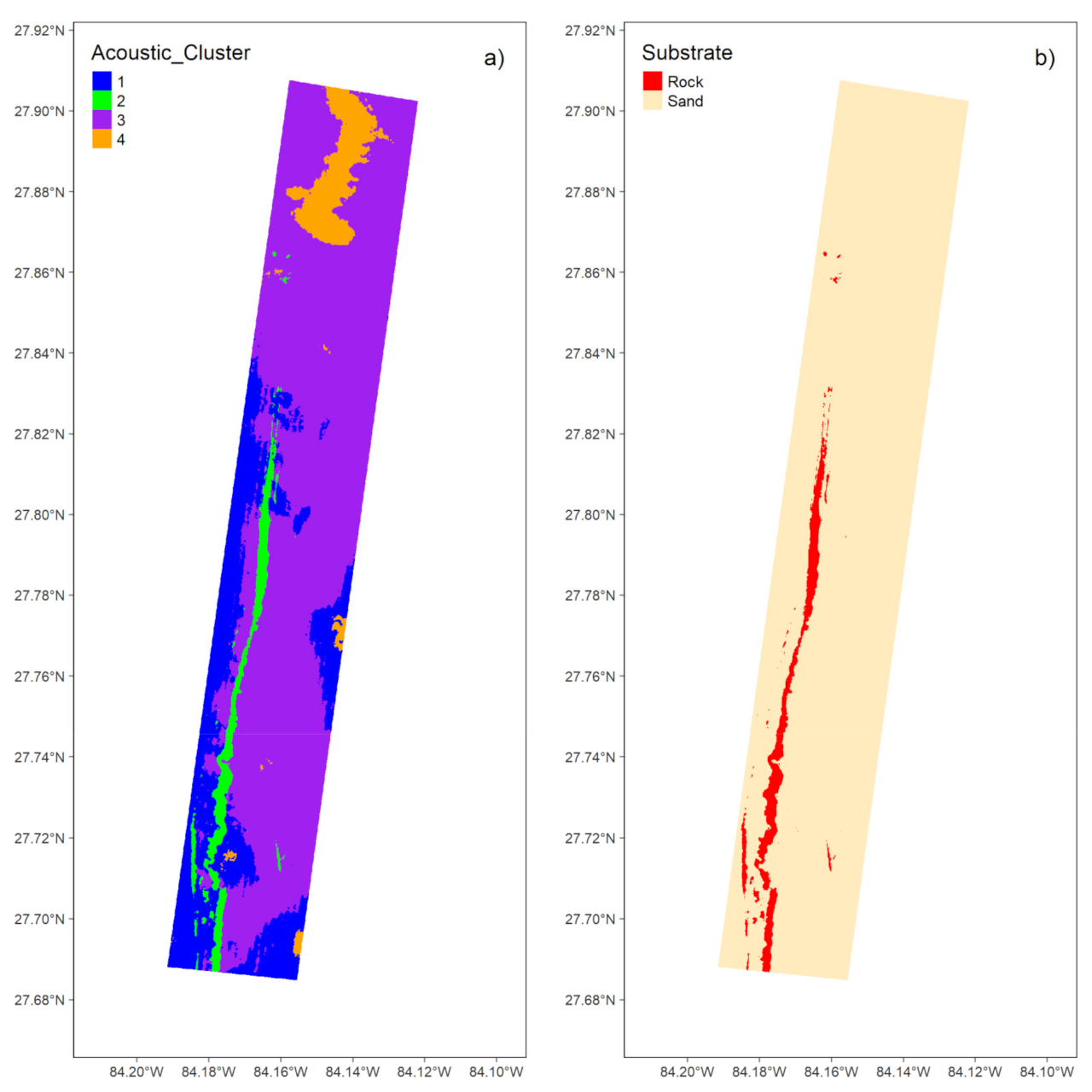
3.1.3. Unsupervised Classification
3.1.4. Map Comparison
3.1.5. Vertical Relief
3.2. Fish Community Analyses and Abundance Estimates
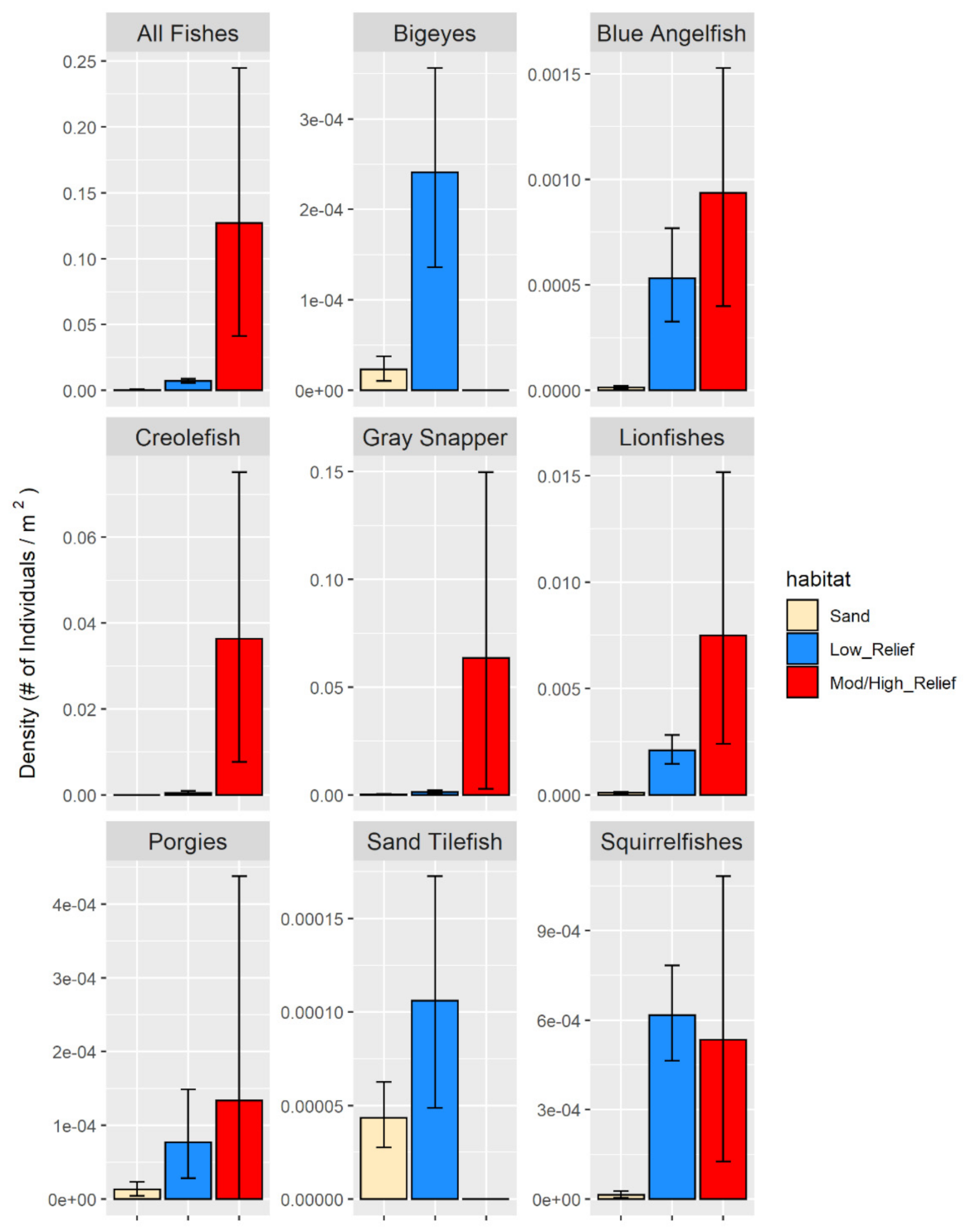
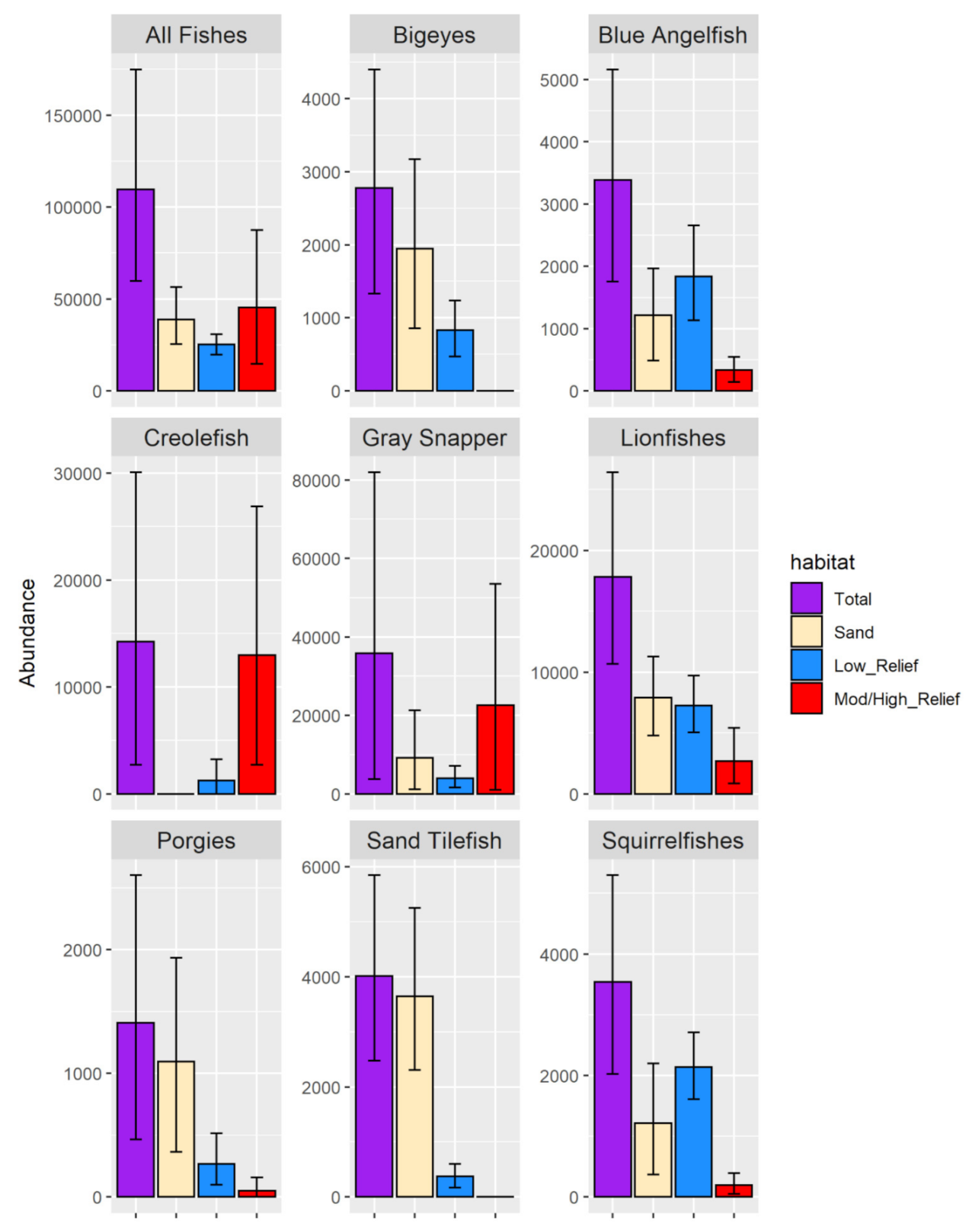
3.2.1. Fish Densities
3.2.2. Multivariate Community Analysis
3.2.3. Fish Abundance Estimates
4. Discussion
4.1. Comparison of the Supervised and Unsupervised Procedures
4.2. Variable Importance for Substrate Mapping
4.3. Fish Community Analysis and Abundance Estimates
4.4. Conclusions, Limitations, and Future Work
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Taxa | Habitat | Lower Bound | Mean | Upper Bound |
|---|---|---|---|---|
| All_Fish | Sand | 0.000305091368 | 0.000464386728 | 0.000675358543 |
| All_Fish | Low_Relief | 0.005654144932 | 0.007306868473 | 0.008924795926 |
| All_Fish | Mod/High_Relief | 0.041204024881 | 0.127203256069 | 0.245024232512 |
| amberjack_spp | Sand | 0.000000000000 | 0.000002902417 | 0.000007282130 |
| amberjack_spp | Low_Relief | 0.000038238047 | 0.000404866063 | 0.000988826791 |
| amberjack_spp | Mod/High_Relief | 0.000000000000 | 0.000133616866 | 0.000390694044 |
| angelfish_blue | Sand | 0.000005804730 | 0.000014512085 | 0.000023436164 |
| angelfish_blue | Low_Relief | 0.000326755837 | 0.000530181749 | 0.000767483569 |
| angelfish_blue | Mod/High_Relief | 0.000399268295 | 0.000935318059 | 0.001528539056 |
| angelfish_gray | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| angelfish_gray | Low_Relief | 0.000009508321 | 0.000057838009 | 0.000126721804 |
| angelfish_gray | Mod/High_Relief | 0.000000000000 | 0.000133616866 | 0.000426533497 |
| angelfish_spp | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| angelfish_spp | Low_Relief | 0.000000000000 | 0.000009639668 | 0.000029689030 |
| angelfish_spp | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| bigeye_spp | Sand | 0.000010242236 | 0.000023219336 | 0.000037805805 |
| bigeye_spp | Low_Relief | 0.000136342357 | 0.000240991704 | 0.000356470592 |
| bigeye_spp | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| boxfish_spp | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| boxfish_spp | Low_Relief | 0.000000000000 | 0.000019279336 | 0.000048421997 |
| boxfish_spp | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| butterflyfish_spp | Sand | 0.000000000000 | 0.000002902417 | 0.000007276943 |
| butterflyfish_spp | Low_Relief | 0.000009548271 | 0.000048198341 | 0.000105886188 |
| butterflyfish_spp | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| eel_spp | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| eel_spp | Low_Relief | 0.000000000000 | 0.000009639668 | 0.000029404678 |
| eel_spp | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| goatfish_spotted | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| goatfish_spotted | Low_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| goatfish_spotted | Mod/High_Relief | 0.000000000000 | 0.000133616866 | 0.000430797429 |
| grouper_black | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| grouper_black | Low_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| grouper_black | Mod/High_Relief | 0.000000000000 | 0.000400850597 | 0.001324759414 |
| grouper_creolefish | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| grouper_creolefish | Low_Relief | 0.000000000000 | 0.000356667722 | 0.000928362228 |
| grouper_creolefish | Mod/High_Relief | 0.007643028251 | 0.036343787448 | 0.075195350579 |
| grouper_gag | Sand | 0.000000000000 | 0.000001451209 | 0.000004381866 |
| grouper_gag | Low_Relief | 0.000000000000 | 0.000019279336 | 0.000059046688 |
| grouper_gag | Mod/High_Relief | 0.000000000000 | 0.000267233731 | 0.000852786071 |
| grouper_goliath | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| grouper_goliath | Low_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| grouper_goliath | Mod/High_Relief | 0.000000000000 | 0.000400850597 | 0.001065084821 |
| grouper_red | Sand | 0.000000000000 | 0.000001451209 | 0.000004381866 |
| grouper_red | Low_Relief | 0.000009425807 | 0.000038558673 | 0.000078463175 |
| grouper_red | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| grouper_scamp | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| grouper_scamp | Low_Relief | 0.000009680947 | 0.000144595023 | 0.000359476725 |
| grouper_scamp | Mod/High_Relief | 0.000000000000 | 0.000133616866 | 0.000430797429 |
| grouper_spp | Sand | 0.000001443556 | 0.000005804834 | 0.000011675402 |
| grouper_spp | Low_Relief | 0.000000000000 | 0.000019279336 | 0.000048726459 |
| grouper_spp | Mod/High_Relief | 0.000000000000 | 0.000133616866 | 0.000426393036 |
| jack_crevalle | Sand | 0.000000000000 | 0.000001451209 | 0.000004375663 |
| jack_crevalle | Low_Relief | 0.000000000000 | 0.000028919005 | 0.000077360701 |
| jack_crevalle | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| jack_rainbowrunner | Sand | 0.000000000000 | 0.000002902417 | 0.000008756979 |
| jack_rainbowrunner | Low_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| jack_rainbowrunner | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| jack_spp | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| jack_spp | Low_Relief | 0.000000000000 | 0.000009639668 | 0.000029677705 |
| jack_spp | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| lionfish_spp | Sand | 0.000056869715 | 0.000094328554 | 0.000134798264 |
| lionfish_spp | Low_Relief | 0.001457130610 | 0.002091807993 | 0.002804478909 |
| lionfish_spp | Mod/High_Relief | 0.002402247287 | 0.007482544475 | 0.015170870783 |
| porgy_spp | Sand | 0.000004360488 | 0.000013060877 | 0.000023075921 |
| porgy_spp | Low_Relief | 0.000028376948 | 0.000077117345 | 0.000148411135 |
| porgy_spp | Mod/High_Relief | 0.000000000000 | 0.000133616866 | 0.000437293986 |
| remora_spp | Sand | 0.000000000000 | 0.000001451209 | 0.000004377825 |
| remora_spp | Low_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| remora_spp | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| shark_spp | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| shark_spp | Low_Relief | 0.000000000000 | 0.000009639668 | 0.000029219565 |
| shark_spp | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| snapper_gray | Sand | 0.000013061514 | 0.000108840639 | 0.000254981252 |
| snapper_gray | Low_Relief | 0.000468269192 | 0.001156760180 | 0.002055718899 |
| snapper_gray | Mod/High_Relief | 0.002742580078 | 0.063468011169 | 0.149759809054 |
| snapper_spp | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| snapper_spp | Low_Relief | 0.000000000000 | 0.000019279336 | 0.000059229663 |
| snapper_spp | Mod/High_Relief | 0.000000000000 | 0.000133616866 | 0.000390694044 |
| snapper_yellowtail | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| snapper_yellowtail | Low_Relief | 0.000000000000 | 0.000038558673 | 0.000116011333 |
| snapper_yellowtail | Mod/High_Relief | 0.000000000000 | 0.000267233731 | 0.000809175808 |
| squirrelfish_spp | Sand | 0.000004354164 | 0.000014512085 | 0.000026267894 |
| squirrelfish_spp | Low_Relief | 0.000464723937 | 0.000616938763 | 0.000782913741 |
| squirrelfish_spp | Mod/High_Relief | 0.000125668348 | 0.000534467462 | 0.001081371358 |
| stingray_spp | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| stingray_spp | Low_Relief | 0.000000000000 | 0.000009639668 | 0.000029666787 |
| stingray_spp | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| surgeonfish_spp | Sand | 0.000000000000 | 0.000001451209 | 0.000004388087 |
| surgeonfish_spp | Low_Relief | 0.000057776382 | 0.000134955354 | 0.000233778550 |
| surgeonfish_spp | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| tilefish_sand | Sand | 0.000027559370 | 0.000043536256 | 0.000062686638 |
| tilefish_sand | Low_Relief | 0.000048742988 | 0.000106036350 | 0.000172505726 |
| tilefish_sand | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| triggerfish_spp | Sand | 0.000000000000 | 0.000004353626 | 0.000010194629 |
| triggerfish_spp | Low_Relief | 0.000009284980 | 0.000038558673 | 0.000078463175 |
| triggerfish_spp | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| wrasse_creole | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| wrasse_creole | Low_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| wrasse_creole | Mod/High_Relief | 0.000000000000 | 0.001336168656 | 0.004307974287 |
| wrasse_hogfish | Sand | 0.000000000000 | 0.000001451209 | 0.000004377401 |
| wrasse_hogfish | Low_Relief | 0.000009404868 | 0.000038558673 | 0.000078650541 |
| wrasse_hogfish | Mod/High_Relief | 0.000000000000 | 0.000133616866 | 0.000418370109 |
| wrasse_pearlyrazorfish | Sand | 0.000000000000 | 0.000001451209 | 0.000004378067 |
| wrasse_pearlyrazorfish | Low_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| wrasse_pearlyrazorfish | Mod/High_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| wrasse_spanishhogfish | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| wrasse_spanishhogfish | Low_Relief | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| wrasse_spanishhogfish | Mod/High_Relief | 0.000000000000 | 0.000267233731 | 0.000654899428 |
| wrasse_spotfinhogfish | Sand | 0.000000000000 | 0.000000000000 | 0.000000000000 |
| wrasse_spotfinhogfish | Low_Relief | 0.000000000000 | 0.000009639668 | 0.000029467959 |
| wrasse_spotfinhogfish | Mod/High_Relief | 0.000000000000 | 0.000133616866 | 0.000421591353 |
| Common Name | Scientific Name | Family | Order | Number of Individuals Observed |
|---|---|---|---|---|
| Amberjack spp. | Seriola spp. | Carangidae | Perciformes | 45 |
| (Jacks) | ||||
| Angelfish spp. | Pomacanthidae | Perciformes | 1 | |
| (Angelfishes) | ||||
| Blue Angelfish | Holacanthus bermudensis | Pomacanthidae | Perciformes | 72 |
| (Angelfishes) | ||||
| Gray Angelfish | Pomacanthus arcuatus | Pomacanthidae | Perciformes | 7 |
| (Angelfishes) | ||||
| Bigeye spp. | Priacanthidae | Perciformes | 41 | |
| (Bigeyes) | ||||
| Boxfish spp. | Ostraciidae | Tetraodontiformes | 2 | |
| (Boxfishes) | ||||
| Butterflyfish spp. | Chaetodontidae | Perciformes | 7 | |
| (Butterflyfishes) | ||||
| Eel spp. | Anguilliformes | 1 | ||
| Spotted Goatfish | Pseudupeneus maculatus | Mullidae | Perciformes | 1 |
| (Goatfishes) | ||||
| Grouper spp. | Serranidae | Perciformes | 7 | |
| (Groupers and Sea Basses) | ||||
| Black Grouper | Mycteroperca bonaci | Serranidae | Perciformes | 3 |
| (Groupers and Sea Basses) | ||||
| Atlantic Creolefish | Paranthias furcifer | Serranidae | Perciformes | 309 |
| (Groupers and Sea Basses) | ||||
| Gag Grouper | Mycteroperca microlepis | Serranidae | Perciformes | 5 |
| (Groupers and Sea Basses) | ||||
| Goliath Grouper | Epinephelus itajara | Serranidae | Perciformes | 3 |
| (Groupers and Sea Basses) | ||||
| Red Grouper | Epinephelus morio | Serranidae | Perciformes | 5 |
| (Groupers and Sea Basses) | ||||
| Scamp | Mycteroperca phenax | Serranidae | Perciformes | 16 |
| (Groupers and Sea Basses) | ||||
| Jack spp. | Carangidae | Perciformes | 1 | |
| (Jacks) | ||||
| Crevalle Jack | Caranx hippos | Carangidae | Perciformes | 4 |
| (Jacks) | ||||
| Rainbow Runner | Elagatis bipinnulata | Carangidae | Perciformes | 2 |
| (Jacks) | ||||
| Lionfish spp. | Pterois spp. | Scorpaenidae | Scorpaeniformes | 338 |
| (Scorpionfishes) | ||||
| Porgy spp. | Sparidae | Perciformes | 18 | |
| (Porgies) | ||||
| Remora spp. | Echeneidae | Perciformes | 1 | |
| (Remoras) | ||||
| Shark spp. | Superorder: Selachimorpha | 1 | ||
| Snapper spp. | Lutjanidae | Perciformes | 3 | |
| (Snappers) | ||||
| Gray Snapper | Lutjanus griseus | Lutjanidae | Perciformes | 670 |
| (Snappers) | ||||
| Yellowtail Snapper | Ocyurus chrysurus | Lutjanidae | Perciformes | 6 |
| (Snappers) | ||||
| Squirrelfish spp. | Holocentridae | Beryciformes | 78 | |
| (squirrelfishes) | ||||
| Whiptail stingray spp. | Dasyatidae | Rajiformes | 1 | |
| (Whiptail Stingrays) | ||||
| Surgeonfish spp. | Acanthuridae | Perciformes | 15 | |
| (Surgeonfishes) | ||||
| Sand Tilefish | Malacanthus plumieri | Malacanthidae | Perciformes | 41 |
| (tilefishes) | ||||
| Triggerfish spp. | Balistidae | Tetraodontiformes | 7 | |
| (Triggerfishes) | ||||
| Creole Wrasse | Clepticus parrae | Labridae | Perciformes | 10 |
| (Wrasses) | ||||
| Hogfish | Lachnolaimus maximus | Labridae | Perciformes | 6 |
| (Wrasses) | ||||
| Pearly Razorfish | Xyrichtys novacula | Labridae | Perciformes | 1 |
| (Wrasses) | ||||
| Spanish Hogfish | Bodianus rufus | Labridae | Perciformes | 2 |
| (Wrasses) | ||||
| Spotfin Hogfish | Bodianus pulchellus | Labridae | Perciformes | 2 |
| (Wrasses) | ||||
| Large No ID | 298 |
| Taxa | Habitat | Lower Bound | Mean | Upper Bound |
|---|---|---|---|---|
| All_Fish | Total | 59,842 | 109,616 | 174,961 |
| All_Fish | Sand | 25,552 | 38,893 | 56,563 |
| All_Fish | Low_Relief | 19,576 | 25,299 | 30,900 |
| All_Fish | Mod/High_Relief | 14,714 | 45,424 | 87,498 |
| amberjack_spp | Total | 132 | 1693 | 4173 |
| amberjack_spp | Sand | 0 | 243 | 610 |
| amberjack_spp | Low_Relief | 132 | 1402 | 3424 |
| amberjack_spp | Mod/High_Relief | 0 | 48 | 140 |
| angelfish_blue | Total | 1760 | 3385 | 5166 |
| angelfish_blue | Sand | 486 | 1215 | 1963 |
| angelfish_blue | Low_Relief | 1131 | 1836 | 2657 |
| angelfish_blue | Mod/High_Relief | 143 | 334 | 546 |
| angelfish_gray | Total | 33 | 248 | 591 |
| angelfish_gray | Sand | 0 | 0 | 0 |
| angelfish_gray | Low_Relief | 33 | 200 | 439 |
| angelfish_gray | Mod/High_Relief | 0 | 48 | 152 |
| angelfish_spp | Total | 0 | 33 | 103 |
| angelfish_spp | Sand | 0 | 0 | 0 |
| angelfish_spp | Low_Relief | 0 | 33 | 103 |
| angelfish_spp | Mod/High_Relief | 0 | 0 | 0 |
| bigeye_spp | Total | 1330 | 2779 | 4401 |
| bigeye_spp | Sand | 858 | 1945 | 3166 |
| bigeye_spp | Low_Relief | 472 | 834 | 1234 |
| bigeye_spp | Mod/High_Relief | 0 | 0 | 0 |
| boxfish_spp | Total | 0 | 67 | 168 |
| boxfish_spp | Sand | 0 | 0 | 0 |
| boxfish_spp | Low_Relief | 0 | 67 | 168 |
| boxfish_spp | Mod/High_Relief | 0 | 0 | 0 |
| butterflyfish_spp | Total | 33 | 410 | 976 |
| butterflyfish_spp | Sand | 0 | 243 | 609 |
| butterflyfish_spp | Low_Relief | 33 | 167 | 367 |
| butterflyfish_spp | Mod/High_Relief | 0 | 0 | 0 |
| eel_spp | Total | 0 | 33 | 102 |
| eel_spp | Sand | 0 | 0 | 0 |
| eel_spp | Low_Relief | 0 | 33 | 102 |
| eel_spp | Mod/High_Relief | 0 | 0 | 0 |
| goatfish_spotted | Total | 0 | 48 | 154 |
| goatfish_spotted | Sand | 0 | 0 | 0 |
| goatfish_spotted | Low_Relief | 0 | 0 | 0 |
| goatfish_spotted | Mod/High_Relief | 0 | 48 | 154 |
| grouper_black | Total | 0 | 143 | 473 |
| grouper_black | Sand | 0 | 0 | 0 |
| grouper_black | Low_Relief | 0 | 0 | 0 |
| grouper_black | Mod/High_Relief | 0 | 143 | 473 |
| grouper_creolefish | Total | 2729 | 14,213 | 30,067 |
| grouper_creolefish | Sand | 0 | 0 | 0 |
| grouper_creolefish | Low_Relief | 0 | 1235 | 3214 |
| grouper_creolefish | Mod/High_Relief | 2729 | 12,978 | 26,852 |
| grouper_gag | Total | 0 | 284 | 876 |
| grouper_gag | Sand | 0 | 122 | 367 |
| grouper_gag | Low_Relief | 0 | 67 | 204 |
| grouper_gag | Mod/High_Relief | 0 | 95 | 305 |
| grouper_goliath | Total | 0 | 143 | 380 |
| grouper_goliath | Sand | 0 | 0 | 0 |
| grouper_goliath | Low_Relief | 0 | 0 | 0 |
| grouper_goliath | Mod/High_Relief | 0 | 143 | 380 |
| grouper_red | Total | 33 | 255 | 639 |
| grouper_red | Sand | 0 | 122 | 367 |
| grouper_red | Low_Relief | 33 | 134 | 272 |
| grouper_red | Mod/High_Relief | 0 | 0 | 0 |
| grouper_scamp | Total | 34 | 548 | 1398 |
| grouper_scamp | Sand | 0 | 0 | 0 |
| grouper_scamp | Low_Relief | 34 | 501 | 1245 |
| grouper_scamp | Mod/High_Relief | 0 | 48 | 154 |
| grouper_spp | Total | 121 | 601 | 1299 |
| grouper_spp | Sand | 121 | 486 | 978 |
| grouper_spp | Low_Relief | 0 | 67 | 169 |
| grouper_spp | Mod/High_Relief | 0 | 48 | 152 |
| jack_crevalle | Total | 0 | 222 | 634 |
| jack_crevalle | Sand | 0 | 122 | 366 |
| jack_crevalle | Low_Relief | 0 | 100 | 268 |
| jack_crevalle | Mod/High_Relief | 0 | 0 | 0 |
| jack_rainbowrunner | Total | 0 | 243 | 733 |
| jack_rainbowrunner | Sand | 0 | 243 | 733 |
| jack_rainbowrunner | Low_Relief | 0 | 0 | 0 |
| jack_rainbowrunner | Mod/High_Relief | 0 | 0 | 0 |
| jack_spp | Total | 0 | 33 | 103 |
| jack_spp | Sand | 0 | 0 | 0 |
| jack_spp | Low_Relief | 0 | 33 | 103 |
| jack_spp | Mod/High_Relief | 0 | 0 | 0 |
| lionfish_spp | Total | 10,666 | 17,815 | 26,417 |
| lionfish_spp | Sand | 4763 | 7900 | 11,290 |
| lionfish_spp | Low_Relief | 5045 | 7242 | 9710 |
| lionfish_spp | Mod/High_Relief | 858 | 2672 | 5418 |
| porgy_spp | Total | 463 | 1409 | 2603 |
| porgy_spp | Sand | 365 | 1094 | 1933 |
| porgy_spp | Low_Relief | 98 | 267 | 514 |
| porgy_spp | Mod/High_Relief | 0 | 48 | 156 |
| remora_spp | Total | 0 | 122 | 367 |
| remora_spp | Sand | 0 | 122 | 367 |
| remora_spp | Low_Relief | 0 | 0 | 0 |
| remora_spp | Mod/High_Relief | 0 | 0 | 0 |
| shark_spp | Total | 0 | 33 | 101 |
| shark_spp | Sand | 0 | 0 | 0 |
| shark_spp | Low_Relief | 0 | 33 | 101 |
| shark_spp | Mod/High_Relief | 0 | 0 | 0 |
| snapper_gray | Total | 3695 | 35,785 | 81,952 |
| snapper_gray | Sand | 1094 | 9116 | 21,355 |
| snapper_gray | Low_Relief | 1621 | 4005 | 7118 |
| snapper_gray | Mod/High_Relief | 979 | 22,664 | 53,479 |
| snapper_spp | Total | 0 | 114 | 345 |
| snapper_spp | Sand | 0 | 0 | 0 |
| snapper_spp | Low_Relief | 0 | 67 | 205 |
| snapper_spp | Mod/High_Relief | 0 | 48 | 140 |
| snapper_yellowtail | Total | 0 | 229 | 691 |
| snapper_yellowtail | Sand | 0 | 0 | 0 |
| snapper_yellowtail | Low_Relief | 0 | 134 | 402 |
| snapper_yellowtail | Mod/High_Relief | 0 | 95 | 289 |
| squirrelfish_spp | Total | 2019 | 3542 | 5297 |
| squirrelfish_spp | Sand | 365 | 1215 | 2200 |
| squirrelfish_spp | Low_Relief | 1609 | 2136 | 2711 |
| squirrelfish_spp | Mod/High_Relief | 45 | 191 | 386 |
| stingray_spp | Total | 0 | 33 | 103 |
| stingray_spp | Sand | 0 | 0 | 0 |
| stingray_spp | Low_Relief | 0 | 33 | 103 |
| stingray_spp | Mod/High_Relief | 0 | 0 | 0 |
| surgeonfish_spp | Total | 200 | 589 | 1177 |
| surgeonfish_spp | Sand | 0 | 122 | 368 |
| surgeonfish_spp | Low_Relief | 200 | 467 | 809 |
| surgeonfish_spp | Mod/High_Relief | 0 | 0 | 0 |
| tilefish_sand | Total | 2477 | 4013 | 5847 |
| tilefish_sand | Sand | 2308 | 3646 | 5250 |
| tilefish_sand | Low_Relief | 169 | 367 | 597 |
| tilefish_sand | Mod/High_Relief | 0 | 0 | 0 |
| triggerfish_spp | Total | 32 | 498 | 1125 |
| triggerfish_spp | Sand | 0 | 365 | 854 |
| triggerfish_spp | Low_Relief | 32 | 134 | 272 |
| triggerfish_spp | Mod/High_Relief | 0 | 0 | 0 |
| wrasse_creole | Total | 0 | 477 | 1538 |
| wrasse_creole | Sand | 0 | 0 | 0 |
| wrasse_creole | Low_Relief | 0 | 0 | 0 |
| wrasse_creole | Mod/High_Relief | 0 | 477 | 1538 |
| wrasse_hogfish | Total | 33 | 303 | 788 |
| wrasse_hogfish | Sand | 0 | 122 | 367 |
| wrasse_hogfish | Low_Relief | 33 | 134 | 272 |
| wrasse_hogfish | Mod/High_Relief | 0 | 48 | 149 |
| wrasse_pearlyrazorfish | Total | 0 | 122 | 367 |
| wrasse_pearlyrazorfish | Sand | 0 | 122 | 367 |
| wrasse_pearlyrazorfish | Low_Relief | 0 | 0 | 0 |
| wrasse_pearlyrazorfish | Mod/High_Relief | 0 | 0 | 0 |
| wrasse_spanishhogfish | Total | 0 | 95 | 234 |
| wrasse_spanishhogfish | Sand | 0 | 0 | 0 |
| wrasse_spanishhogfish | Low_Relief | 0 | 0 | 0 |
| wrasse_spanishhogfish | Mod/High_Relief | 0 | 95 | 234 |
| wrasse_spotfinhogfish | Total | 0 | 81 | 253 |
| wrasse_spotfinhogfish | Sand | 0 | 0 | 0 |
| wrasse_spotfinhogfish | Low_Relief | 0 | 33 | 102 |
| wrasse_spotfinhogfish | Mod/High_Relief | 0 | 48 | 151 |
References
- Cogan, C.B.; Todd, B.J.; Lawton, P.; Noji, T.T. The Role of Marine Habitat Mapping in Ecosystem-Based Management. ICES J. Mar. Sci. 2009, 66, 2033–2042. [Google Scholar] [CrossRef]
- Kurland, J.; Woodby, D. What Is Marine Habitat Mapping and Why Do Managers Need It? University of Alaska Fairbanks: Fairbanks, AK, USA, 2008. [Google Scholar]
- Baker, E.K.; Harris, P.T. Habitat Mapping and Marine Management. In Seafloor Geomorphology as Benthic Habitat: GeoHab Atlas of Seafloor Geomorphic Features and Benthic Habitats, 2nd ed.; Harris, P.T., Baker, E.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 17–33. [Google Scholar] [CrossRef]
- Harris, P.T.; Baker, E.K. Why Map Benthic Habitats? In Seafloor Geomorphology as Benthic Habitat: GeoHab Atlas of Seafloor Geomorphic Features and Benthic Habitats, 2nd ed.; Harris, P.T., Baker, E.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 3–15. [Google Scholar] [CrossRef]
- Copeland, A.; Edinger, E.; Devillers, R.; Bell, T.; LeBlanc, P.; Wroblewski, J. Marine Habitat Mapping in Support of Marine Protected Area Management in a Subarctic Fjord: Gilbert Bay, Labrador, Canada. J. Coast. Conserv. 2013, 17, 225–237. [Google Scholar] [CrossRef]
- Buhl-Mortensen, L.; Buhl-Mortensen, P.; Dolan, M.; Gonzalez-Mirelis, G. Habitat Mapping as a Tool for Conservation and Sustainable Use of Marine Resources: Some Perspectives from the MAREANO Programme, Norway. J. Sea Res. 2015, 100, 46–61. [Google Scholar] [CrossRef]
- Kostylev, V.E.; Todd, B.J.; Fader, G.B.; Courtney, R.; Cameron, G.D.; Pickrill, R.A. Benthic Habitat Mapping on the Scotian Shelf Based on Multibeam Bathymetry, Surficial Geology and Sea Floor Photographs. Mar. Ecol. Prog. Ser. 2001, 219, 121–137. [Google Scholar] [CrossRef]
- Kenny, A.; Cato, I.; Desprez, M.; Fader, G.; Schüttenhelm, R.; Side, J. An Overview of Seabed-Mapping Technologies in the Context of Marine Habitat Classification. ICES J. Mar. Sci. 2003, 60, 411–418. [Google Scholar] [CrossRef]
- Vassallo, P.; Bianchi, C.N.; Paoli, C.; Holon, F.; Navone, A.; Bavestrello, G.; Vietti, R.C.; Morri, C. A Predictive Approach to Benthic Marine Habitat Mapping: Efficacy and Management Implications. Mar. Pollut. Bull. 2018, 131, 218–232. [Google Scholar] [CrossRef]
- Brown, C.J.; Smith, S.J.; Lawton, P.; Anderson, J.T. Benthic Habitat Mapping: A Review of Progress Towards Improved Understanding of the Spatial Ecology of the Seafloor Using Acoustic Techniques. Estuar. Coast. Shelf Sci. 2011, 92, 502–520. [Google Scholar] [CrossRef]
- Lamarche, G.; Orpin, A.R.; Mitchell, J.S.; Pallentin, A. Benthic Habitat Mapping. In Biological Sampling in the Deep Sea; Clark, M.R., Consalvey, M., Rowden, A.A., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2016; pp. 80–102. [Google Scholar] [CrossRef]
- Goff, J.; Olson, H.; Duncan, C. Correlation of Side-Scan Backscatter Intensity with Grain-Size Distribution of Shelf Sediments, New Jersey Margin. Geo Mar. Lett. 2000, 20, 43–49. [Google Scholar] [CrossRef]
- Collier, J.; Brown, C. Correlation of Sidescan Backscatter with Grain Size Distribution of Surficial Seabed Sediments. Mar. Geol. 2005, 214, 431–449. [Google Scholar] [CrossRef]
- McGonigle, C.; Collier, J.S. Interlinking Backscatter, Grain Size and Benthic Community Structure. Estuar. Coast. Shelf Sci. 2014, 147, 123–136. [Google Scholar] [CrossRef]
- Brizzolara, J. Characterizing Benthic Habitat Using Multibeam Sonar and Towed Underwater Video in Two Marine Protected Areas on the West Florida Shelf, US. Master’s Thesis, University of South Florida, Tampa, FL, USA, 2017. [Google Scholar]
- Brizzolara, J.L.; Grasty, S.E.; Ilich, A.R.; Gray, J.W.; Naar, D.F.; Murawski, S.A. Characterizing Benthic Habitats in Two Marine Protected Areas on the West Florida Shelf. In Seafloor Geomorphology as Benthic Habitat: GeoHab Atlas of Seafloor Geomorphic Features and Benthic Habitats, 2nd ed.; Harris, P.T., Baker, E.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 605–618. [Google Scholar] [CrossRef]
- Costa, B. Multispectral Acoustic Backscatter: How Useful Is it for Marine Habitat Mapping and Management? J. Coast. Res. 2019, 35, 1062–1079. [Google Scholar] [CrossRef]
- Stevens, T.; Connolly, R.M. Testing the Utility of Abiotic Surrogates for Marine Habitat Mapping at Scales Relevant to Management. Biol. Conserv. 2004, 119, 351–362. [Google Scholar] [CrossRef]
- Harris, P.T. Surrogacy. In Seafloor Geomorphology as Benthic Habitat: GeoHab Atlas of Seafloor Geomorphic Features and Benthic Habitats, 2nd ed.; Harris, P.T., Baker, E.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 97–113. [Google Scholar] [CrossRef]
- Diesing, M.; Green, S.L.; Stephens, D.; Lark, R.M.; Stewart, H.A.; Dove, D. Mapping Seabed Sediments: Comparison of Manual, Geostatistical, Object-Based Image Analysis and Machine Learning Approaches. Continent. Shelf Res. 2014, 84, 107–119. [Google Scholar] [CrossRef]
- Lecours, V.; Devillers, R.; Simms, A.E.; Lucieer, V.L.; Brown, C.J. Towards a Framework for Terrain Attribute Selection in Environmental Studies. Environ. Model. Softw. 2017, 89, 19–30. [Google Scholar] [CrossRef]
- Wilson, M.F.; O’Connell, B.; Brown, C.; Guinan, J.C.; Grehan, A.J. Multiscale Terrain Analysis of Multibeam Bathymetry Data for Habitat Mapping on the Continental Slope. Mar. Geod. 2007, 30, 3–35. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, 610–621. [Google Scholar] [CrossRef]
- Hall-Beyer, M. GLCM Textrure: A Tutorial; University of Calgary: Calgary, AL, Canada, 2017. [Google Scholar]
- Porskamp, P.; Rattray, A.; Young, M.; Ierodiaconou, D. Multiscale and Hierarchical Classification for Benthic Habitat Mapping. Geosciences 2018, 8, 119. [Google Scholar] [CrossRef]
- Hasan, R.C.; Ierodiaconou, D.; Laurenson, L.; Schimel, A. Integrating Multibeam Backscatter Angular Response, Mosaic and Bathymetry Data for Benthic Habitat Mapping. PLoS ONE 2014, 9, e97339. [Google Scholar] [CrossRef]
- Prampolini, M.; Blondel, P.; Foglini, F.; Madricardo, F. Habitat Mapping of the Maltese Continental Shelf Using Acoustic Textures and Bathymetric Analyses. Estuar. Coast. Shelf Sci. 2018, 207, 483–498. [Google Scholar] [CrossRef]
- Montereale-Gavazzi, G.; Roche, M.; Lurton, X.; Degrendele, K.; Terseleer, N.; Van Lancker, V. Seafloor Change Detection Using Multibeam Echosounder Backscatter: Case Study on the Belgian Part of the North Sea. Mar. Geophys. Res. 2018, 39, 229–247. [Google Scholar] [CrossRef]
- Trzcinska, K.; Janowski, L.; Nowak, J.; Rucinska-Zjadacz, M.; Kruss, A.; von Deimling, J.S.; Pocwiardowski, P.; Tegowski, J. Spectral Features of Dual-Frequency Multibeam Echosounder Data for Benthic Habitat Mapping. Mar. Geol. 2020, 427, 106239. [Google Scholar] [CrossRef]
- Smith, S.G.; Ault, J.S.; Bohnsack, J.A.; Harper, D.E.; Luo, J.; McClellan, D.B. Multispecies Survey Design for Assessing Reef-Fish Stocks, Spatially Explicit Management Performance, and Ecosystem Condition. Fish. Res. 2011, 109, 25–41. [Google Scholar] [CrossRef]
- Bryan, M.D.; McCarthy, K. Standardized Catch Rates for Red Grouper from the United States Gulf of Mexico Vertical Line and Longline Fisheries; SEDAR42-AW-02; SEDAR: North Charleston, SC, USA, 2015; p. 32. [Google Scholar]
- Smith, M.W.; Goethel, D.; Rios, A.; Isley, J. Standardized Catch Rate Indices for Gulf of Mexico Gray Triggerfish (Balistes capriscus) Landed During 1993–2013 by the Commercial Handline Fishery; SEDAR43-WP-05; SEDAR: North Charleston, SC, USA, 2015; p. 16. [Google Scholar]
- SEDAR. SEDAR 51: Gulf of Mexico Gray Snapper Stock Assessment Report; SEDAR: North Charleston, SC, USA, 2018; p. 428. [Google Scholar]
- Switzer, T.S.; Tyler-Jedlund, A.J.; Keenan, S.F.; Weather, E.J. Benthic Habitats, as Derived from Classification of Side-Scan-Sonar Mapping Data, Are Important Determinants of Reef-Fish Assemblage Structure in the Eastern Gulf of Mexico. Mar. Coast. Fish. Dynam. Manag. Ecosyst. Sci. 2020, 12, 21–32. [Google Scholar] [CrossRef]
- Keenan, S.F.; Switzer, T.S.; Thompson, K.A.; Tyler-Jedlund, A.J.; Knapp, A.R. Comparison of Reef-Fish Assemblages Between Artificial and Geologic Habitats in the Northeastern Gulf of Mexico: Implications for Fishery-Independent Surveys. In Marine Artificial Reef Research and Development: Integrating Fisheries Management Objectives; Bortone, S.A., Ed.; American Fisheries Society: Bethesda, MD, USA, 2018; p. 24. [Google Scholar]
- Switzer, T.S.; Keenan, S.; Purtlebaugh, C. Exploring the Utility of Side-Scan Sonar and Experimental Z-Traps in Improving the Efficiency of Fisheries-Independent Surveys of Reef Fishes on the West Florida Shelf; F4031-11-14-F, MARFIN Final Report; Florida Fish and Wildlife Research Institute: St. Petersburg, FL, USA, 2014. [Google Scholar]
- Pope, K.L.; Lochmann, S.E.; Young, M.K. Methods for Assessing Fish Populations. In Inland Fisheries Management in North America, 3rd ed.; Hubert, W.A., Quist, M.C.., Eds.; American Fisheries Society: Bethesda, MD, USA, 2010; pp. 325–351. [Google Scholar]
- National Research Council. Robust Methods for the Analysis of Images and Videos for Fisheries Stock Assessment: Summary of a Workshop; National Academies Press: Washington, DC, USA, 2015; p. 88. [Google Scholar] [CrossRef]
- Stunz, G.; Patterson, W., III; Powers, S.; Cowan, J., Jr.; Rooker, J.; Ahrens, R.; Boswell, K.; Carleton, L.; Catalano, M.; Drymon, J.; et al. Estimating the Absolute Abundance of Age-2+ Red Snapper (Lutjanus campechanus) in the U.S. Gulf of Mexico. Mississippi-Alabama Sea Grant Consortium; NOAA Sea Grant: Silver Spring, MD, USA, 2021; p. 439. [Google Scholar]
- Moe, M.A., Jr. A Survey of Offshore Fishing in Florida; SEDAR28-RD05; Marine Laboratory: St. Petersburg, FL, USA, 1963. [Google Scholar]
- Hine, A.C.; Locker, S.D. Florida Gulf of Mexico Continental Shelf: Great Contrasts and Significant Transitions. In Gulf of Mexico: Origin, Waters, and Marine Life; Buster, N.A., Holmes, C.W., Eds.; Texas A&M University Press: College Station, TX, USA, 2011; Volume 3, pp. 101–127. [Google Scholar]
- Cockrell, M.L. Spatial Dynamics and Productivity of a Gulf of Mexico Commercial Reef Fish Fishery Following Large Scale Disturbance and Management Change. Ph.D. Dissertation, University of South Florida, Tampa, FL, USA, 2018. [Google Scholar]
- Cockrell, M.L.; O’Farrell, S.; Sanchirico, J.; Murawski, S.A.; Perruso, L.; Strelcheck, A. Resilience of a Commercial Fishing Fleet Following Emergency Closures in the Gulf of Mexico. Fish. Res. 2019, 218, 69–82. [Google Scholar] [CrossRef]
- Applanix. POS MV Oceanmaster Specifications; Trimble: Vaughan, ON, Canada, 2017. [Google Scholar]
- Calder, B.R.; Wells, D.E. CUBE User’s Manual; University of New Hampshire Center for Coastal and Ocean Mapping: Durham, NH, USA, 2007; p. 54. [Google Scholar]
- Lembke, C.; Silverman, A.; Butcher, S.; Murawski, S.; Grasty, S.; Shi, X. Development and Sea Trials of a New Camera-Based Assessment Survey System for Reef Fish Stocks Assessment. In Proceedings of the 2013 Oceans-San Diego, San Diego, CA, USA, 23–27 September 2013. [Google Scholar]
- Lembke, C.; Grasty, S.; Silverman, A.; Broadbent, H.; Butcher, S.; Murawski, S. The Camera-Based Assessment Survey System (C-BASS): A Towed Camera Platform for Reef Fish Abundance Surveys and Benthic Habitat Characterization in the Gulf of Mexico. Continent. Shelf Res. 2017, 151. [Google Scholar] [CrossRef]
- Bowden, D.A.; Jones, D.O.B. Towed Camera Systems. In Biological Sampling in the Deep Sea; Clark, M.R., Consalvey, M., Rowden, A.A., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2016; pp. 260–284. [Google Scholar] [CrossRef]
- Kilborn, J.P. Investigating Marine Resources in the Gulf of Mexico at Multiple Spatial and Temporal Scales of Inquiry. Ph.D. Thesis, University of South Florida, Tampa, FL, USA, 2017. [Google Scholar]
- Federal Geographic Data Committee. Coastal and Marine Ecological Classification Standard; FGDC-STD-018-2012; Federal Geographic Data Committee: Reston, VA, USA, 2012. [Google Scholar]
- Sheehan, E.; Cousens, S.; Nancollas, S.; Stauss, C.; Royle, J.; Attrill, M. Drawing Lines at the Sand: Evidence for Functional vs. Visual Reef Boundaries in Temperate Marine Protected Areas. Mar. Pollut. Bull. 2013, 76, 194–202. [Google Scholar] [CrossRef]
- Woodward, B.; Takahashi, J. Tator: The Video and Image Annotator; CVision: Medford, MA, USA, 2017; Available online: https://github.com/cvisionai/Tator-Native/releases (accessed on 11 April 2021).
- Grasty, S. Use of a Towed Camera System for Estimating Reef Fish Population Dynamics on the West Florida Shelf. Master’s Thesis, University of South Florida, Tampa, FL, USA, 2014. [Google Scholar]
- Foody, G.M. Status of Land Cover Classification Accuracy Assessment. Remote Sens. Environ. 2002, 80, 185–201. [Google Scholar] [CrossRef]
- Stehman, S.V.; Czaplewski, R.L. Design and Analysis for Thematic Map Accuracy Assessment: Fundamental Principles. Remote Sens. Environ. 1998, 64, 331–344. [Google Scholar] [CrossRef]
- Diesing, M.; Mitchell, P.; Stephens, D. Image-Based Seabed Classification: What Can We Learn from Terrestrial Remote Sensing? ICES J. Mar. Sci. 2016, 73, 2425–2441. [Google Scholar] [CrossRef]
- Horning, N.; Leutner, B.; Wegmann, M. Land Cover or Image Classification Approaches. In Remote Sensing and GIS for Ecologists: Using Open Source Software; Wegmann, M., Leutner, B., Dech, S., Eds.; Pelagic Publishing: Exeter, UK, 2016; pp. 166–196. [Google Scholar]
- Misiuk, B.; Lecours, V.; Bell, T. A Multiscale Approach to Mapping Seabed Sediments. PLoS ONE 2018, 13, e0193647. [Google Scholar] [CrossRef]
- Lecours, V.; Devillers, R.; Schneider, D.C.; Lucieer, V.L.; Brown, C.J.; Edinger, E.N. Spatial Scale and Geographic context in Benthic Habitat Mapping: Review and Future Directions. Mar. Ecol. Progress Ser. 2015, 535, 259–284. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, 3.6.3.; R Core Team: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 11 April 2021).
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling, 3.4-5. 2020. Available online: https://CRAN.R-project.org/package=raster (accessed on 5 April 2021).
- Weiss, A. Topographic Position and Landforms Analysis. In Proceedings of the ESRI User Conference, San Diego, CA, USA, 9–13 July 2001. [Google Scholar]
- Horn, B.K. Hill Shading and the Reflectance Map. Proc. IEEE 1981, 69, 14–47. [Google Scholar] [CrossRef]
- Dolan, M.F. Calculation of Slope Angle from Bathymetry Data Using GIS-Effects of Computation Algorithms, Data Resolution and Analysis Scale; 2012.041; Geological Survey of Norway: Trondheim, Norway, 2012. [Google Scholar]
- Hyndman, R.J.; Fan, Y. Sample Quantiles in Statistical Packages. Am. Statist. 1996, 50, 361–365. [Google Scholar] [CrossRef]
- Foy, J.J.; Robinson, K.R.; Li, H.; Giger, M.L.; Al-Hallaq, H.; Armato, S.G. Variation in Algorithm Implementation Across Radiomics Software. J. Med. Imag. 2018, 5, 044505. [Google Scholar] [CrossRef] [PubMed]
- Dove, D.; Acoba, T.; DesRochers, A. Seafloor Substrate Characterization from Shallow Reefs to the Abyss: Spatially-Continuous Seafloor Mapping Using Multispectral Satellite Imagery, and Multibeam Bathymetry and Backscatter Data Within the Pacific Remote Islands Marine National Monument and the Main Hawaiian Islands; IR-18-018; NOAA Pacific Islands Fisheries Science Center: Honolulu, HI, USA, 2018; p. 37. [Google Scholar]
- Ilich, A.R. GLCMTextures; 0.1. 2021. Available online: https://github.com/ailich/GLCMTextures (accessed on 5 April 2021). [CrossRef]
- HYPACK. Common HYPACK® Drivers: Interfacing Notes; Xylem: Middletown, CT, USA, 2017. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chen, C.; Liaw, A.; Breiman, L. Using Random Forest to Learn Imbalanced Data; 666; University of California: Berkeley, CA, USA, 2004. [Google Scholar]
- Leutner, B.; Horning, N.; Schwalb-Willmann, J.; Hijmans, R.J. RStoolbox: Tools for Remote Sensing Data Analysis, 0.2.6. 2020. Available online: https://github.com/bleutner/RStoolbox (accessed on 5 April 2021).
- Wright, M.N.; Ziegler, A. ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J. Stat. Softw. 2017, 77. [Google Scholar] [CrossRef]
- Hasan, R.C.; Ierodiaconou, D.; Monk, J. Evaluation of Four Supervised Learning Methods for Benthic Habitat Mapping Using Backscatter From Multi-beam Sonar. Remote Sens. 2012, 4, 3427–3443. [Google Scholar] [CrossRef]
- Ierodiaconou, D.; Schimel, A.C.; Kennedy, D.; Monk, J.; Gaylard, G.; Young, M.; Diesing, M.; Rattray, A. Combining Pixel and Object Based Image Analysis of Ultra-High Resolution Multibeam Bathymetry and Backscatter for Habitat Mapping in Shallow Marine Waters. Mar. Geophys. Res. 2018, 39, 271–288. [Google Scholar] [CrossRef]
- Lucieer, V.; Hill, N.A.; Barrett, N.S.; Nichol, S. Do Marine Substrates ‘Look’ and ‘Sound’ the Same? Supervised Classification of Multibeam Acoustic Data Using Autonomous Underwater Vehicle Images. Estuar. Coast. Shelf Sci. 2013, 117, 94–106. [Google Scholar] [CrossRef]
- Stephens, D.; Diesing, M. A Comparison of Supervised Classification Methods for the Prediction of Substrate Type using Multibeam Acoustic and Legacy Grain-Size Data. PLoS ONE 2014, 9, e93950. [Google Scholar] [CrossRef]
- De’ath, G.; Fabricius, K.E. Classification and Regression Trees: A Powerful yet Simple Technique for Ecological Data Analysis. Ecology 2000, 81, 3178–3192. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef]
- Sousa, S.; Caeiro, S.; Painho, M. Assessment of Map Similarity of Categorical Maps Using Kappa Statistics; ISEGI: Lisbon, Portugal, 2002. [Google Scholar]
- MacQueen, J. Some Methods for Classification and Analysis of Multivariate Observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Oakland, CA, USA, 1 January 1967; pp. 281–297. [Google Scholar]
- Green, E.P.; Clark, C.D.; Edwards, A.J. Image Classification and Habitat Mapping. In Remote Sensing Handbook for Tropical Coastal Management; Edwards, A.J., Ed.; UNESCO: Paris, France, 2000; pp. 141–154. [Google Scholar]
- Kursa, M.B.; Rudnicki, W.R. Feature Selection with the Boruta Package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef]
- Strobl, C.; Zeileis, A. Danger: High Power!—Exploring the Statistical Properties of a Test for Random Forest Variable Importance; 017; University of Munich: Munich, Germany, 2008. [Google Scholar]
- Breiman, L.; Cutler, A. Random Forests-Classification Manual. Available online: https://www.stat.berkeley.edu/~breiman/RandomForests/cc_manual.htm (accessed on 11 April 2021).
- Legendre, P. Acoustic Seabed Classification Methodology: A User’s Statistical Comparison; Université de Montréal: Montreal, QC, Canada, 2002. [Google Scholar]
- Frontier, S. Étude de la Décroissance des Valeurs Propres Dans Une Analyse en Composantes Principales: Comparaison Avec le Moddle du Bâton Brisé. J. Exp. Mar. Biol. Ecol. 1976, 25, 67–75. [Google Scholar] [CrossRef]
- Jackson, D.A. Stopping Rules in Principal Components Analysis: A Comparison of Heuristical and Statistical Approaches. Ecology 1993, 74, 2204–2214. [Google Scholar] [CrossRef]
- King, J.R.; Jackson, D.A. Variable Selection in Large Environmental Data Sets Using Principal Components Analysis. Environmetrics 1999, 10, 67–77. [Google Scholar] [CrossRef]
- Congalton, R.G. A Review of Assessing the Accuracy of Classifications of Remotely Sensed Data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- McKenzie, D.P.; Mackinnon, A.J.; Péladeau, N.; Onghena, P.; Bruce, P.C.; Clarke, D.M.; Harrigan, S.; McGorry, P.D. Comparing Correlated Kappas by Resampling: Is One Level of Agreement Significantly Different from Another? J. Psychiatric Res. 1996, 30, 483–492. [Google Scholar] [CrossRef]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 623–656. [Google Scholar] [CrossRef]
- Lecours, V. On the Use of Maps and Models in Conservation and Resource Management (Warning: Results May Vary). Front. Mar. Sci. 2017, 4, 288. [Google Scholar] [CrossRef]
- Pontius, R., Jr. Quantification Error Versus Location Error in Comparison of Categorical Maps. Photogram. Eng. Remote Sens. 2000, 66, 1011–1016. [Google Scholar]
- Hagen, A. Multi-Method Assessment of Map Similarity. In Proceedings of the 5th AGILE Conference on Geographic Information Science, Mallorca, Spain, 25–27 April 2002; pp. 171–182. [Google Scholar]
- Allee, R.J.; David, A.W.; Naar, D.F. Two Shelf-Edge Marine Protected Areas in the Eastern Gulf of Mexico. In Seafloor Geomorphology as Benthic Habitat: GeoHAB Atlas of Seafloor Geomorphic Features and Benthic Habitats, 1st ed.; Harris, P.T., Baker, E.K., Eds.; Elsevier: Amsterdam, The Netherlands, 2012; pp. 435–448. [Google Scholar] [CrossRef]
- Parker, R.O., Jr.; Chester, A.J.; Nelson, R.S. A Video Transect Method for Estimating Reef Fish Abundance, Composition, and Habitat Utilization at Gray’s Reef National Marine Sanctuary, Georgia. Fish. Bull. 1994, 92, 787–799. [Google Scholar]
- Gratwicke, B.; Speight, M. The Relationship Between Fish Species Richness, Abundance and Habitat Complexity in a Range of Shallow Tropical Marine Habitats. J. Fish Biol. 2005, 66, 650–667. [Google Scholar] [CrossRef]
- Kendall, M.S.; Bauer, L.J.; Jeffrey, C.F. Influence of Hard Bottom Morphology on Fish Assemblages of the Continental Shelf off Georgia, Southeastern USA. Bull. Mar. Sci. 2009, 84, 265–286. [Google Scholar]
- Logan, J.M.; Young, M.A.; Harvey, E.S.; Schimel, A.C.; Ierodiaconou, D. Combining Underwater Video Methods Improves Effectiveness of Demersal Fish Assemblage Surveys Across Habitats. Mar. Ecol. Prog. Ser. 2017, 582, 181–200. [Google Scholar] [CrossRef]
- McCollough, E. Photographic Topography. Industry: A Monthly Magazine Devoted to Science, Engineering and Mechanic Arts; Industrial Publishing Company: San Francisco, CA, USA, 1893; pp. 399–406. [Google Scholar]
- Anderson, M.J. A New Method for Non-Parametric Multivariate Analysis of Variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Anderson, M.J. Distance-Based Tests for Homogeneity of Multivariate Dispersions. Biometrics 2006, 62, 245–253. [Google Scholar] [CrossRef]
- Anderson, M.J.; Willis, T.J. Canonical Analysis of Principal Coordinates: A Useful Method of Constrained Ordination for Ecology. Ecology 2003, 84, 511–525. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a Package of R Functions for Community Ecology. J. Vegetat. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, 2.5-7. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 5 April 2021).
- Kindt, R.; Coe, R. Tree Diversity Analysis: A Manual and Software for Common Statistical Methods for Ecological and Biodiversity Studies; World Agroforestry Centre: Nairobi, Kenya, 2005. [Google Scholar]
- Kindt, R. BiodiversityR, 2.12-3. 2020. Available online: http://www.worldagroforestry.org/output/tree-diversity-analysis (accessed on 5 April 2021).
- Clarke, K.; Warwick, R. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation, 2nd ed.; PRIMER-E Ltd: Plymouth, UK, 2001. [Google Scholar]
- Holm, S. A Simple Sequentially Rejective Multiple Test Procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Rice, W.R. Analyzing Tables of Statistical Tests. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef]
- Efron, B.; Tibshirani, R. The Bootstrap Method for Assessing Statistical Accuracy. Behaviormetrika 1985, 12, 1–35. [Google Scholar] [CrossRef]
- Lecours, V.; Brown, C.J.; Devillers, R.; Lucieer, V.L.; Edinger, E.N. Comparing Selections of Environmental Variables for Ecological Studies: A Focus on Terrain Attributes. PLoS ONE 2016, 11, e0167128. [Google Scholar] [CrossRef] [PubMed]
- Foody, G. Local Characterization of Thematic Classification Accuracy Through Spatially Constrained Confusion Matrices. Int. J. Remote Sens. 2005, 26, 1217–1228. [Google Scholar] [CrossRef]
- Steele, B.M.; Winne, J.C.; Redmond, R.L. Estimation and Mapping of Misclassification Probabilities for Thematic Land Cover Maps. Remote Sens. Environ. 1998, 66, 192–202. [Google Scholar] [CrossRef]
- Kyriakidis, P.C.; Dungan, J.L. A Geostatistical Approach for Mapping Thematic Classification Accuracy and Evaluating the Impact of Inaccurate Spatial Data on Ecological Model Predictions. Environ. Ecol. Stat. 2001, 8, 311–330. [Google Scholar] [CrossRef]
- Ierodiaconou, D.; Laurenson, L.; Burq, S.; Reston, M. Marine Benthic Habitat Mapping Using Multibeam Data, Georeferenced Video and Image Classification Techniques in Victoria, Australia. J. Spat. Sci. 2007, 52, 93–104. [Google Scholar] [CrossRef]
- Borland, H.P.; Gilby, B.L.; Henderson, C.J.; Leon, J.X.; Schlacher, T.A.; Connolly, R.M.; Pittman, S.J.; Sheaves, M.; Olds, A.D. The Influence of Seafloor Terrain on Fish and Fisheries: A Global Synthesis. Fish Fish. 2021. [Google Scholar] [CrossRef]
- Rivoirard, J.; Simmonds, J.; Foote, K.; Fernandes, P.; Bez, N. Geostatistics for Estimating Fish Abundance; Wiley-Blackwell: Bodmin, Cornwakk, UK, 2000. [Google Scholar]
- Switzer, T.S.; (Florida Fish and Wildlife Conservation Commission Fish and Wildlife Research Institute, St. Petersburg, FL, USA); Ilich, A.R.; (University of South Florida College of Marine Science, St. Petersburg, FL, USA). Personal communication, 2021.
- Cailliet, G.M.; Andrews, A.H.; Wakefield, W.W.; Moreno, G.; Rhodes, K.L. Fish Faunal and Habitat Analyses Using Trawls, Camera Sleds and Submersibles in Benthic Deep-Sea Habitats off Central California. Oceanol. Acta 1999, 22, 579–592. [Google Scholar] [CrossRef][Green Version]
- Stoner, A.W.; Ryer, C.H.; Parker, S.J.; Auster, P.J.; Wakefield, W.W. Evaluating the Role of Fish Behavior in Surveys Conducted with Underwater Vehicles. Can. J. Fish. Aquat. Sci. 2008, 65, 1230–1243. [Google Scholar] [CrossRef]
- Petitgas, P. Geostatistics and Their Applications to Fisheries Survey Data. In Computers in Fisheries Research, 1st ed.; Megrey, B.A., Moksness, E., Eds.; Chapman & Hall: London, UK, 1996; pp. 113–142. [Google Scholar] [CrossRef]
- Rattray, A.; Ierodiaconou, D.; Monk, J.; Laurenson, L.; Kennedy, P. Quantification of Spatial and Thematic Uncertainty in the Application of Underwater Video for Benthic Habitat Mapping. Mar. Geod. 2014, 37, 315–336. [Google Scholar] [CrossRef]
- Collins, D.L.; Langlois, T.J.; Bond, T.; Holmes, T.H.; Harvey, E.S.; Fisher, R.; McLean, D.L. A Novel Stereo-Video Method to Investigate Fish-Habitat Relationships. Methods Ecol. Evol. 2017, 8, 116–125. [Google Scholar] [CrossRef]
- Wiens, J.A. Spatial Scaling in Ecology. Funct. Ecol. 1989, 3, 385–397. [Google Scholar] [CrossRef]
- Levin, S.A. The Problem of Pattern and Scale in Ecology. Ecology 1992, 73, 1943–1967. [Google Scholar] [CrossRef]
- Pittman, S.J. Seascape Ecology: A New science for the Spatial Information Age. Mar. Sci. 2013, 44, 20–23. [Google Scholar]







| Feature | Source | Description/Algorithm |
|---|---|---|
| Local mean of bathymetry | Bathymetry | Mean of bathymetry in a given window |
| Local standard deviation of bathymetry | Bathymetry | Standard deviation of bathymetry in a given window (a measure of rugosity) |
| Eastness | Bathymetry | |
| Northness | Bathymetry | |
| Slope | Bathymetry | Measure of the rate of change in bathymetry |
| Topographic Position Index | Bathymetry | Indicates whether a location is a local high or low |
| Local mean of backscatter | Backscatter | Mean of backscatter in a given window |
| Local standard deviation of backscatter | Backscatter | Standard deviation of backscatter in a given window (a measure of heterogeneity) |
| GLCM Mean (µ) | Backscatter | |
| GLCM Variance (σ2) | Backscatter | |
| GLCM Homogeneity | Backscatter | |
| GLCM Contrast | Backscatter | |
| GLCM Dissimilarity | Backscatter | |
| GLCM Entropy | Backscatter | |
| GLCM Angular Second Moment | Backscatter | |
| GLCM Correlation | Backscatter |
| Rock | Sand | User’s Accuracy | ||
|---|---|---|---|---|
| Rock | 23 | 9 | 71.9% | |
| Sand | 5 | 575 | 99.1% | |
| Producer’s Accuracy | 82.1% | 98.5% | Overall Accuracy | 97.7% |
| κ | 0.75 |
| Cluster | Rock Observations | Sand Observations | Assigned Substrate |
|---|---|---|---|
| 1 | 21 | 229 | Sand |
| 2 | 179 | 75 | Rock |
| 3 | 10 | 1447 | Sand |
| 4 | 0 | 194 | Sand |
| Rock | Sand | User’s Accuracy | ||
|---|---|---|---|---|
| Rock | 10 | 0 | 100.0% | |
| Sand | 18 | 585 | 97.0% | |
| Producer’s Accuracy | 35.7% | 100.0% | Overall Accuracy | 97.1% |
| κ | 0.51 |
| Degrees of Freedom | Sum of Squares | Mean Square | F | p | |
|---|---|---|---|---|---|
| Habitat | 3 | 4.762 | 1.58737 | 4.4454 | 0.001 |
| Residual | 134 | 47.849 | 0.35708 | ||
| Total | 137 | 52.611 |
| Comparison | F | p | Adjusted p |
|---|---|---|---|
| Sand vs. Low Relief Rock | 8.685 | 0.0001 | 0.0006 |
| Sand vs. Moderate Relief Rock | 3.114 | 0.0006 | 0.0024 |
| Sand vs. High Relief Rock | 3.513 | 0.0002 | 0.0010 |
| Low Relief Rock vs. Moderate Relief Rock | 2.727 | 0.0066 | 0.0198 |
| Low Relief Rock vs. High Relief Rock | 2.410 | 0.0165 | 0.0330 |
| Moderate Relief Rock vs. High Relief Rock | 0.839 | 0.5773 | 0.5773 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ilich, A.R.; Brizzolara, J.L.; Grasty, S.E.; Gray, J.W.; Hommeyer, M.; Lembke, C.; Locker, S.D.; Silverman, A.; Switzer, T.S.; Vivlamore, A.; et al. Integrating Towed Underwater Video and Multibeam Acoustics for Marine Benthic Habitat Mapping and Fish Population Estimation. Geosciences 2021, 11, 176. https://doi.org/10.3390/geosciences11040176
Ilich AR, Brizzolara JL, Grasty SE, Gray JW, Hommeyer M, Lembke C, Locker SD, Silverman A, Switzer TS, Vivlamore A, et al. Integrating Towed Underwater Video and Multibeam Acoustics for Marine Benthic Habitat Mapping and Fish Population Estimation. Geosciences. 2021; 11(4):176. https://doi.org/10.3390/geosciences11040176
Chicago/Turabian StyleIlich, Alexander R., Jennifer L. Brizzolara, Sarah E. Grasty, John W. Gray, Matthew Hommeyer, Chad Lembke, Stanley D. Locker, Alex Silverman, Theodore S. Switzer, Abigail Vivlamore, and et al. 2021. "Integrating Towed Underwater Video and Multibeam Acoustics for Marine Benthic Habitat Mapping and Fish Population Estimation" Geosciences 11, no. 4: 176. https://doi.org/10.3390/geosciences11040176
APA StyleIlich, A. R., Brizzolara, J. L., Grasty, S. E., Gray, J. W., Hommeyer, M., Lembke, C., Locker, S. D., Silverman, A., Switzer, T. S., Vivlamore, A., & Murawski, S. A. (2021). Integrating Towed Underwater Video and Multibeam Acoustics for Marine Benthic Habitat Mapping and Fish Population Estimation. Geosciences, 11(4), 176. https://doi.org/10.3390/geosciences11040176