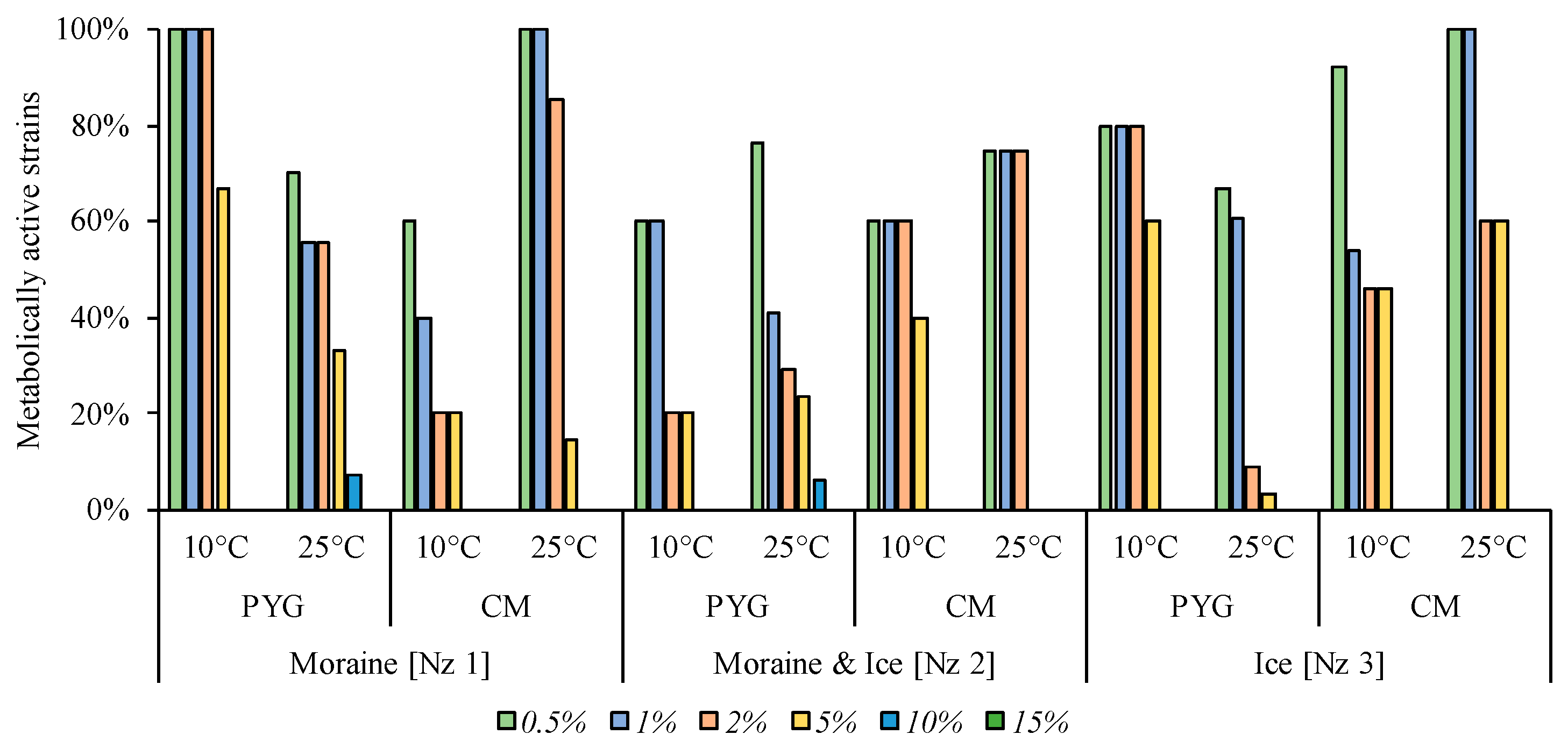Abstract
The study of bacterial communities associated with extreme ecosystems is one of the most important tasks in modern microbial ecology. Despite a large number of studies being performed, the ecosystems that have not been sufficiently explored from the microbiological point of view still exist. Such research is needed for improving the understanding of the limits and mechanisms of bacterial survival under extreme conditions, and for revealing previously undescribed species and their role in global biospheric processes and their functional specifics. The results of the complex microbiological characteristics of permafrost and ice—collected on the Severniy Island in the northern part of the Novaya Zemlya archipelago—which have not previously been described from microbiological point of view, are presented in this article. The analysis included both culture-independent and culture-dependent methods, in particular, the spectra of metabolic activity range analysis in vitro under different temperature, pH and salinity conditions. High values for the total number of prokaryotes in situ (1.9 × 108–3.5 × 108 cells/g), a significant part of which was able to return to a metabolically active state after thawing, and moderate numbers of culturable bacteria (3.3 × 106–7.8 × 107 CFU/g) were revealed. Representatives of Proteobacteria, Actinobacteria, and Bacteroidetes were dominant in situ; Actinobacteria, Firmicutes, Proteobacteria, and Bacteroidetes were the most abundant phyla in vitro. Physiological assays revealed the mesophilic and neutrophilic optima of temperature and pH of culturing conditions, respectively, and wide temperature and pH ranges of culturable communities’ reproduction activity. Isolated strains were characterized by moderate halotolerant properties and antibiotic resistance, including multiple antibiotic resistance. It was found that almost all cultured bacterial diversity revealed (not just a few resistant species) had extremotolerant properties regarding a number of stress factors. This indicates the high adaptive potential of the studied microbial communities and their high sustainability and capability to retain functional activity under changing environmental (including climatic) conditions in wide ranges.
1. Introduction
The study of microorganisms associated with extreme ecological niches of the biosphere is one of the most important tasks of modern microbial ecology [1]. Such investigations have both fundamental and applied importance; they provide new data on microbial biodiversity, functioning of ecosystems, global biogeochemical substances cycles, evolutionary paths of microorganisms, resistance mechanisms (including drug-resistance), new metabolic processes and genetic systems, previously undescribed biologically active compounds, and so forth [2,3,4,5]. Simultaneously, many of modern extreme ecotopes are considered as analogs of some Solar System space bodies because of their list of characteristics and, for this reason, attract researchers’ attention as model objects in different astrobiological investigations [6,7,8]. To date, microbiological studies have been performed for a wide spectrum of ecosystems, characterized by different types and levels of extreme influences. Every year the amount of such research grows, which is partially caused by development and distribution of molecular-genetic methods, sterile sampling techniques, increasing availability of geographically remote territories, and other reasons [9,10,11].
Studies of microbial communities associated with hot and cold desert soils [1,6,12,13,14,15], different saline [16,17], acid, and alkali soils and sediments [18,19], polar and non-polar glaciers [20,21] have been performed. They revealed high taxonomic and functional diversity of bacterial communities, associated with extreme environments. Unique properties of bacteria isolated from extreme environments such as high resistance to different stress impacts, the presence of unique metabolic processes, production of enzymes with unusual properties, and so forth, has been found [4,6,15,22,23,24].
Despite the wide geography of the performed studies, ecosystems barely described from a microbiological perspective are still present. In particular, one of these ecosystems is the Novaya Zemlya archipelago, located in the Arctic Ocean between the Barents and the Kara seas.
Many diverse microbiological studies in the northern regions of both hemispheres have been performed previously [2,10,25,26,27]. A high total number of prokaryotic cells in different frozen soils and sediments at a level of 105–108 cells per gram of dry soil was found using molecular-genetic approaches [2]. Culturable bacteria were detected in different frozen samples in amounts from 101 to 107 colony forming units (CFU) per gram [2,10,27,28]. Most of the culturable bacteria isolated from different permafrost and cold-environments’ samples were affiliated to Firmicutes, Actinobacteria, Bacteroidetes, and Proteobacteria phyla, for representatives of which different mechanisms of survivability under extreme conditions were described [2,28]. Representatives of Bacillus, Arthrobacter, Micrococcus, Cellulomonas, Rhodococcus, Flavobacterium, Pseudomonas, Aeromonas, Myxococcus, Exiguobacterium, Nitrobacter, Nitrosomonas, Nitrospira, Streptomyces, Brevundimonas, Staphylococcus, Halomonas, Herbaspirillum, Hymenobacter, Azotobacter, Promicromonospora, and other genera were identified among culturable isolates [7,10,27,28,29]. Representatives of different physiological groups, such as nitrogen-fixing, nitrifying, proteolytic, urea-decomposing [27], high halotolerant, oligotrophic, psychrotolerant [30], methanogenic, methanotrophic, sulphate-reducing, iron-reducing, denitrifying bacteria [10,31] were isolated from frozen samples.
Microorganisms’ preservation of metabolic activity in permafrost has been shown previously [31,32]. Bacteria isolated from Arctic and Antarctic permafrost samples were able to grow at temperatures from −10 to 50 °C [27]. Bacterial growth in laboratory conditions at temperatures down to −15 °C [33], and preservation of metabolic activity (including in situ) at −20 °C and lower [31,32,34] were revealed. Several authors noted that bacteria isolated from polar regions are mainly characterized by mesophilic temperature optima, but are also able to grow psychrotrophically [27,35,36,37]. A low percentage of culturable cells regarding the total number of prokaryotic cells, close to 0.001%–1%, is characteristic to permafrost associated prokaryotic communities [10,27]. The formation of unculturable, resting, and dormant cell forms is supposedly one of the most important survivability mechanisms of bacteria in cold environments [38].
Many studies were performed on Canadian Arctic samples. In particular, metagenomic analysis of permafrost soils and sediments revealed Proteobacteria, Firmicutes, Chloroflexi, Acidobacteria, Actinobacteria, and Bacteroidetes to be the most common phyla and detected the representatives of previously undescribed phyla [39,40,41]. A predominance of Bacteria and low relative abundance of Archaea and Fungi in Canadian Arctic microbial communities was detected using the quantitative polymerase chain reaction (PCR) approach [40]. The representatives of Euryarchaeota, Crenarchaeota, and Thaumarchaeota were observed among the Archaea in permafrost [39,41]. Analysis of functional genes in the permafrost revealed wide distribution of ammonia- and methane-oxidizing enzyme genes [42]. A high abundance of carbon and nitrogen cycle genes, genes coding chitinase, cellobiose, β-glucosidase, β-galactosidase, osmoprotector sugars, and nitrogen fixing enzymes, were detected in microbial communities of Canadian High Arctic, Alaska, and Sweden permafrost ([2] and references therein). Besides the mentioned gene clusters, a high number of genes encoding pathogenicity and different stress responses were found [40]. A prevalence of different drug-resistance genes in microbial communities of these regions was observed using functional metagenomic analysis [43].
Many culture-dependent and culture-independent studies of frozen samples collected in the different regions of Kolyma lowland (Siberia, Russia) revealed the dominance of Proteobacteria in the microbial communities and a high relative abundance of Actinobacteria and Firmicutes [10,36,44,45,46,47]. Bacterial cultures isolated from cryopegs (lenses of perennially overcooled water brines forming part of the permafrost, freezing of which is prevented by freezing-point depression due to the dissolved solids content or pore water [48]) of this region were affiliated to Psychrobacter, Arthrobacter, Frigoribacterium, Paenibacillus, Bacillus, Subtercola, Microbacterium, Rhodococcus, and Erwinia genera and were characterized by psychrophilic properties—all isolates were able to reproduce at 10 °C and the majority of them were able to metabolise at 4 °C. For the Psychrobacter sp. St1 strain isolated from this region the metabolic activity at −10 °C was shown [49].
A study of the microbial communities of Mammoth mountain (central Yakutiya, Siberia, Russia) sediment performed by phospholipid fatty acid analysis revealed the predominance of bacteria over fungi, and among the bacteria Gram-negative was double the number of Gram-positive. The total abundance of prokaryotic cells in permafrost sediment was at the level of 108 cells per gram of dry sediment. The authors discovered that the number of bacteria cultured at 0 °C and 10–25 °C was about 104 CFU/g and 105 CFU/g of dry sediment, respectively. Among the isolated strains the representatives of Arthrobacter phenanthrenivorans, Subtercola frigoramans, and Glaciimonas immobilis were identified. In vitro tests revealed the weak resistance of isolated bacteria to antibiotics and heavy metals, but high resistance to salts [50].
Summarizing previously obtained data on microbial communities of glaciers and snows, Miteva in her review [20] points out the structure typical for different ice-associated prokaryotic communities, in which dominant positions are represented by Proteobacteria, followed by Gram-positive bacteria and Citophaga-Flavobacteria-Bacteroidetes bacteria group. The most common bacteria, cultured from such ecotopes, were the representatives of Arthrobacter, Frigoribacterium, Rhodococcus, Exiguobacterium, Sphingomonas, Methylobacterium, Acinetobacter, Janthinobacterium, and Chryseobacterium genera.
This way, the preformed studies of permafrost sediments and other cold ecotopes testify to the reservoir role of these environments, containing high functional diversity of microorganisms, actively participating in global processes of substance transformation, as well as the high biodiversity of microorganisms, characterized by high adaptive potential and variability, including rare, insufficiently explored or previously not described prokaryotes. Physiological and phylogenetical diversity of prokaryotes, associated with cold environments, is still insufficiently described and could play a significant role in the future development of different areas of scientific knowledge, including biosphere processes investigation, biotechnology, medicine, genetics, and others.
The objective of this study was to investigate the structure and stress tolerance of bacterial communities associated with virtually microbiologically unexplored Arctic sediments and ice collected in the northern part of Novaya Zemlya archipelago. The present study is a part of a research project that aims to reveal the structure and physiological properties of bacterial communities (especially culturable bacteria) associated with different extreme environments [6,15].
2. Materials and Methods
2.1. Sampling
The samples were collected in the northern part of the Severniy Island (Figure 1) of Novaya Zemlya archipelago (76.896389N, 67.561389E). This location was chosen due to it being the most northern permanent glacier in the archipelago. Three samples were collected—shungite (black carbon-consisting mineraloid) moraine [Nz 1], moraine and glacier ice [Nz 2], and glacier ice [Nz 3]. The presented coordinates indicate the moraine sample collection place. The altitude of Nz 1 and Nz 2 collection points was 200 m, Nz 3 collection point altitude was 209 m. The moraine and ice sample [Nz 2] was collected approximately 200 m to the South of the moraine sample [Nz 1] collection point; the glacier ice sample was collected at approximately 70 m in the same direction from the Nz 2 sample collection point. Each sample was collected from two coring specimens at a distance of 10 m to each other. Mineral or ice materials from the 1–5 cm depth layer were mixed and taken into sterile polypropylene containers using sterile instruments. The surface layer (0–1 cm) was discarded to avoid contamination by dust- and atmosphere-associated microorganisms. The analysed samples were mixed from two specimens taken. The total mass of each mixed sample was approximately 50 g.
Figure 1.
Sampling region map. Red circle indicates sampling site location. The map in more detailed scale is presented on Figure A1.
The moraine [Nz 1] sample was represented by coarse gravel particles (5–7 mm in diameter); the moraine and ice [Nz 2] sample was represented by an equal mixture of the same moraine coarse gravel mineral particles and glacier ice; ice sample [Nz 3] was presented by clear glacier ice. Samples were collected in September 2013 and were stored at −18 ± 2 °C without thawing and refreezing until the analysis.
The archipelago includes two large islands—Severniy and Yuzhniy (literal translation—Northern and Southern, respectively), divided by the Matochkin Shar channel, and several small islands. The area of all islands of the archipelago exceeds 83,000 km². The archipelago is characterised by an arctic climate type; winter is long, cold, with strong winds (wind speed may reach 40–50 m/s) and blizzards. The air temperature may fall to −40 °C, the average temperature of the warmest month (August) is from 2.5 °C in the North of the Archipelago to 6.5 °C in the South. Approximately half of Severniy Island’s area is covered by glaciers, characterized by an ice depth of above 300 m. The total area covered by glaciation is 29,767 km² [51].
The chemical composition was analyzed using the emission atomic spectroscopy system ICP-OES 5110 (Agilent Technologies, CA, USA) for the moraine and ice sample [Nz 2] in the research laboratory MSU-Lab (Available in Reference [52]) and is presented in Table 1. This sample was chosen for analysis because it was located between the Nz 1 and the Nz 2 samples and contained both mineral material and ice. The pH of the samples was measured in soil-water (1:1) solution and stood at 5.1, 5.4, and 5.0 for [Nz 1], [Nz 2], and [Nz 3] samples, respectively.

Table 1.
Chemical characteristics of the moraine and ice sample [Nz 2].
2.2. Epifluorescence Microscopy
The total number of prokaryotes in all the collected samples was determined with epifluorescence microscopy (EFM) with acridine orange dye. Cell desorption was carried out by vortexing using Heidolph Multi Reax vortex for 30 min at 2000 rpm. The preparations were produced in six analytical replicates, fixed by heating, then stained with water solution of acridine orange (1:10,000) for 3 min, washed in distilled water for 20 min, dried at room temperature, and viewed under the Primo Star (Zeiss, Germany) microscope with fluorescence epi-illumination system (AmScope, Irvine, CA, USA) at 700× magnification for 20 fields of vision for each replication. Cells with green fluorescence were counted. The water used for the dilution preparation was simultaneously examined as control.
The prokaryotic cells number was calculated using Equation (1)
where N is the number of cells per gram of sample; S1 is the area of the preparation (μm2); a is the number of cells in the field of view; n is the dilution index; V is the volume of the sample suspension drop placed on the glass (mL); S2 is the field of view of the microscope (μm2); and c is the sample weight (g) [15].
N = (S1 × a × n)/(V × S2 × c),
2.3. Total DNA Extraction and 16S rRNA Genes Illumina Sequencing
Genomic DNA (gDNA) extraction and Illumina sequencing were performed for the sample containing both moraine and ice [Nz 2]. gDNA was extracted from 0.5 g of the sample using the DNeasy PowerLyzer PowerSoil Kit (MoBio Laboratories, Inc., Carlsbad, CA) according to the manufacturer’s instructions.
An amplicon of V3–V4 variable regions of 16S rRNA genes of bacteria was obtained using the 341f + 805r primer system [53]. After obtaining the amplicon, the libraries (including Nz 2 and other samples, not described in current paper) were purified and equimolarly mixed using SequalPrep™ Normalization Plate Kit (ThermoFisher, MA, USA; Cat # A10510-01). Libraries pool was sequenced on the Illumina MiSeq sequinator (301 cycles for each side of the fragment) using MiSeq Reagent Kit v3 (600 cycles). FASTQ files were obtained using MiSeq Reporter software (Illumina, San Diego, CA, USA).
The reference round of data processing was carried out for the operational taxonomical units (OTU)-picking—the sequences obtained after the quality control procedure (FastQC), reads trimming (trimmomatic) and reads pairs assembly (vsearch) were charted to GreenGenes 13.8 database using cd-hit-est-2d protocol in exact mode with similarity threshold of 97%. Further on, random statistical samples without return were created—initial cover was 2500 reads, finite cover was 100,000 reads, step was 2500 reads, and the number of replicates was 5.
Further, the indices of alpha-diversity (Shennon, Simpson, Pielou, and Chao indices) were calculated for the statistical samples using the scikit-bio package for Python programming language.
High-throughput sequencing was performed by the research company Evrogen (Moscow, Russia), primary data processing was carried out by the research company Ksivalue (Moscow, Russia).
Raw data produced in this study were deposited in National Center for Biotechnology Information (NCBI) under the BioProject accession PRJNA554636.
2.4. Fluorescence in situ Hybridization
Analysis of metabolically active prokaryotes was performed for the moraine and ice sample [Nz 2], incubated prior analysis in sterile phosphate buffer saline (PBS) for 12 h at 25 °C.
The sample was placed in sterile PBS (pH 7.4) in dilution 1:10. Prokaryotic cells were desorbed from mineral particles using sonification with a Soniprep 150 ultrasonic disintegrator (MSE, Heathfield, UK) for 2 min at an amperage of 0.4 A and a frequency of 22 kHz. Separation of prokaryotic cells and mineral particles was then performed using the centrifugation at 2000 rpm for 10 min. The supernatant was taken and centrifugated at 14,500 rpm for 15 min for the cell precipitation. Cell fixation was carried out with formaldehyde, for which cells were resuspended in phosphate buffer saline (PBS), containing 3% formaldehyde and then vortexed at 2000 rpm for 90 min. After that, the cells were precipitated by centrifugation at 14,500 rpm for 15 min and after double washing by PBS were resuspended in sterile PBS. Finally, the cells were fixed on slides with Teflon coating and consistently washed in the ethanol concentrations of 50%, 80%, and 96% for three minutes for each concentration.
Hybridisation was performed according to the methodics described by Amann [54], with RNA-specific probes with Cy3 fluorescent dye for detection of Acidobacteria, Actinobacteria, Bacteroidetes, Firmicutes, α-Proteobacteria, β-Proteobacteria, γ-Proteobacteria, δ-Proteobacteria, Planctomyces, Verrucomicrobia, Euryarchaeota, Crenoarchaeota, and Thaumarchaeota representatives. Hybridisation and flushing buffer solutions compositions, as well as a detailed protocol, are described in Reference [55].
The preparations were produced in four replicates, viewed under the Primo Star (Zeiss, Germany) microscope with fluorescence epi-illumination system (AmScope, Irvine, CA, USA) at 1000× magnification for 50 fields of vision for each replication. Cells with red fluorescence were counted.
The prokaryotic cells numbers were calculated using Equation (1).
2.5. Multisubstrate Testing
To assess the potential metabolic activity of the [Nz 2] sample microbial community, the multisubstrate testing (MST) was performed ([6] and references therein). The testing was carried out with 47 substrates including sugars, alcohols, amino acids, salts of organic acids, and polymers (all substrates are listed in Reference [56]; all tests were performed in two replicates). The sample was diluted (1:100) in sterile phosphate saline buffer (pH 7.4) and vortexed for 15 min at 2000 rpm using Heidolph Multi Reax vortex. The mineral particles were precipitated by centrifugation (2000 rpm, 2 min). The hydrogenase activity indicator (triphenyltetrazolium bromide) was added to the supernatant, and, after mixing, a 200 μL aliquot was added to each well of a 96-well plate containing a set of 47 test substrates in two replicates. The plates were incubated at 10 °C and 25 °C for 96 h. After the incubation, the optical density of the solutions was measured photometrically at a wavelength of λ = 510 nm using a Sunrise (Tecan, Menendov, Switzerland) plate photometer.
2.6. Isolation of Culturable Bacterial Communities
Peptone Yeast Glucose (PYG) medium rich in various carbon sources and nutrients (glucose: 1 g/L, peptone: 2 g/L, yeast extract: 1 g/L, tryptone: 1 g/L, agar: 20 g/L) and modified Czapek (CM) medium (glucose: 2 g/L, sucrose: 2 g/L, starch: 2 g/L, NaNO3: 2 g/L, KH2PO4: 1 g/L, MgSO4: 0.5 g/L, KCl: 0.2 g/L, agar: 20 g/L) [6] were used for bacteria culturing and isolation. These media were used to increase the proportion of culturable bacteria analysed from the samples studied. The difference between the diversity of culturable bacteria on these media was expected due to different carbon sources, several of which require (sucrose and starch in the CM medium) pre-hydrolysis. Dilutions of the samples in sterile 0.01 M phosphate-saline buffer solution (pH 7.4, 0.137 M NaCl, 0.0027M KCl) with tenfold dilution-step were plated on the solid nutrient media in three replicates for each dilution [15].
For the more complete characterization of the cultured bacterial community, besides the two different media used, culturing was performed at temperatures of 10, 25, and 50 °C (characteristic optima of psychrophilic, mesophilic, and thermophilic bacteria [57]) until the formation of macro- and microcolonies was completed (typically for 14 days at 25 and 50 °C and 30 days at 10 °C). Colonies’ morphotypes were distinguished by the colour and morphology of the colonies as well as cell morphology. Isolated bacterial strains (Table A1) were deposited in the Astrobiological Collection of Microorganisms of the National Depository Bank of Live Systems “Noah’s Ark” (Available at [58]).
2.7. Physiological Assays of Isolated Strains
The resistance spectra of all isolated cultures (134 strains) to various factor influences were obtained by culturing isolates on the same liquid medium that was used to isolate each particular strain from a natural sample; for each of the investigated factors the experiments were conducted in three replicates. To determine resistance to temperature, cultures were incubated in thermostats at temperatures of 2, 4, 10, 25, 37, 45, and 50 °C. Phosphate (KH2PO4 + H3PO4 final concentration of 100 mM, pH 2–6) and tris-alkaline (Tris + HCl + NaOH final concentration of 100 mM, pH 7–12) buffer systems were used to determine resistance to various pH levels. Bacterial strains’ resistance to the presence of salts was determined by aseptic preparation and subsequent dilution of 20% (w/v) stock solutions of NaCl, KCl, NaHCO3, MgSO4 to final concentrations of 2%, 5%, 10%, 15%, and 20%; the series of magnesium perchlorate concentrations were prepared similarly by diluting a 20% (w/v) sterile stock solution to final concentrations of 0.5%, 1%, 2%, 5%, 10%, and 15%. This set of tests was selected to reveal the effects on bacteria of salt compounds, different to cation and anion composition and biological effects. Sodium and potassium chlorides were chosen because of widespread representation in natural environments and different osmotic properties of this salts. Sodium bicarbonate was chosen due to its high toxicity for the living organisms and alkaline nature of its solutions. Magnesium sulphate was chosen due to its insufficient explored effect on bacterial cells. Magnesium perchlorate was chosen as an oxidative compound, in which solutions both osmotic and oxidising effects are revealed. Antibiotic resistance was determined on nutrient media containing ampicillin (Amp), cephalexin (Ceph), chloramphenicol (Chl), tetracycline (Tet), doxycycline (Dox), kanamycin (Kan), or rifampicin (Rif) (Belmed, Russia) with a final concentration of 100 μg/mL. All tests (except for resistance to temperature) were carried out at a temperature of 25 °C. The growth was registered on the tenth day after inoculation for the cultures cultivated at temperatures of 10, 25, 37, 45, and 50 °C; for cultures growing at temperatures of 2 and 4 °C the incubation period was 60 and 30 days, respectively. Growth registering was performed using a Sunrise (Tecan, Menendov, Switzerland) plate photometer at a wavelength λ = 620 nm. The plates were measured immediately after inoculation and after incubation period. Detailed protocol of physiological assays is described in Reference [15].
2.8. Pure Cultures Identification
For isolated strains identification, the amplification of the 16S rRNA gene was performed. The DNA of each strain was extracted by pure bacterial culture biomass boiling in Tris-EDTA buffer, (pH 8.0) which contained 5% Triton X-100, and further homogenization by mechanical destruction with sterile glass beads (250–300 μm in diameter) using a Homogenizer Minilys (Bertin Instruments, Montigny-le-Bretonneux, France) at 5000 rpm for 30 s. The resulting homogenate was centrifuged, and the resulting supernatant was used as a DNA template for PCR [59].
For amplification of the gene of interest, the primer systems 27f + Un1492r [60,61], 63f + 1387r [59], 341f + 805r [62], and 27f + 537r [60,62] were used in order of priority. The amplification process was performed with cell lysate, prepared as described above. If the procedure failed to obtain the PCR product by the first primer system, the next primer system with the same DNA matrix was used and so on until a product that can be used for further sequencing was acquired. The detailed protocol is described in Reference [6].
The PCR products were purified and sequenced by the Research and Production Company Evrogen (Moscow, Russia) using the 1100r [62], 805r, or 537r [53] primers (Table S1). The editing of the nucleotide sequences was carried out using Chromas Lite 2.01 [63]. For the alignment, comparison, and identification of the nucleotide sequences, the Clustal Omega [64] and the BLAST algorithm from the GenBank database [65] were used. The sequences were deposited in GenBank under the accession numbers MN093986–MN094107.
The phylogenetic trees were constructed using MEGA 7 software [66]. The sequences were then aligned using the Clustal Omega online alignment tool [64]. The type material sequences from GenBank were predominantly used for phylogenetic trees construction. Strain identification was based upon phylogenetic analysis and similarity of the sequences with GenBank data.
2.9. Data Analysis
Statistical data processing was performed using the Microsoft Office Excel 2011 software package (Redmond, Washington, USA). For data on cell numbers, the confidence limits of the mean were calculated by standard error of the mean multiplied by Student’s t-criteria for 95% probability. Culturable communities’ structures were reconstructed based on three analytical replicates; percentage of each strain in the community was calculated as a mean value. When determining the physiological spectra, the results obtained in three replicates were considered a positive reaction.
3. Results
3.1. Total Numbers of Prokaryotic Cells and Numbers of Culturable Bacteria
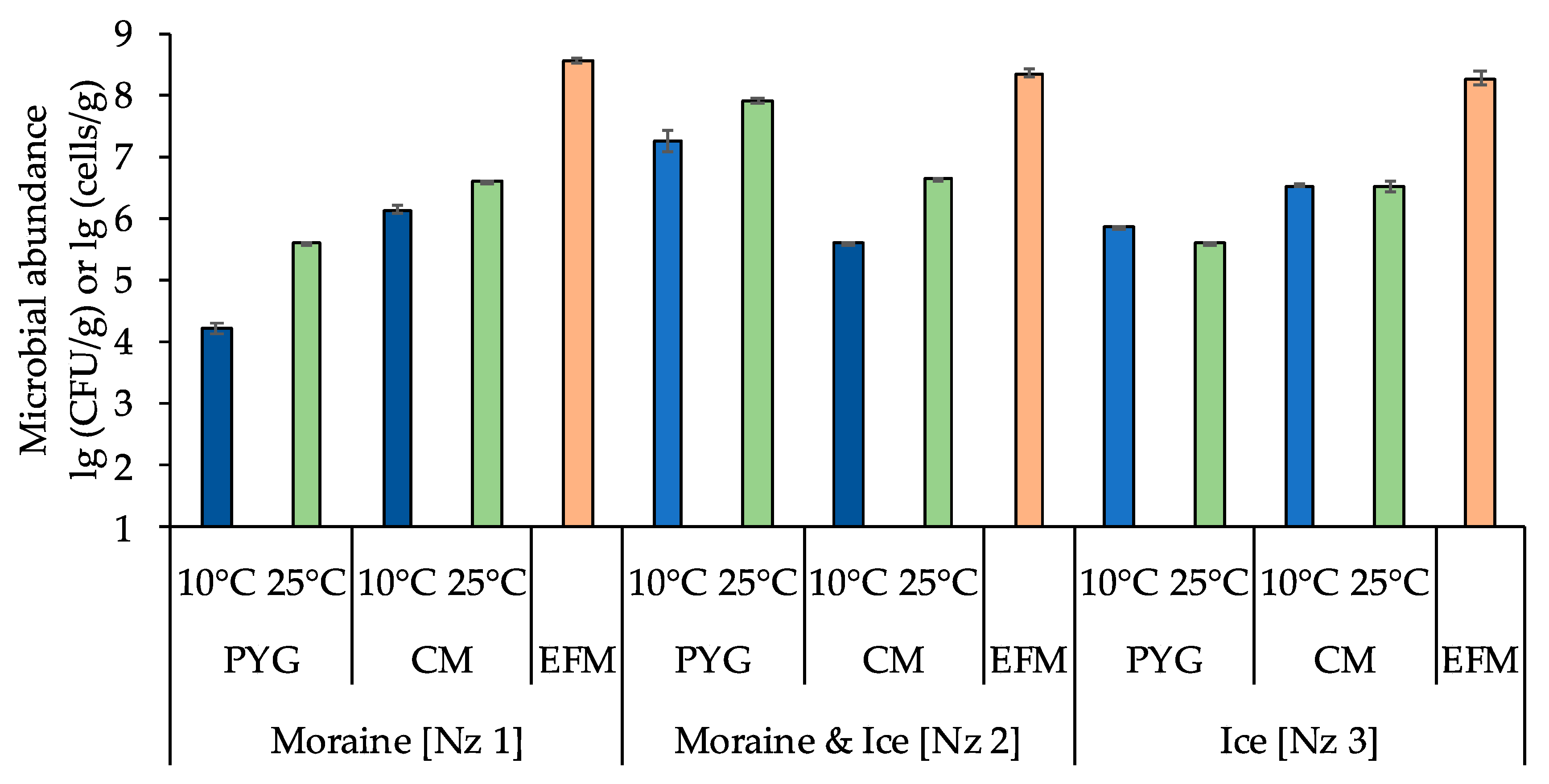
The total numbers of prokaryotic cells in all samples, obtained by epifluorescence microscopy, were about 108 cells/g and decreased in consecutively from moraine to ice (Figure 2, Table A1).
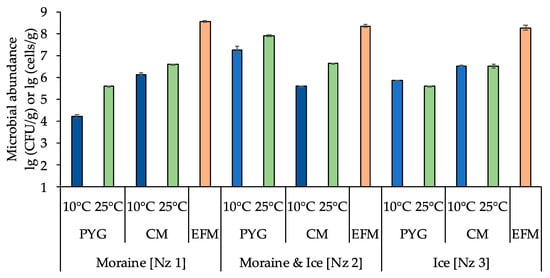
Figure 2.
Total numbers of prokaryotic cells and numbers of culturable cells in the samples. The blue columns indicate the communities incubated at 10 °C, the green columns indicate the communities incubated at 25 °C, the orange columns indicate the total numbers of prokaryotic cells obtained by epifluorescence microscopy. PYG—peptone yeast glucose medium, CM—modified Czapek medium, EFM—the total numbers of prokaryotic cells obtained by epifluorescence microscopy. Error bars are corresponding with the 95% confidence limits.
The numbers of culturable bacteria in the samples ranged from ~104 to ~107 CFU/g. For moraine [Nz 1] and moraine and ice [Nz 2] samples, an increase of culturable bacteria count on both media at 25 °C was detected. For the ice sample [Nz 3] the situation was different: the number of culturable bacteria at 10 °C was higher than at 25 °C on peptone yeast glucose (PYG) media, and was approximately the same on modified Czapek (CM) media (Figure 2, Table A1).
3.2. 16S rRNA Illumina Sequencing Based Bacterial Community Structure
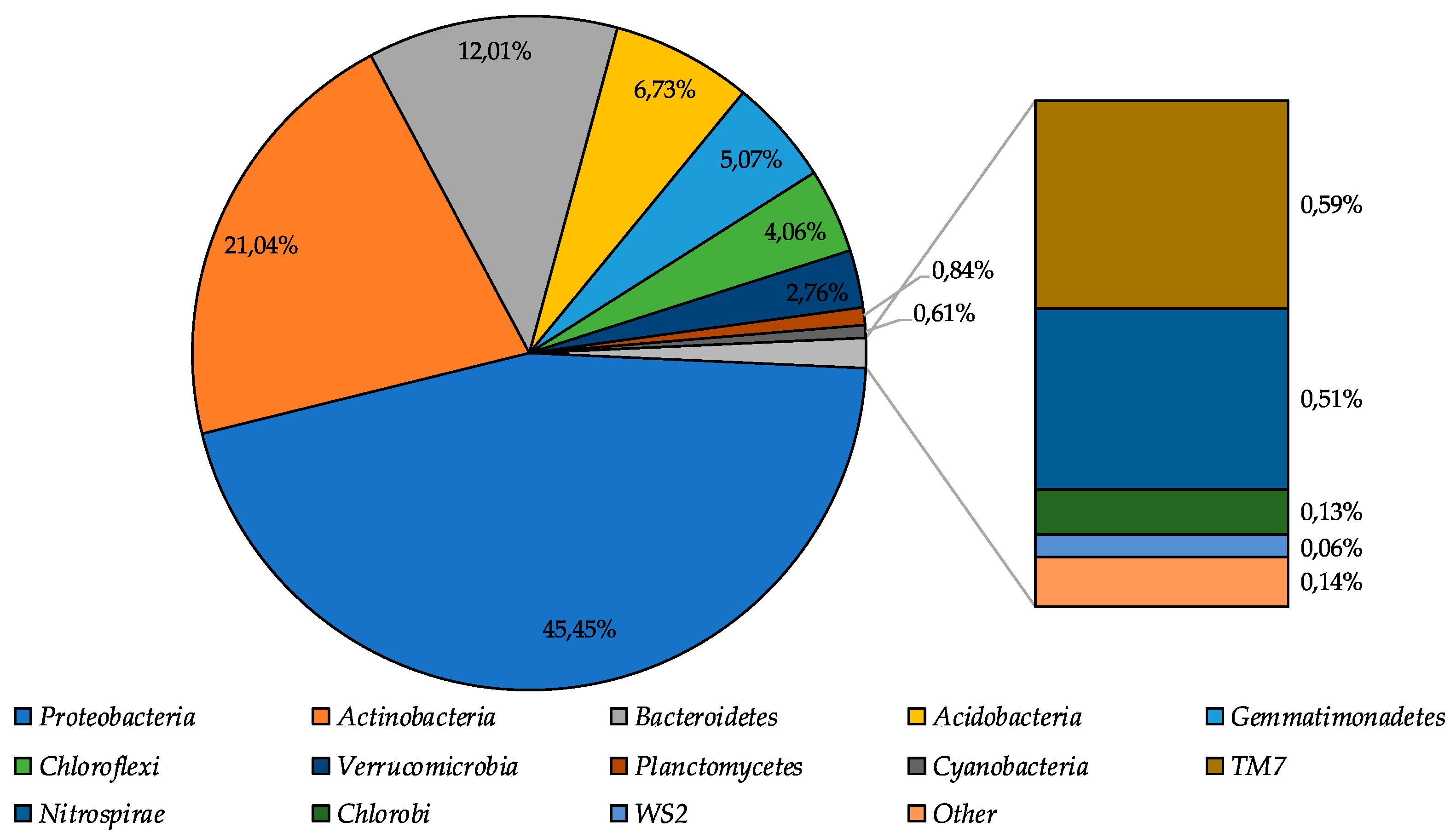
During the high-throughput sequencing of the moraine and ice sample [Nz 2] 322,714 reads were obtained; after removing the poor-quality reads and joining procedure, the 173,906 reads were mapped. Bacterial community structure was reconstructed based on the analysis of these reads. Community structure diagram is represented in Figure 3.
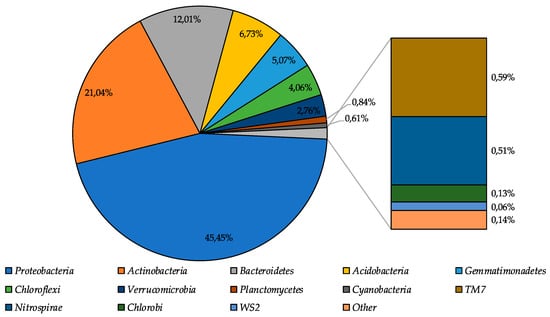
Figure 3.
Index obtained was 1109.1; the Pielou index (evenness)—0.79, the Shannon index—7.99, the Simpson index—0.99. The shape of rarefaction curves (Figure S1) built for these indices indicates the high coverage of biodiversity of the sample analysed.
Proteobacteria were identified as dominant group in the community (45.5%), subdominant positions in the community were represented by Actinobacteria, Bacteroidetes, and Acidobacteria phyla (39.8% in total), and representatives of Gemmatimonadetes, Chloroflexi, and Verrucomicrobia phyla were the minor components of the community (11.9% in total). The percentage of other bacterial phyla representatives was less than 1%.
3.3. Culturable Bacterial Communities’ Structure
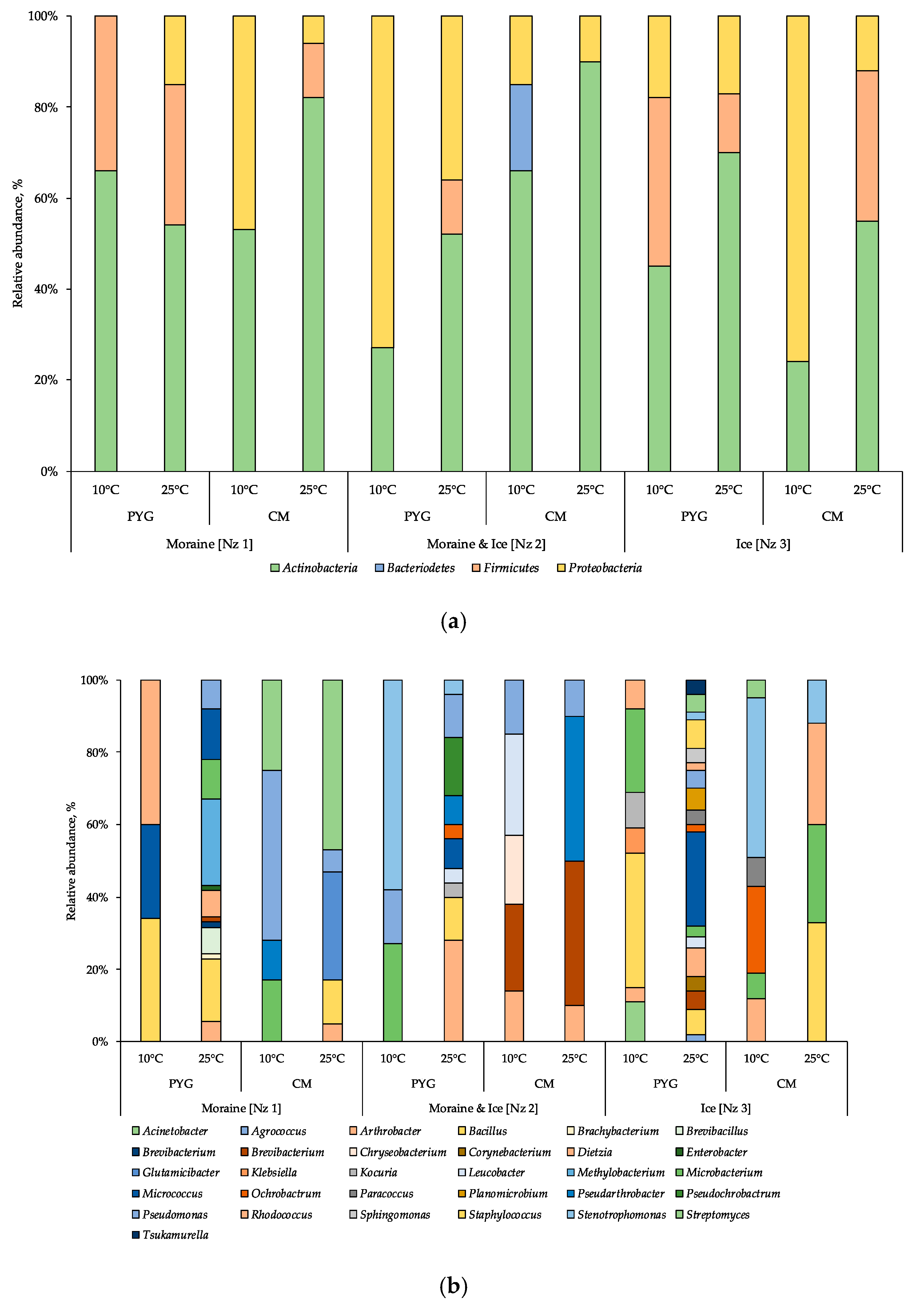
In total, 134 strains of aerobic heterotrophic bacteria were isolated from investigated samples (Table 2). In culturable aerobic heterotrophic bacterial communities of the samples analysed the representatives of Actinobacteria, Firmicutes, Proteobacteria, and Bacteroidetes phyla were revealed (Figure 4a). In the majority of the communities the Actinobacteria were the dominant phylum, excluding the ones isolated from the moraine and ice sample [Nz 2] at 10 °C on the PYG medium and isolated from the ice sample [Nz 3] at 25 °C on the CM medium, in which Proteobacteria were dominant.

Table 2.
Number of isolated bacterial strains in investigated culturable communities.
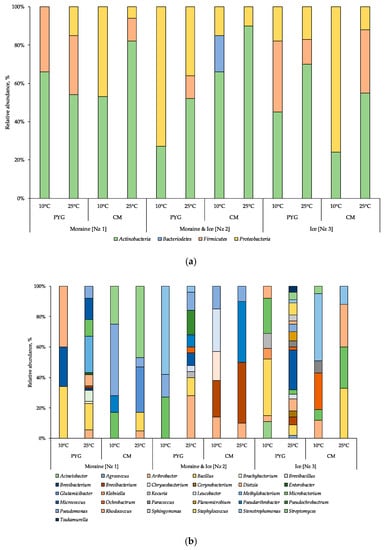
Figure 4.
Culturable bacterial communities’ structure at phylum (a) and at genus (b) level, obtained by 16S rRNA genes sequencing. The horizontal axis indicates the culturing conditions for community isolation. PYG—peptone yeast glucose medium, CM—modified Czapek medium.
In the bacterial complex (Figure 4b) isolated from the moraine sample [Nz 1] at 10 °C on the PYG medium, representatives of Rhodococcus, Bacillus, and Micrococcus genera (here and further on listed in decreasing order of percentage in community) were identified. The community cultured on the same medium at 25 °C was characterised by Bacillus and Micrococcus as dominant genera. From the same sample on the CM medium at 10 °C, the community was dominated by Pseudomonas oryzihabitans; at 25 °C Streptomyces and Glutamicibacter were the most abundant genera.
In the community cultured from the moraine and ice sample [Nz 2] on the PYG medium at 10 °C Stemotrophomonas were the most abundant genus; at 25 °C Arthrobacter was the dominant genus in the bacterial complex. On the CM medium at 10 °C Leucobacter and Brevibacterium constituted 28% and 24% of all cultured colonies, respectively; at 25 °C, 80% of all the isolated community was represented by Brevibacterium and Pseudarthrobacter.
In the community isolated from the ice sample [Nz 3] on the PYG medium at 10 °C, Bacillus and Microbacterium were dominant; at 25 °C Micrococcus was the most abundant of isolated bacteria. On the CM medium at 10 °C Stenotrophomonas and Ochrobactrum dominated; at 25 °C, Bacillus, Rhodococcus, and Microbacterium were the dominants.
For all samples studied, an increase in the culturable community’s biodiversity at mesophilic temperatures (25 °C) as opposed to psychrophilic (10 °C) was detected. Simultaneously, at low incubation temperatures, the biodiversity of communities isolated on CM medium was higher compared to PYG, excluding the communities isolated from the ice sample [Nz 3]. At mesophilic conditions on the PYG medium, the biodiversity of the studied communities was twice or more times higher than in other culturing conditions. Comparing the biodiversity of all culturable communities, it increases in the following sequence: moraine and ice [Nz 2], moraine [Nz 1] and ice [Nz 3] sample. No fungi were isolated from the studied samples, despite of the fact that no antifungal compounds were added to the nutrient media.
3.4. Metabolically Active Communities’ Structure
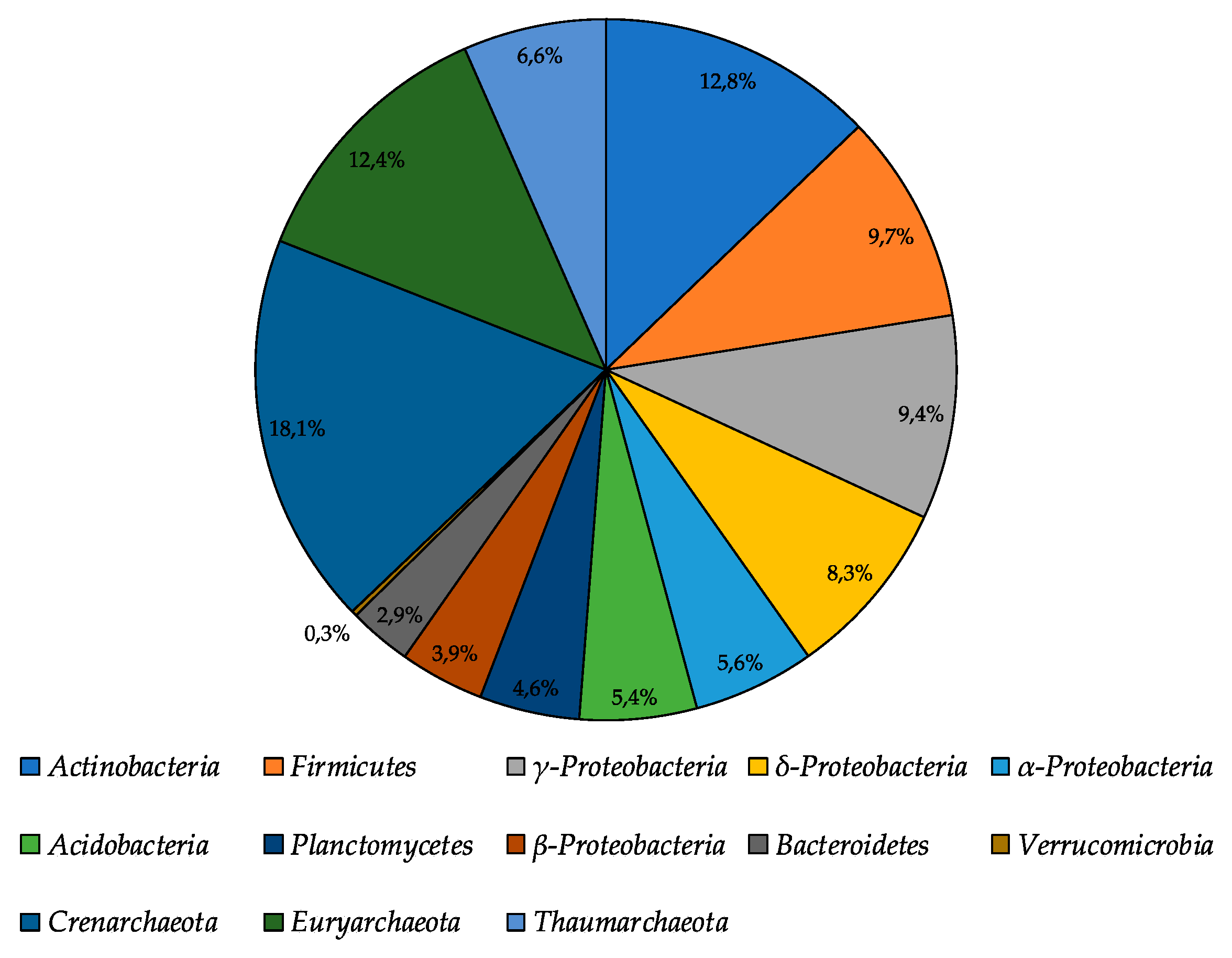
The total number of metabolically active prokaryotes revealed using fluorescence in situ hybridization (FISH) in the moraine and ice sample [Nz 2] was found to be at a level of (3.79 ± 0.96) × 107 cells/g. Archaea constituted 37.1% of all metabolically active prokaryotic cells (Figure 5); among metabolically active Archaea, the most abundant were Crenarchaeota representatives (18.1% of all metabolically active prokaryotes). Among the metabolically active Bacteria, the most abundant were Actinobacteria, Firmicutes, and γ-Proteobacteria, which constituted 12.8%, 9.7%, and 9.1% of all metabolically active prokaryotes, respectively. In general, FISH results were consistent with the high-throughput sequencing data. Proteobacteria and Actinobacteria were dominating, and Acidobacteria and Bacteroidetes constituted a significant part of the bacterial community. At the same time, Firmicutes representatives were in minority according to the Illumina sequencing.
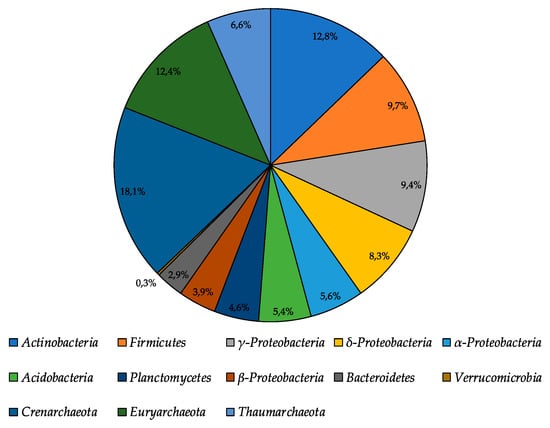
Figure 5.
The structure of metabolically active prokaryotic community of moraine and ice [Nz 2] sample, obtained by fluorescence in situ hybridization analysis.
3.5. Multisubstrate Testing
Multisubstrate testing of the moraine and ice sample [Nz 2] revealed a high functional activity of microbial community at 10 °C: assimilation of 17 substrates (predominantly amino acids, polymers, organic acid salts and oligosaccharides) was observed, while at 25 °C a weak reaction was detected: assimilation of 7 substrates was detected, the most intensive consumption was observed for amino acids, (approximately 2.7 times less intense, compared with their consumption intensity at 10 °C) (Table 3).

Table 3.
Multisubstrate testing results.
3.6. Physiological Assays of Isolated Strains
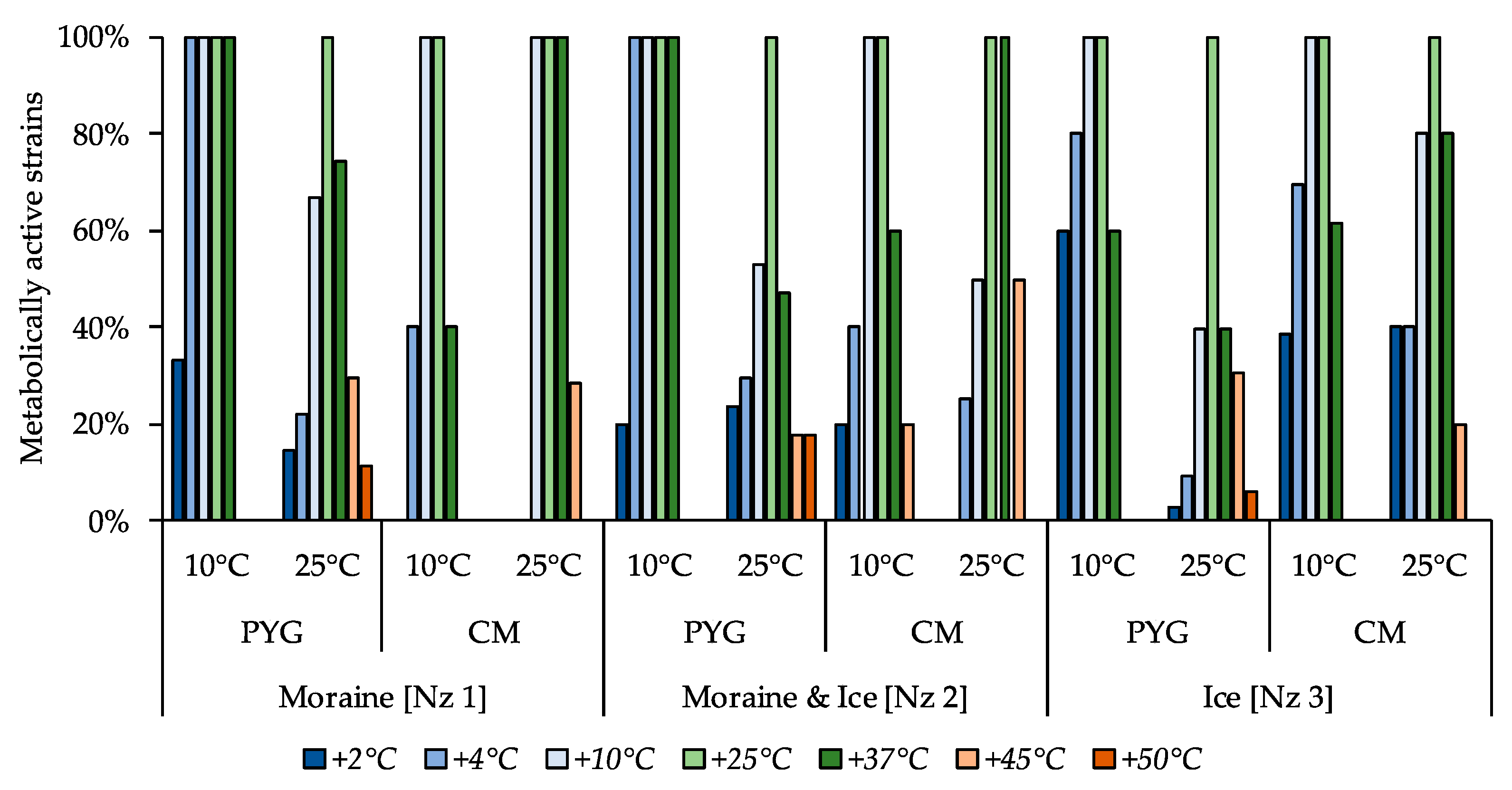
The majority of the isolated strains were able to reproduce in a temperature range from 10 to 37 °C (Figure 6). The ability to reproduce at temperatures above 37 °C was expressed predominantly among the cultures isolated at 25 °C. Simultaneously, the proportion of cultures able to reproduce at temperatures in the range of 2–4 °C was much higher among the strains isolated at 10 °C as opposed to the strains isolated at 25 °C. Nevertheless, the most thermotolerant properties were revealed for the communities isolated from all samples on the PYG medium at 25 °C (which were characterized by complex structure and high biodiversity compared to other obtained culturable communities). The strain Pseudarthrobacter sp. KBP.AS.487, isolated from moraine and ice sample [Nz 2] at 25 °C was able to reproduce in all the studied temperature conditions.
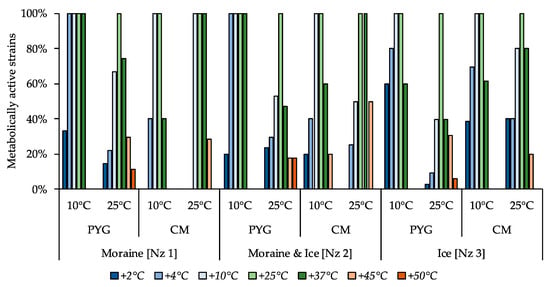
Figure 6.
Isolated strains’ resistance to cultivation temperature. Vertical axis: proportion of the strains in the culturable community able to reproduce in considered conditions. Horizontal axis: the tested communities. Temperature and medium abbreviations indicate the isolation conditions. PYG and CM—peptone yeast glucose and modified Czapek media, respectively.
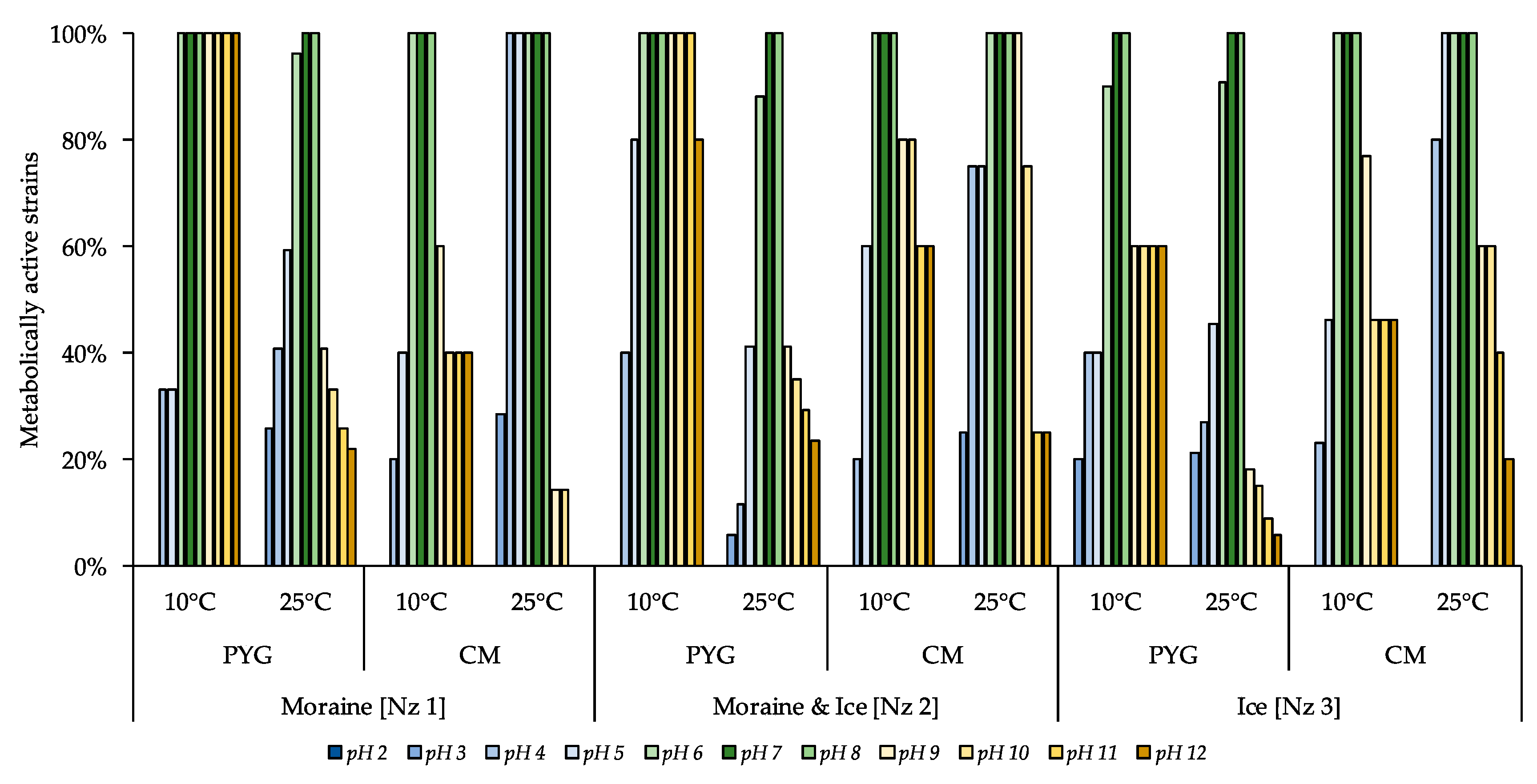
The following pH-resistance profiles of the isolated bacteria were obtained (Figure 7)—no strains able to reproduce on the medium with pH 2 were found; all isolates were able to metabolise on the media with pH 7–8. All culturable communities analysed were characterized by wide ranges of pH-values (pH 3–12) suitable for metabolic activity in vitro. The higher proportion of pH tolerant strains in the communities were found for communities isolated on PYG medium at 10 °C in following decreasing sequence—moraine and ice [Nz 2], moraine [Nz 1], and ice [Nz 3] samples.
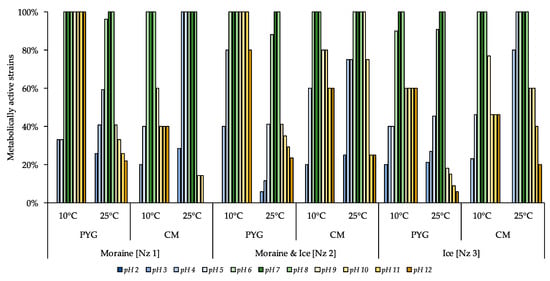
Figure 7.
Isolated strains’ resistance to pH of the media. Vertical axis: proportion of the strains in the culturable community able to reproduce in considered conditions. Horizontal axis: the tested communities. Temperature and medium abbreviations indicate the isolation conditions. PYG and CM—peptone yeast glucose and modified Czapek media, respectively.
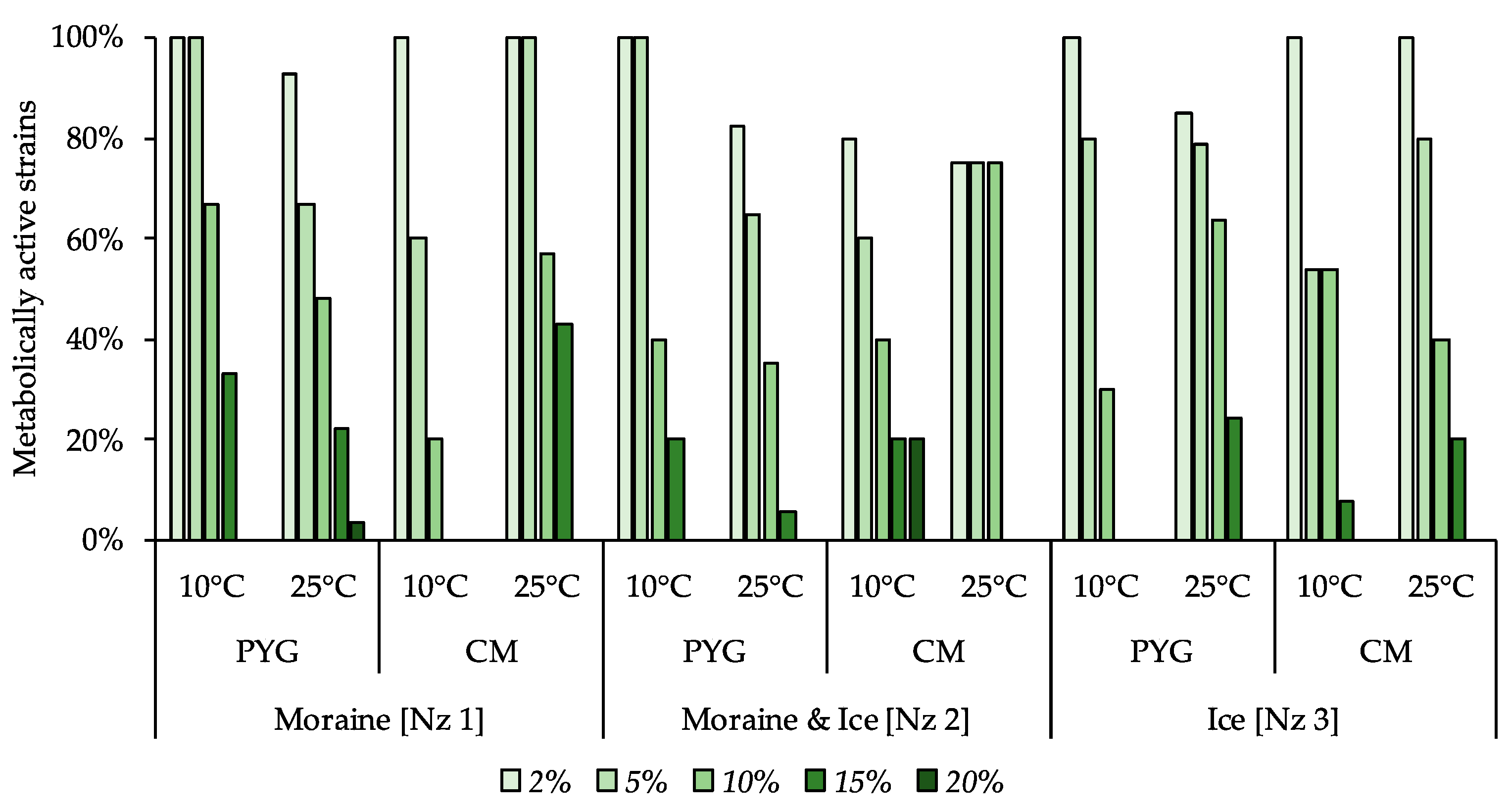
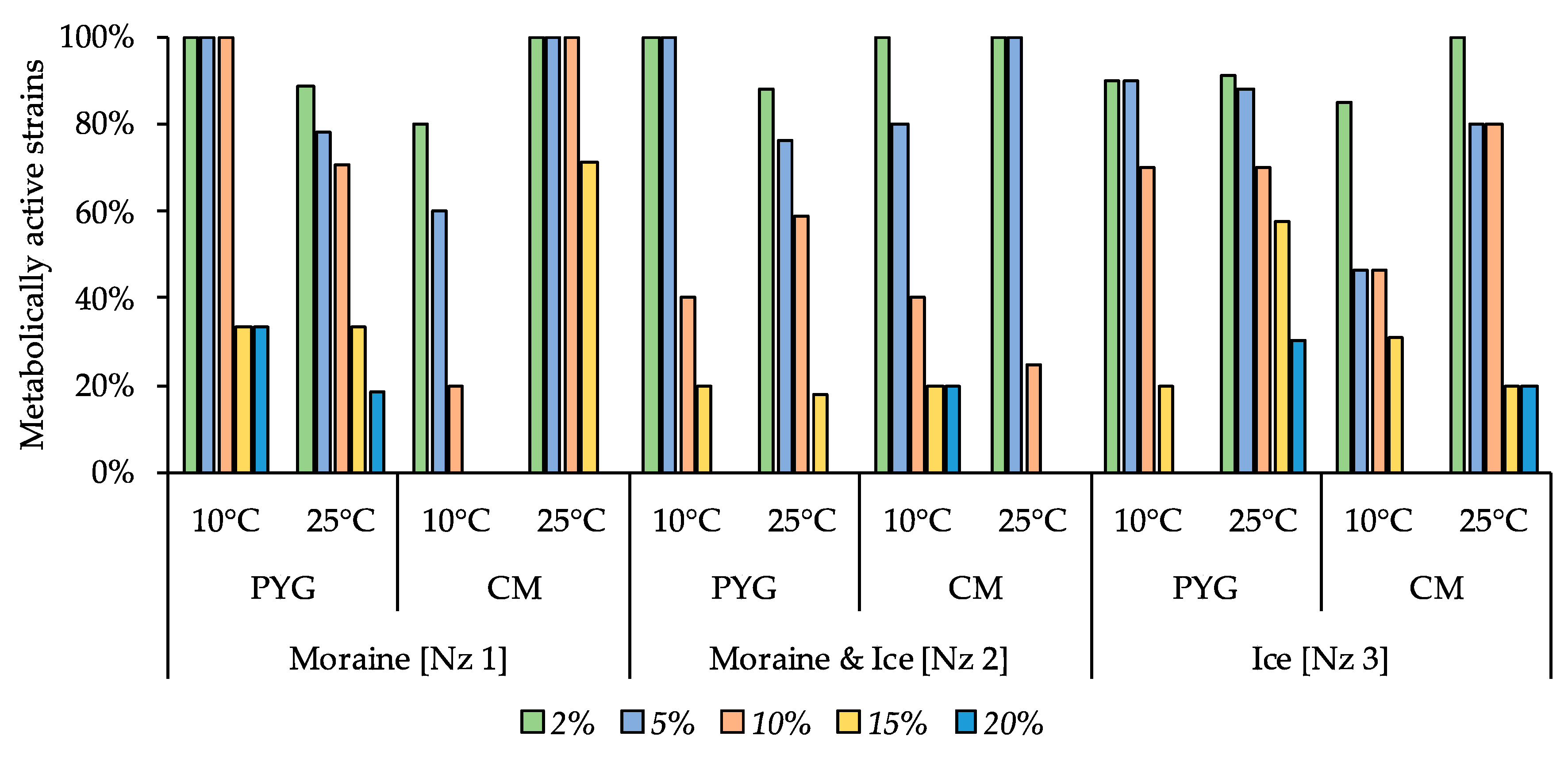
Moderate halotolerant properties were revealed for the investigated culturable bacterial communities (Figure 8). In all the communities, 54% or more of strains were able to reproduce on the media containing 5% of NaCl and from 20% to 75% of the studied communities’ strains were able to reproduce in presence of 10% of this salt. The communities isolated on the PYG medium at both temperature conditions demonstrated a higher proportion of resistant strains compared to the strains isolated on the CM medium. Extremely halotolerant strains, which were able to reproduce on the medium containing 20% of sodium chloride, were found in communities isolated from moraine [Nz 1] on the PYG media at 25 °C and from moraine and ice [Nz 2] on the CM media at 10 °C.
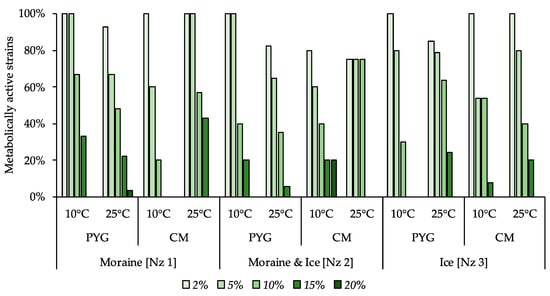
Figure 8.
Isolated strains’ resistance to the presence of sodium chloride in nutrient medium. Vertical axis: proportion of the strains in the culturable community able to reproduce in the considered conditions. Horizontal axis: the tested communities. Temperature and medium abbreviations indicate the isolation conditions. PYG and CM—peptone yeast glucose and modified Czapek media, respectively.
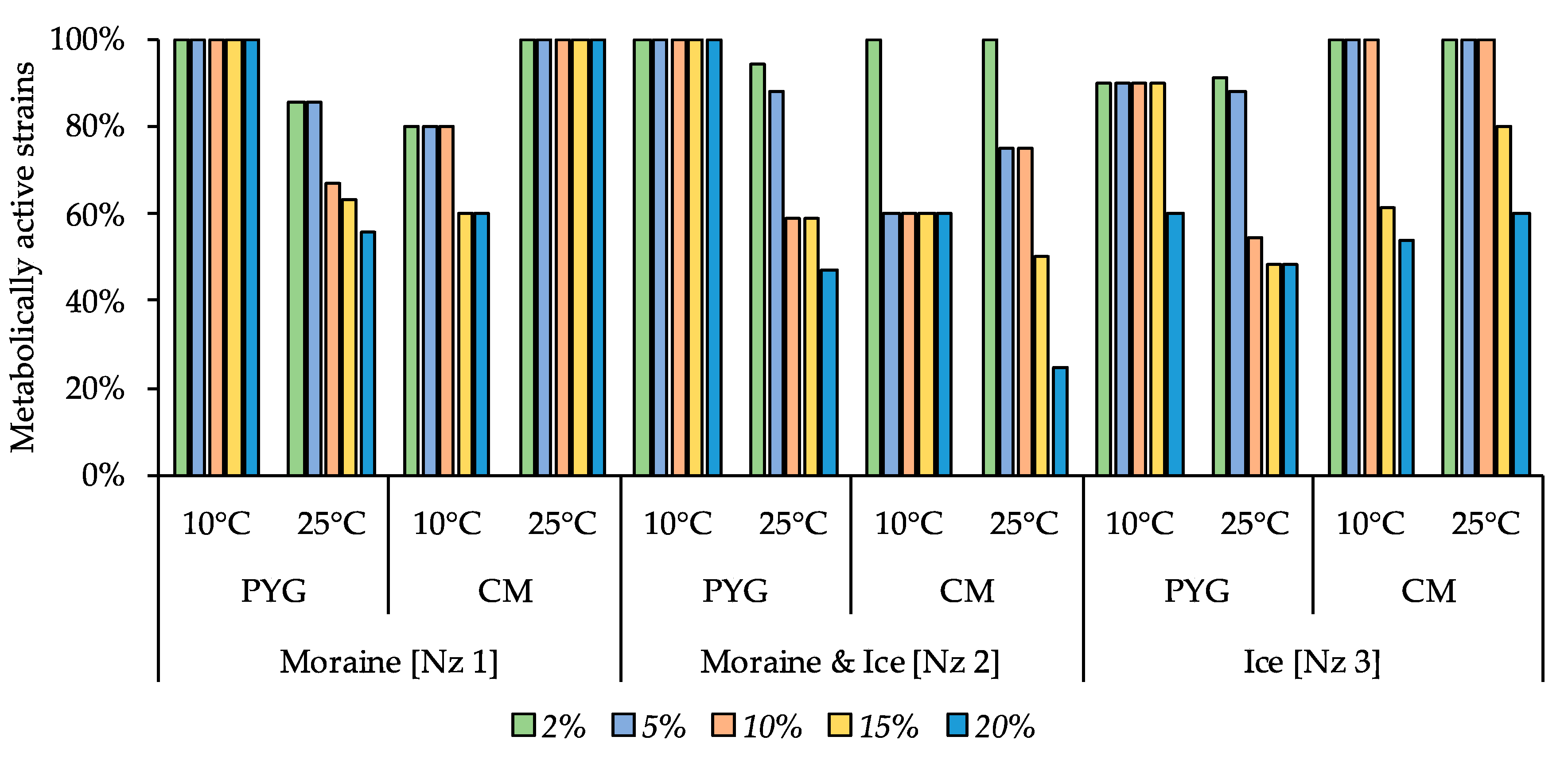
Among all the communities tested, strains able to reproduce on the medium containing 10% KCl were found (Figure 9). The most resistant strains were cultured from the moraine [Nz 1] sample on the PYG medium at 25 °C, from moraine and ice [Nz 2] sample on the CM medium at 10 °C, and from ice [Nz 3] sample at 25 °C on the PYG and CM media both.
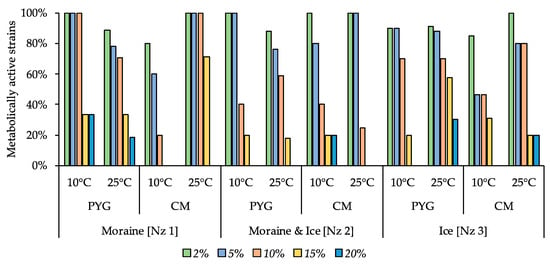
Figure 9.
Isolated strains’ resistance to the presence of potassium chloride in nutrient medium. Vertical axis: proportion of the strains in the culturable community able to reproduce in the considered conditions. Horizontal axis: the tested communities. Temperature and medium abbreviations indicate the isolation conditions. PYG and CM—peptone yeast glucose and modified Czapek media, respectively.
A minimal inhibitory effect among all salts tested was observed for magnesium sulphate (Figure 10): all strains isolated from the moraine [Nz 1] sample on PYG at 10 °C and on the CM medium at 25 °C, as well as all bacteria isolated from moraine and ice [Nz 2] sample at 10 °C on the PYG medium, were able to metabolise in all MgSO4 concentrations tested. In the other communities, apart from those isolated from moraine and ice [Nz 2] sample at 25 °C on the PYG and CM media, half or more of the strains were able to metabolise on the medium containing 20% of magnesium sulphate.
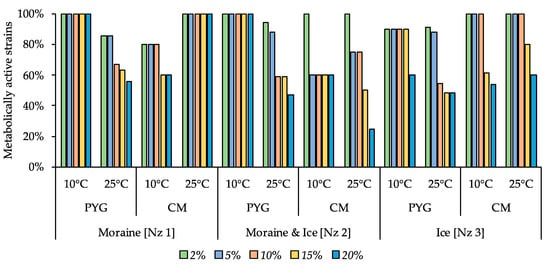
Figure 10.
Isolated strains’ resistance to the presence of magnesium sulphate in nutrient medium. Vertical axis: proportion of the strains in the culturable community able to reproduce in considered conditions. Horizontal axis: the tested communities. Temperature and medium abbreviations indicate the isolation conditions. PYG and CM—peptone yeast glucose and modified Czapek media, respectively.
In contrast with the low inhibitory effect caused by the magnesium sulphate addition, sodium bicarbonate inhibited the majority of the isolated strains (Figure 11) even at low concentrations. Among all strains tested, the strains able to reproduce in the presence of 2% sodium bicarbonate were the most common. High-resistant bacterial strain was found in the community isolated from the moraine and ice sample [Nz 2]. The strain Stenotrophomonas sp. KBP.AS.220 was able to reproduce on the media containing from 2% to 20% of NaHCO3.
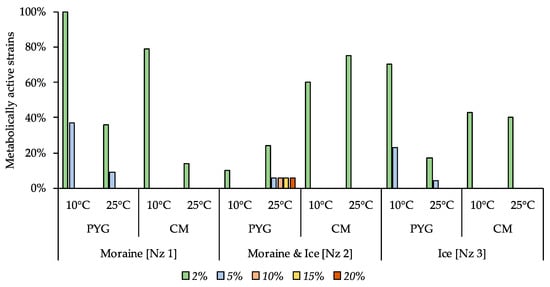
Figure 11.
Isolated strains’ resistance to the presence of sodium bicarbonate in nutrient medium. Vertical axis: proportion of the strains in the culturable community able to reproduce in considered conditions. Horizontal axis: the tested communities. Temperature and medium abbreviations indicate the isolation conditions. PYG and CM—peptone yeast glucose and modified Czapek media, respectively.
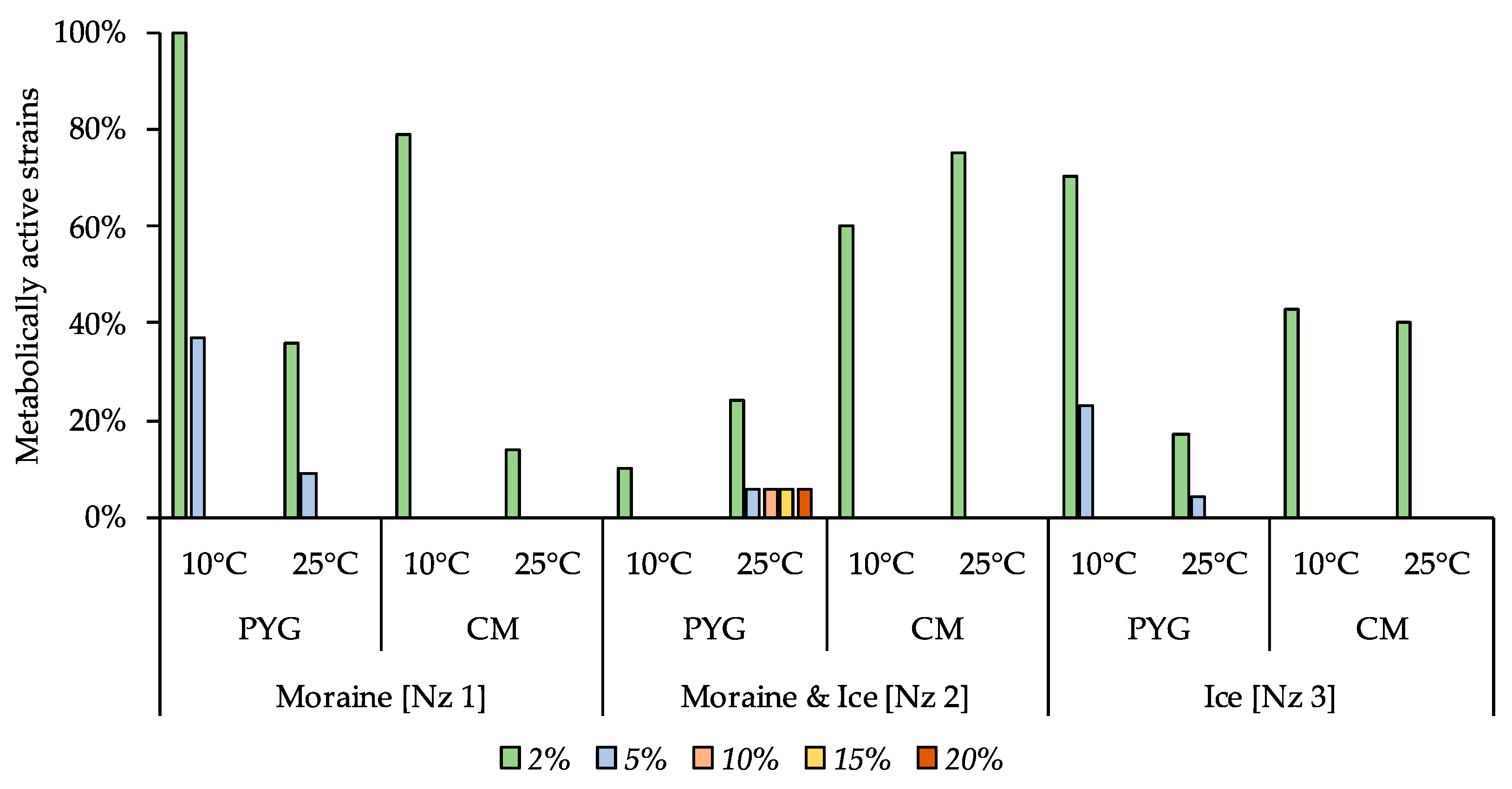
Magnesium perchlorate as an oxidative agent added to the media at 10% concentration inhibited the growth in the majority of strains. Only 7% and 6% of the strains isolated on the PYG medium at 25 °C from the [Nz 1] and [Nz 2] samples, respectively, were able to reproduce in the presence of the given concentration of perchlorate. From 3% to 67% of all strains of the investigated communities were able to metabolise on the medium containing 5% of this salt. Simultaneously, from 9% to 100% of the strains of the communities were active in the presence of 2% perchlorate. With 0.5% of magnesium perchlorate in the media virtually no inhibition of the isolated bacteria took place—the minimal percentage of the strains in the communities that were able to reproduce in such conditions was 60% (detains on the Figure 12).
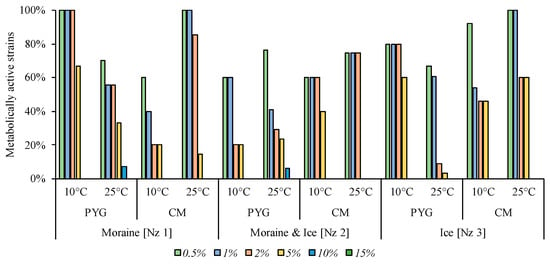
Figure 12.
Isolated strains’ resistance to the presence of magnesium perchlorate in nutrient medium. Vertical axis: proportion of the strains in the culturable community able to reproduce in considered conditions. Horizontal axis: the tested communities. Temperature and medium abbreviations indicate the isolation conditions. PYG and CM—peptone yeast glucose and modified Czapek media, respectively.
Detailed data on the individual strain resistance to temperature, pH, and salts are presented in the Table S1.
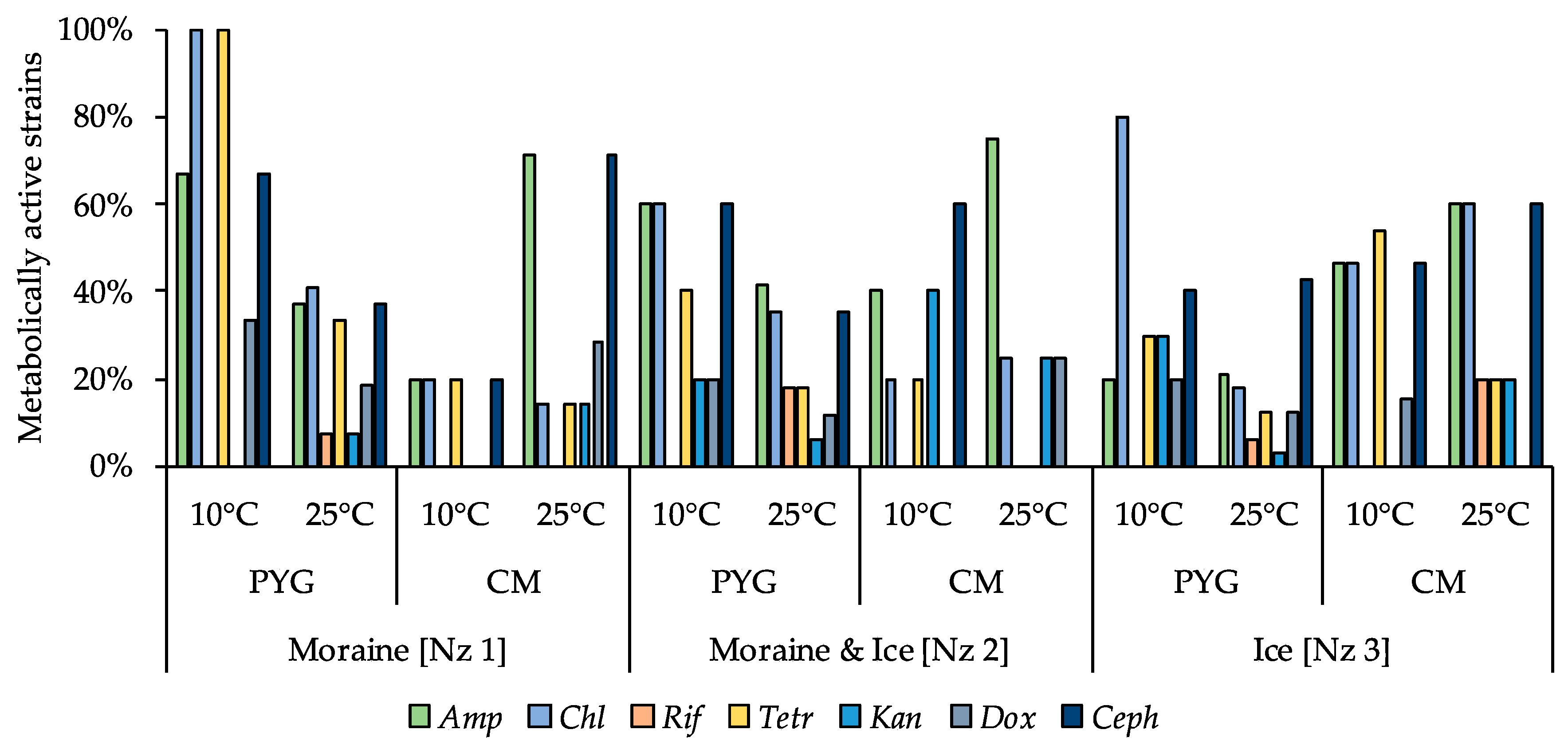
Antibiotic-resistant strains were observed among all isolated communities. All strains in the community, isolated from the moraine sample [Nz 1] on the PYG medium at 10 °C, were resistant to chloramphenicol and tetracycline, and 80% of isolates from the ice sample [Nz 3] obtained at the same conditions were resistant to chloramphenicol. Among the investigated strains isolated from the Novaya Zemlya archipelago samples, the six isolates from different samples isolated in different conditions were resistant to five out of seven studied antibiotics—Ochrobactrum sp. KBP.AS.113, Bacillus sp. KBP.AS.262, Microbacterium paraoxydans KBP.AS.275, Stenotrophomonas maltophilia KBP.AS.402, Brachybacterium sp. KBP.AS.478, Brevibacillus brevis KBP.AS.483. More than a half of the isolated strains (66%) were antibiotic resistant—besides aforementioned high-resistant strains, 18% of all isolated bacteria were resistant to 4 different antibiotics, 13% were resistant to 3 antibiotics, 11% were resistant to two investigated antibiotics, and 20% were characterised as being resistant to one antibiotic. Detailed characteristics are presented in the Figure 13 and in the Table S2.
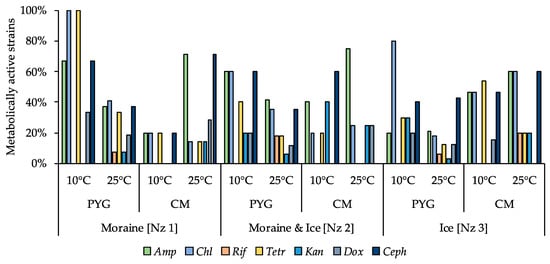
Figure 13.
Isolated strains’ resistance to antibiotics. Vertical axis: proportion of the strains in the culturable community able to reproduce in considered conditions. Horizontal axis: the tested communities. Temperature and medium abbreviations indicate the isolation conditions. PYG and CM—peptone yeast glucose and modified Czapek media, respectively. Amp—ampicillin; Chl—chloramphenicol; Rif—rifampicin; Tetr—tetracycline; Kan—kanamycin; Dox—doxycycline; Ceph—cephalexin.
4. Discussion
The revealed total numbers of prokaryotic cells at the level of ~108 cells per gram of sediment or ice and numbers of culturable bacteria about 106–108 CFU/g are well consistent with the data of previously performed studies on the microbial communities of cold environments [2,10,27]. The increase in the number of culturable bacteria at 25 °C (excluding the ice sample [Nz 3], where the highest number of culturable bacteria was observed at 10 °C) is well consistent with the previously published data on the other cold ecotopes, in which the number of culturable bacteria at mesophilic temperature was higher than in other temperature conditions [27,35,36,37].
The dominant phyla found both using 16S rRNA genes Illumina sequencing and by culturing, were typical for frozen soil and ice samples. Many representatives of isolated taxa are well-known for their ability to survive the effects of low temperatures [20]. The difference between percentage of Firmicutes in the prokaryotic community according to Illumina sequencing, the FISH-analysis, and the culturing procedures could be due to high speed of transition to the metabolically active state during FISH-analysis preparation as well as at culturing procedures. It also could be caused by primer bias as well as by the Firmicutes phylum including several anaerobic classes that could not be revealed by the applied culture technique. The changes of dominant phyla representation in communities’ structure obtained by molecular-genetic vs. culture-based methods are due to Proteobacteria percentage changes, which, presumably, are caused by the presence of difficult-to-culture or unculturable species [67] or by inappropriate culturing conditions.
The alpha-diversity indices obtained by Illumina sequencing indicates the high biodiversity of the studied bacterial community that was previously shown for the other bacterial communities associated with frozen samples [20]. The obtained Chao1 index value is comparable with the corresponding indices, obtained for the Greenland ice samples [68], and for the Svalbard permafrost samples [69]. Representatives of all genera isolated by us were previously isolated from permafrost or frozen environments [10,46,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84].
It is also remarkable that representatives of Agrococcus, Arthrobacter, Bacillus, Brachybacterium, Brevibacillus, Brevibacterium, Glutamicibacter, Leucobacter, Microbacterium, Micrococcus, Planomicrobium, Pseudoarthrobacter, Pseudochrobactrum, Rhodococcus, Sphingomonas, Streptomyces genera and Leucobacter aridicollis, Microbacterium oxydans, Planomicrobium okeanokoites, Pseudomonas putida, Stenotrophomonas maltophilia species were isolated under the identical culturing conditions from non-freezing semi-arid and arid desert soils earlier [6,15] as well as from ice and permafrost samples in the current study. On the one hand, it could be caused by the same culturing conditions performed in these studies. On the other hand, however, representatives of these genera were previously isolated by other authors, using other culturable techniques on different nutrient media. So, this can indicate that community structure was determined by the external influence of high and low temperatures, both of which can lead, for instance, to a decrease of water availability for the microbial cell [85].
The increase in the CFU number and biodiversity of bacterial communities isolated at mesophilic optimum temperature (25 °C) indicates the low percentage of strictly psychrophilic bacteria in the communities, which corresponds with the previously obtained data on bacteria associated with cold environments [86].
The results of fluorescent in situ hybridisation (FISH) analysis indicates the weak metabolic rest level of the prokaryotes in the studied community (the number of metabolically active prokaryotic cells was only ten times less than the total number of prokaryotic cells in the sample). At the same time, high taxonomic diversity, revealed during FISH analysis, could indicate the complex structure of the prokaryotic community and potential wide spectra of geochemical processes, performed by the community and, hence, its high physiological mobility and participation in biospheric processes [87].
High functional activity of the studied communities was also indicated by the results of multisubstrate testing. Relatively higher indices of the community’s metabolic activity on different substrates at low temperature (10 °C) are probably caused by the climate of the studied region [51]—temperatures close to 10 °C are much more regular during the year, compared to higher temperatures, which cause the microbial community adaptations to low-temperature conditions of metabolic activity. However, higher numbers of culturable bacteria and an increase of their morphological and taxonomic diversity were detected at mesophilic temperatures. Based on this, the multisubstrate testing results could be explained by the predominant proliferation of quickly growing bacteria, which, apparently, could pass to metabolically active state faster at low temperatures, compared to 25 °C.
The hypothesis about the adaptation of studied communities’ bacteria to low-temperature functioning is indirectly confirmed by the results of physiological assays of the strains, which revealed the ability of investigated strains to reproduce on the nutrient media right up to the temperature of 2 °C, regardless of the temperature conditions of primary isolation from the native sample. Apart from the ability to reproduce at low temperatures, many of the isolated strains were able to reproduce in high temperature conditions (45–50 °C), which allows to assume the thermotolerant, predominantly mesophilic [57] properties of culturable communities. In regard to the pH of the media, the investigated communities demonstrated neutrophilic moderate acido and high alkalitolerant properties, which were previously shown for the culturable bacterial communities of extreme ecosystems [1,6,9,15]. 40% and more of the strains in the communities studied were able to reproduce at the pH of native sample (close to pH 5).
It was previously suggested that cold and freeze tolerance could be associated with salt tolerance [30]. Investigated strains’ resistance to the presence of different salts in the media could be explained by the linkage of physiological mechanisms responsible to cold- and salt- tolerance mentioned above. However, explicit correlations between the resistance to low temperature and high salt concentrations, based on the data obtained, were not observed (data not shown). Furthermore, high resistance to high concentrations of soluble salts in the media could be caused by studied strains’ adaptations to low water activity (aw) conditions, which originates both at lower temperatures and high concentration of soluble substances [85]. Inhibitory effect rising in the following sequence: MgSO4, KCl, NaCl, Mg(ClO4)2, NaHCO3 was observed previously for the culturable bacterial communities associated with hot arid desert soils [6,15].
Isolation of antibiotic resistant bacteria from frozen samples of Novaya Zemlya archipelago corresponds to previously performed studies of bacteria found in permafrost environments [43,88]. The high morphological (data not shown) and physiological (Tables S1 and S2) variability within the representatives of one species is the notable feature of isolated strains. This phenomenon could be interpreted as the stressogenic dissociation [89] of the bacterial population in situ with the aim of increasing the physiological heterogeneity and, hence, population sustainability. The same mechanism, it seems, is primary for microbial community adaptation in extreme environments.
5. Conclusions
In this article, we have characterised the samples collected in an ecotope that has been almost unexplored from the bacteriological perspective—the Severniy Island of the Novaya Zemlya archipelago. A high total number of prokaryotic cells, a significant proportion of which could pass to a metabolically active state after thawing and incubation at mesophilic temperatures, was revealed. Representatives of Proteobacteria, Actinobacteria, Bacteroidetes, and Acidobacteria phyla were found to be dominant in situ. The typical for cold environments bacteria were cultured on laboratory media. Representatives of Actinobacteria and Firmicutes dominated in culturable communities. Isolated bacterial strains, regardless of the isolation conditions, were characterised by mesophilic and neutrophilic temperature and pH optima, respectively, and by moderate salt tolerance. For many of the strains, the antibiotic resistance was revealed, including multiple resistance.
Thereby, the considered communities were characterized by moderate taxonomical and functional diversity; their culturable segment was metabolically active in vitro in a wide range of physico-chemical growth conditions. According to the physiological properties and taxonomical composition considered communities are close to culturable bacterial communities isolated from hot arid deserts [6,15]. The similarities between studied hot arid desert communities and permafrost samples of this investigation are probably related to the low water availability caused by both low and high temperatures. Further studies on the taxonomical structure and eco-physiological properties of prokaryotes associated with extreme environments could be useful for the exploration of survival limits and physiological adaptations of bacteria to stress impacts as well as for further astrobiological investigations.
The obtained results allow to suggest that the bacterial communities of the investigated samples are able to retain metabolic activity in a wide range of external influences and thereby to contribute to global biospheric processes. The results obtained indicate a high adaptive potential of the studied microbial communities to temperature fluctuations, different pH and different concentrations of soluble compounds. It could testify on the retention of functional activity of the communities in conditions that are changing and non-typical for the Arctic ecosystem.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3263/10/2/67/s1, Figure S1. Illumina sequencing-based biodiversity indices rarefaction curves: a—Chao 1 index, b—Pielou index, c—Shannon index, d—Simpson index. Figure S2. Phylogenetic analysis tree of 16S rRNA genes of the strains isolated from the Novaya Zemlya archipelago samples, which were sequenced using the 537r primer. Figure S3. Phylogenetic analysis tree of 16S rRNA genes of the strains isolated from the Novaya Zemlya archipelago samples, which were sequenced using the 805r primer. Figure S4. Phylogenetic analysis tree of 16S rRNA genes of the strains isolated from the Novaya Zemlya archipelago samples, which were sequenced using the 1100r primer. Table S1. Isolated strains catalogue. Table S2. Taxonomic affiliation of isolated bacteria. Table S3. Physiological characteristics of isolated strains. Table S4. Antibiotics resistance spectra of isolated strains.
Author Contributions
A.A.B. and V.S.C. designed the study, all authors performed the study, A.A.B. and V.S.C. prepared the draft manuscript, all authors edited and commented on the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Russian Foundation for Basic Research according to the research project No 18-34-00331.
Acknowledgments
The authors express their respect to Elena A. Vorobyova and Dmitry D. Badukov, who provided the samples from the Novaya Zemlya archipelago. The authors thank Timofey E. Samsonov for the topography map design. The authors are deeply grateful to the anonymous reviewers for the help in improving the article. The authors thank George A. Abashin for the help in language improvement.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing the manuscript, and in the decision to publish the results.
Appendix A


Figure A1.
The samples collection points; (a) large-scale photo, (b) small-scale photo (images were obtained by Google Earth service).
Figure A1.
The samples collection points; (a) large-scale photo, (b) small-scale photo (images were obtained by Google Earth service).

Table A1.
Numbers of prokaryotic cells and numbers of culturable bacteria in the studied samples.
Table A1.
Numbers of prokaryotic cells and numbers of culturable bacteria in the studied samples.
| Sample | Media | Temperature of Culturing | Cells/g or CFU/g |
|---|---|---|---|
| Moraine [Nz 1] | PYG | 10 °C | (1.60 ± 0.35) × 104 |
| 25 °C | (3.90 ± 0.15) × 105 | ||
| CM | 10 °C | (1.40 ± 0.17) × 106 | |
| 25 °C | (3.90 ± 0.14) × 106 | ||
| EFM | (3.51± 0.44)×108 | ||
| Moraine & Ice [Nz 2] | PYG | 10 °C | (1.77 ± 0.85) × 107 |
| 25 °C | (7.81 ± 0.99) × 107 | ||
| CM | 10 °C | (4.00 ± 0.16) × 105 | |
| 25 °C | (4.27 ± 0.39)×106 | ||
| EFM | (2.25 ± 0.36) × 108 | ||
| Ice [Nz 3] | PYG | 10 °C | (7.33 ± 0.21) × 105 |
| 25 °C | (3.90 ± 0.15) × 105 | ||
| CM | 10 °C | (3.35 ± 0.17) × 106 | |
| 25 °C | (3.28 ± 0.68) × 106 | ||
| EFM | (1.85 ± 0.46) × 108 | ||
Appendix B

Table A2.
Culturable communities’ structure.
Table A2.
Culturable communities’ structure.
| Taxonomic Affiliation | Moraine [Nz 1] | Moraine & Ice [Nz 2] | Ice [Nz 3] | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PYG | CM | PYG | CM | PYG | CM | |||||||
| 10 °C | 25 °C | 10 °C | 25 °C | 10 °C | 25 °C | 10 °C | 25 °C | 10 °C | 25 °C | 10 °C | 25 °C | |
| Acinetobacter pittii | - | - | - | - | - | - | - | - | 11% | - | - | - |
| Agrococcus jenensis | - | - | - | - | - | - | - | - | - | 2% | - | - |
| Arthrobacter agilis | - | - | - | - | - | 4% | - | - | - | - | - | - |
| Arthrobacter crystallopoietes | - | - | - | 5% | - | - | - | - | - | - | - | - |
| Arthrobacter ginsengisoli | - | 5% | - | - | - | 24% | - | 10% | - | - | 12% | - |
| Arthrobacter spp. | - | 2% | - | - | - | - | 14% | - | - | - | - | - |
| Arthrobacter sulfonivorans | - | - | - | - | - | - | - | - | 4% | - | - | - |
| Bacillus infantis | - | - | - | - | - | - | - | - | - | 1% | - | - |
| Bacillus megaterium | - | - | - | - | - | - | - | - | - | - | - | 21% |
| Bacillus pumilus | - | - | - | - | - | 4% | - | - | - | - | - | - |
| Bacillus simplex | - | 2% | - | - | - | - | - | - | 31% | - | - | - |
| Bacillus spp. | 34% | 20% | - | 12% | - | 8% | - | - | 6% | 5% | - | 12% |
| Bacillus subtilis | - | - | - | - | - | - | - | - | - | 1% | - | - |
| Brachybacterium spp. | - | 2% | - | - | - | - | - | - | - | - | - | - |
| Brevibacillus brevis | - | 9% | - | - | - | - | - | - | - | - | - | - |
| Brevibacterium aureum | - | 2% | - | - | - | - | - | - | - | - | - | - |
| Brevibacterium epidermidis | - | - | - | - | - | - | - | 40% | - | - | - | - |
| Brevibacterium linens | - | - | - | - | - | - | - | - | - | 2% | - | - |
| Brevibacterium luteolum | - | 2% | - | - | - | - | - | - | - | - | - | - |
| Brevibacterium spp. | - | - | - | - | - | - | 24% | - | - | 3% | - | - |
| Chryseobacterium kwangjuense | - | - | - | - | - | - | 19% | - | - | - | - | - |
| Corynebacterium mucifaciens | - | - | - | - | - | - | - | - | - | 4% | - | - |
| Dietzia cinnamea | - | 9% | - | - | - | - | - | - | - | 8% | - | - |
| Enterobacter spp. | - | 2% | - | - | - | - | - | - | - | - | - | - |
| Glutamicibacter spp. | - | - | - | 30% | - | - | - | - | - | - | - | - |
| Klebsiella spp. | - | - | - | - | - | - | - | - | 7% | - | - | - |
| Kocuria rosea | - | - | - | - | - | 4% | - | - | 10% | - | - | - |
| Leucobacter aridicollis | - | - | - | - | - | 4% | - | - | - | 3% | - | - |
| Leucobacter spp. | - | - | - | - | - | - | 28% | - | - | - | - | - |
| Methylobacterium spp. | - | 3% | - | - | - | - | - | - | - | - | - | - |
| Microbacterium flavescens | - | - | - | - | - | - | - | - | 9% | - | - | - |
| Microbacterium oxydans | - | 9% | 17% | - | - | - | - | - | 9% | - | - | - |
| Microbacterium paraoxydans | - | - | - | - | - | - | - | - | - | - | 7% | 27% |
| Microbacterium pumilum | - | 5% | - | - | - | - | - | - | - | - | - | - |
| Microbacterium spp. | - | - | - | - | 27% | - | - | - | 5% | 3% | - | - |
| Micrococcus luteus | - | 2% | - | - | - | - | - | - | - | - | - | - |
| Micrococcus spp. | 26% | 16% | - | - | - | 8% | - | - | - | 15% | - | - |
| Micrococcus terreus | - | - | - | - | - | - | - | - | - | 11% | - | - |
| Ochrobactrum spp. | - | - | - | - | - | - | - | - | - | 2% | - | - |
| Ochrobactrum thiophenivorans | - | - | - | - | - | 4% | - | - | - | - | 24% | - |
| Paracoccus marcusii | - | - | - | - | - | - | - | - | - | 4% | - | - |
| Paracoccus spp. | - | - | - | - | - | - | - | - | - | - | 8% | - |
| Planomicrobium okeanokoites | - | - | - | - | - | - | - | - | - | 2% | - | - |
| Planomicrobium spp. | - | - | - | - | - | - | - | - | - | 4% | - | - |
| Pseudarthrobacter scleromae | - | - | - | - | - | - | - | 40% | - | - | - | - |
| Pseudarthrobacter siccitolerans | - | - | 11% | - | - | - | - | - | - | - | - | - |
| Pseudarthrobacter spp. | - | - | - | - | - | 8% | - | - | - | - | - | - |
| Pseudochrobactrum spp. | - | - | - | - | - | 16% | - | - | - | - | - | - |
| Pseudomonas frederiksbergensis | - | 4% | - | 6% | - | - | - | - | - | - | - | - |
| Pseudomonas oryzihabitans | - | - | 47% | - | - | - | - | - | - | - | - | - |
| Pseudomonas putida | - | - | - | - | 15% | - | - | - | - | - | - | - |
| Pseudomonas spp. | - | 6% | - | - | - | 12% | 15% | 10% | - | 5% | - | - |
| Rhodococcus spp. | 40% | - | - | - | - | - | - | - | 8% | 2% | - | 28% |
| Sphingomonas spp. | - | - | - | - | - | - | - | - | - | 4% | - | - |
| Staphylococcus epidermidis | - | - | - | - | - | - | - | - | - | 8% | - | - |
| Stenotrophomonas maltophilia | - | - | - | - | 43% | - | - | - | - | 2% | 39% | 12% |
| Stenotrophomonas rhizophila | - | - | - | - | 15% | - | - | - | - | - | - | - |
| Stenotrophomonas spp. | - | - | - | - | - | 4% | - | - | - | - | 5% | - |
| Streptomyces spp. | - | - | 25% | 47% | - | - | - | - | - | 5% | 5% | - |
| Tsukamurella tyrosinosolvens | - | - | - | - | - | - | - | - | - | 4% | - | - |
References
- Makhalanyane, T.P.; Valverde, A.; Gunnigle, E.; Frossard, A.; Ramond, J.B.; Cowan, D.A. Microbial ecology of hot desert edaphic systems. FEMS Microbiol. Rev. 2015, 39, 203–221. [Google Scholar] [CrossRef] [PubMed]
- Jansson, J.K.; Taş, N. The microbial ecology of permafrost. Nat. Rev. Microbiol. 2014, 12, 414. [Google Scholar] [CrossRef] [PubMed]
- Mikucki, J.A.; Han, S.K.; Lanoil, B.D. Ecology of psychrophiles: Subglacial and permafrost environments. In Extremophiles Handbook; Horikoshi, K., Antranikian, G., Bull, A.T., Robb, F.T., Stetter, K.O., Eds.; Springer: Berlin, Germany, 2011; pp. 755–775. ISBN 978-4-431-53899-8. [Google Scholar]
- Goodfellow, M.; Nouioui, I.; Sanderson, R.; Xie, F.; Bull, A.T. Rare taxa and dark microbial matter: Novel bioactive actinobacteria abound in Atacama Desert soils. Anton. Leeuw. Int. J. G. 2018, 111, 1315–1332. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Golyshina, O.; Beloqui, A.; Golyshin, P.N. Mining enzymes from extreme environments. Curr. Opin. Microbiol. 2007, 10, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Belov, A.A.; Cheptsov, V.S.; Vorobyova, E.A.; Manucharova, N.A.; Ezhelev, Z.S. Stress-Tolerance and Taxonomy of Culturable Bacterial Communities Isolated from a Central Mojave Desert Soil Sample. Geosciences 2019, 9, 166. [Google Scholar] [CrossRef]
- Musilova, M.; Wright, G.; Ward, J.M.; Dartnell, L.R. Isolation of radiation-resistant bacteria from Mars analog Antarctic Dry Valleys by preselection, and the correlation between radiation and desiccation resistance. Astrobiology 2015, 15, 1076–1090. [Google Scholar] [CrossRef]
- Chyba, C.F.; Hand, K.P. Astrobiology: The study of the living universe. Annu. Rev. Astron. Astrophys. 2005, 43, 31–74. [Google Scholar] [CrossRef]
- Rampelotto, P. Extremophiles and extreme environments. Life 2013, 3, 482–485. [Google Scholar] [CrossRef]
- Gilichinsky, D.; Vishnivetskaya, T.; Petrova, M.; Spirina, E.; Mamykin, V.; Rivkina, E. Bacteria in permafrost. In Psychrophiles: From Biodiversity to Biotechnology; Margesin, R., Schinner, F., Marx, J.C., Eds.; Springer: Berlin, Heidelberg, 2008; ISBN 978-3-319-57056-3. [Google Scholar]
- Lindgren, A.R.; Buckley, B.A.; Eppley, S.M.; Reysenbach, A.L.; Stedman, K.M.; Wagner, J.T. Life on the edge—the biology of organisms inhabiting extreme environments: An introduction to the symposium. Integr. Comp. Biol. 2016, 56, 493–499. [Google Scholar] [CrossRef][Green Version]
- Smith, J.J.; Tow, L.A.; Stafford, W.; Cary, C.; Cowan, D.A. Bacterial diversity in three different Antarctic cold desert mineral soils. Microb. Ecol. 2006, 51, 413–421. [Google Scholar] [CrossRef]
- Yadav, A.N.; Sachan, S.G.; Verma, P.; Tyagi, S.P.; Kaushik, R.; Saxena, A.K. Culturable diversity and functional annotation of psychrotrophic bacteria from cold desert of Leh Ladakh (India). World J. Microb. Biot. 2015, 31, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.; Van Nostrand, J.D.; Zhou, J.; Pointing, S.B.; Farrell, R.L. Functional ecology of an Antarctic dry valley. Proc. Natl. Acad. Sci. USA 2013, 110, 8990–8995. [Google Scholar] [CrossRef] [PubMed]
- Belov, A.A.; Cheptsov, V.S.; Vorobyova, E.A. Soil bacterial communities of Sahara and Gibson deserts: Physiological and taxonomical characteristics. AIMS Microbiol. 2018, 4, 685–710. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. Molecular ecology of extremely halophilic Archaea and Bacteria. FEMS Microbiol. Ecol. 2002, 39, 1–7. [Google Scholar] [CrossRef]
- Aanderud, Z.T.; Vert, J.C.; Lennon, J.T.; Magnusson, T.W.; Breakwell, D.P.; Harker, A.R. Bacterial dormancy is more prevalent in freshwater than hypersaline lakes. Front. Microbiol. 2016, 7, 853. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B. Biodiversity and ecology of acidophilic microorganisms. FEMS Microbiol. Ecol. 1998, 27, 307–317. [Google Scholar] [CrossRef]
- Zhang, G.; Ma, X.; Niu, F.; Dong, M.; Feng, H.; An, L.; Cheng, G. Diversity and distribution of alkaliphilic psychrotolerant bacteria in the Qinghai–Tibet Plateau permafrost region. Extremophiles 2007, 11, 415–424. [Google Scholar] [CrossRef]
- Miteva, V. Bacteria in snow and glacier ice. In Psychrophiles: From Biodiversity to Biotechnology; Margesin, R.., Schinner, F., Marx, J.C., Eds.; Springer: Berlin, Germany, 2008; pp. 31–50. ISBN 978-3-319-57057-0. [Google Scholar]
- Anesio, A.M.; Laybourn-Parry, J. Glaciers and ice sheets as a biome. Trends. Ecol. Evol. 2012, 27, 219–225. [Google Scholar] [CrossRef]
- Gaboyer, F.; Le Milbeau, C.; Bohmeier, M.; Schwendner, P.; Vannier, P.; Beblo-Vranesevic, K.; Rabbow, E.; Foucher, F.; Gautret, P.; Guégan, R.; et al. Mineralization and preservation of an extremotolerant bacterium isolated from an early Mars analog environment. Sci. Rep. 2017, 7, 8775. [Google Scholar] [CrossRef]
- Takai, K. Limits of Terrestrial Life and Biosphere. In Astrobiology: From the Origins of Life to the Search for Extraterrestrial Intelligence; Yamagishi, A., Kakegawa, T., Usui, T., Eds.; Springer: Singapore, 2019; pp. 323–344. ISBN 978-981-13-3639-3. [Google Scholar]
- Tse, C.; Ma, K. Growth and Metabolism of Extremophilic Microorganisms. In Biotechnology of Extremophiles; Rampelotto, P.H., Ed.; Springer: Cham, Germany, 2016; pp. 1–46. ISBN 978-3-319-13521-2. [Google Scholar]
- Gilichinsky, D.A.; Wagener, S.; Vishnevetskaya, T.A. Permafrost microbiology. Permafrost. Periglac. 1995, 6, 281–291. [Google Scholar] [CrossRef]
- Margesin, R.; Collins, T. Microbial ecology of the cryosphere (glacial and permafrost habitats): Current knowledge. Appl. Microbiol. Biotechnol. 2019, 103, 2537–2549. [Google Scholar] [CrossRef] [PubMed]
- Vorobyova, E.; Soina, V.; Gorlenko, M.; Minkovskaya, N.; Zalinova, N.; Mamukelashvili, A.; Gilichinsky, D.; Rivkina, E.; Vishnivetskaya, T. The deep cold biosphere: Facts and hypothesis. FEMS Microbiol. Rev. 1997, 20, 277–290. [Google Scholar] [CrossRef]
- Gilichinsky, D.A.; Wilson, G.S.; Friedmann, E.I.; McKay, C.P.; Sletten, R.S.; Rivkina, E.M.; Vishnivetskaya, T.A.; Erokhina, L.G.; Ivanushkina, N.E.; Kochkina, G.A.; et al. Microbial populations in Antarctic permafrost: Biodiversity, state, age, and implication for astrobiology. Astrobiology 2007, 7, 275–311. [Google Scholar] [CrossRef] [PubMed]
- Dartnell, L.R.; Hunter, S.J.; Lovell, K.V.; Coates, A.J.; Ward, J.M. Low-temperature ionizing radiation resistance of Deinococcus radiodurans and Antarctic Dry Valley bacteria. Astrobiology 2010, 10, 717–732. [Google Scholar] [CrossRef]
- Vishnivetskaya, T.; Kathariou, S.; McGrath, J.; Gilichinsky, D.; Tiedje, J.M. Low-temperature recovery strategies for the isolation of bacteria from ancient permafrost sediments. Extremophiles 2000, 4, 165–173. [Google Scholar] [CrossRef]
- Rivkina, E.M.; Laurinavichus, K.S.; Gilichinsky, D.A.; Shcherbakova, V.A. Methane generation in permafrost sediments. In Doklady Biological Sciences; MAIK Nauka/Interperiodica: Moscow, Russia, 2002; Volume 383, pp. 179–181. ISSN 1608-3105. [Google Scholar] [CrossRef]
- Rivkina, E.M.; Friedmann, E.I.; McKay, C.P.; Gilichinsky, D.A. Metabolic activity of permafrost bacteria below the freezing point. Appl. Environ. Microbiol. 2000, 66, 3230–3233. [Google Scholar] [CrossRef]
- Mykytczuk, N.C.; Foote, S.J.; Omelon, C.R.; Southam, G.; Greer, C.W.; Whyte, L.G. Bacterial growth at −15 °C; molecular insights from the permafrost bacterium Planococcus halocryophilus Or1. ISME J. 2013, 7, 1211–1226. [Google Scholar] [CrossRef]
- Panikov, N.S.; Sizova, M.V. Growth kinetics of microorganisms isolated from Alaskan soil and permafrost in solid media frozen down to− 35 C. FEMS Microbiol. Ecol. 2007, 59, 500–512. [Google Scholar] [CrossRef]
- Antony, R.; Krishnan, K.P.; Laluraj, C.M.; Thamban, M.; Dhakephalkar, P.K.; Engineer, A.S.; Shivaji, S. Diversity and physiology of culturable bacteria associated with a coastal Antarctic ice core. Microbiol. Res. 2012, 167, 372–380. [Google Scholar] [CrossRef]
- Cheptsov, V.S.; Vorobyova, E.A.; Manucharova, N.A.; Gorlenko, M.V.; Pavlov, A.K.; Vdovina, M.A.; Lomasov, V.N.; Bulat, S.A. 100 kGy gamma-affected microbial communities within the ancient Arctic permafrost under simulated Martian conditions. Extremophiles 2017, 21, 1057–1067. [Google Scholar] [CrossRef] [PubMed]
- Vishniac, H.S. The microbiology of Antarctic soils. In Antarctic Microbiology; Friedmann, E.I., Thistle, A.B., Eds.; Wiley-Liss: New York, NY, USA, 1993; pp. 297–341. ISBN 0-471-50776-8. [Google Scholar]
- Soina, V.S.; Mulyukin, A.L.; Demkina, E.V.; Vorobyova, E.A.; El-Registan, G.I. The structure of resting bacterial populations in soil and subsoil permafrost. Astrobiology 2004, 4, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, R.C.; Radtke, K.J.; Mykytczuk, N.C.; Greer, C.W.; Whyte, L.G. Life at the wedge: The activity and diversity of Arctic ice wedge microbial communities. Astrobiology 2012, 12, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Yergeau, E.; Hogues, H.; Whyte, L.G.; Greer, C.W. The functional potential of high Arctic permafrost revealed by metagenomic sequencing, qPCR and microarray analyses. ISME J. 2010, 4, 1206. [Google Scholar] [CrossRef]
- Steven, B.; Briggs, G.; McKay, C.P.; Pollard, W.H.; Greer, C.W.; Whyte, L.G. Characterization of the microbial diversity in a permafrost sample from the Canadian high Arctic using culture-dependent and culture-independent methods. FEMS Microbiol. Ecol. 2007, 59, 513–523. [Google Scholar] [CrossRef]
- Frank-Fahle, B.A.; Yergeau, E.; Greer, C.W.; Lantuit, H.; Wagner, D. Microbial functional potential and community composition in permafrost-affected soils of the NW Canadian Arctic. PLoS ONE 2014, 9, e84761. [Google Scholar] [CrossRef]
- Perron, G.G.; Whyte, L.; Turnbaugh, P.J.; Goordial, J.; Hanage, W.P.; Dantas, G.; Desai, M.M. Functional characterization of bacteria isolated from ancient arctic soil exposes diverse resistance mechanisms to modern antibiotics. PLoS ONE 2015, 10, e0069533. [Google Scholar] [CrossRef]
- Zhou, J.; Davey, M.E.; Figueras, J.B.; Rivkina, E.; Gilichinsky, D.; Tiedje, J.M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 1997, 143, 3913–3919. [Google Scholar] [CrossRef]
- Neufeld, J.D.; Mohn, W.W. Unexpectedly high bacterial diversity in arctic tundra relative to boreal forest soils, revealed by serial analysis of ribosomal sequence tags. Appl. Environ. Microbiol. 2005, 71, 5710–5718. [Google Scholar] [CrossRef]
- Vishnivetskaya, T.A.; Petrova, M.A.; Urbance, J.; Ponder, M.; Moyer, C.L.; Gilichinsky, D.A.; Tiedje, J.M. Bacterial community in ancient Siberian permafrost as characterized by culture and culture-independent methods. Astrobiology 2006, 6, 400–414. [Google Scholar] [CrossRef]
- Rivkina, E.; Petrovskaya, L.; Vishnivetskaya, T.; Krivushin, K.; Shmakova, L.; Tutukina, M.; Meyers, A.; Kondrashov, F. Metagenomic analyses of the late Pleistocene permafrost–additional tools for reconstruction of environmental conditions. Biogeosciences 2016, 13, 2207–2219. [Google Scholar] [CrossRef]
- Pradeep Kumar, P. Cryopeg. In Encyclopedia of Snow, Ice and Glaciers. Encyclopedia of Earth Sciences Series; Singh, V.P., Singh, P., Haritashya, U.K., Eds.; Springer: Dordrecht, The Netherlands, 2011; ISBN 978-90-481-2642-2_90. [Google Scholar]
- Bakermans, C.; Tsapin, A.I.; Souza-Egipsy, V.; Gilichinsky, D.A.; Nealson, K.H. Reproduction and metabolism at −10 C of bacteria isolated from Siberian permafrost. Environ. Microbiol. 2003, 5, 321–326. [Google Scholar] [CrossRef]
- Zhang, D.C.; Brouchkov, A.; Griva, G.; Schinner, F.; Margesin, R. Isolation and characterization of bacteria from ancient Siberian permafrost sediment. Biology 2013, 2, 85–106. [Google Scholar] [CrossRef]
- Goryachkin, S.V. The Soils of the North (Structure, Genesis, Ecology, Evolution); Russian Academy of Science: Moscow, Russia, 2010; p. 414, [Book in Russian]; ISBN 978-5-89118-508-1. [Google Scholar]
- MSU-Lab Research Laboratory. Available online: https://www.msulab.ru/ (accessed on 27 September 2019).
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W. Ribosomal RNA-Targeted Nucleic Acid Probes for Studies in Microbial Ecology. FEMS Microbiol. Rev. 2000, 24, 555–565. [Google Scholar] [CrossRef]
- Manucharova, N.A.; Vlasenko, A.N.; Men’ko, E.V.; Zvyagintsev, D.G. Specificity of the chitinolytic microbial complex of soils incubated at different temperatures. Microbiology 2011, 80, 205–215. [Google Scholar] [CrossRef]
- Cheptsov, V.S.; Vorobyova, E.A.; Osipov, G.A.; Manucharova, N.A.; Polyanskaya, L.M.; Gorlenko, M.V.; Pavlov, A.K.; Rosanova, M.S.; Lomasov, V.N. Microbial activity in Martian analog soils after ionizing radiation: Implications for the preservation of subsurface life on Mars. AIMS Microbiol. 2018, 4, 541–562. [Google Scholar] [CrossRef]
- Rampelotto, P.H. Resistance of microorganisms to extreme environmental conditions and its contribution to astrobiology. Sustainability 2010, 2, 1602–1623. [Google Scholar] [CrossRef]
- Depository of Live Systems. Available online: https://depo.msu.ru (accessed on 27 September 2019).
- Marchesi, J.R.; Sato, T.; Weightman, A.J.; Martin, T.A.; Fry, J.C.; Hiom, S.J.; Wade, W.G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 1998, 64, 795–799. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- DeLong, E.F. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 1992, 89, 5685–5689. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematic; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 1991; pp. 115–175. [Google Scholar]
- Chromas and ChromasPro DNA Sequencing Software—Technelysium Pty Ltd. Available online: http: //www.technelysium.com.au (accessed on 27 September 2019).
- Clustal Omega <Multiple Sequence Alignment <EMBL-EBI. Available online: http://www.ebi.ac.uk/ Tools/msa/clustalo/ (accessed on 27 September 2019).
- BLAST: Basic Local Alignment Search Tool. Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 27 September 2019).
- Mega Home. Available online: https://www.megasoftware.net/ (accessed on 27 September 2019).
- Cottrell, M.T.; Wood, D.N.; Yu, L.; Kirchman, D.L. Selected chitinase genes in cultured and uncultured marine bacteria in the α-and γ-subclasses of the proteobacteria. Appl. Environ. Microbiol. 2000, 66, 1195–1201. [Google Scholar] [CrossRef]
- Miteva, V.; Rinehold, K.; Sowers, T.; Sebastian, A.; Brenchley, J. Abundance, viability and diversity of the indigenous microbial populations at different depths of the NEEM Greenland ice core. Polar Res. 2015, 34, 25057. [Google Scholar] [CrossRef]
- Muller, O.; Bang-Andreasen, T.; White III, R.A.; Elberling, B.; Tas, N.; Kneafsey, T.; Jansson, J.K.; Ovreas, L. Disentangling the complexity of permafrost soil by using high resolution profiling of microbial community composition, key functions and respiration rates. Environ. Microbiol. 2018, 20, 4328–4342. [Google Scholar] [CrossRef]
- Behrendt, U.; Schumann, P.; Ulrich, A. Agrococcus versicolor sp. nov., an actinobacterium associated with the phyllosphere of potato plants. Int. J. Syst. Evol. Microbiol. 2008, 58, 2833–2838. [Google Scholar] [CrossRef]
- Manucharova, N.A.; Trosheva, E.V.; Kol’tsova, E.M.; Demkina, E.V.; Karaevskaya, E.V.; Rivkina, E.M.; Mardanov, A.V. Characterization of the structure of the prokaryotic complex of Antarctic permafrost by molecular genetic techniques. Microbiology 2016, 85, 102–108. [Google Scholar] [CrossRef]
- Bajerski, F.; Ganzert, L.; Mangelsdorf, K.; Padur, L.; Lipski, A.; Wagner, D. Chryseobacterium frigidisoli sp. nov., a psychrotolerant species of the family Flavobacteriaceae isolated from sandy permafrost from a glacier forefield. Int. J. Syst. Evol. Microbiol. 2013, 63, 2666–2671. [Google Scholar] [CrossRef]
- Ponder, M.; Vishnivetskaya, T.; McGrath, J.; Tiedje, J. Microbial life in permafrost: Extended times in extreme conditions. In Life in the Frozen State; Fuller, B.J., Lane, N., Benson, E.E., Eds.; CRC Press: Boca Raton, FL, USA, 2004; ISBN 0-203-67332-8. [Google Scholar] [CrossRef]
- Steven, B.; Pollard, W.H.; Greer, C.W.; Whyte, L.G. Microbial diversity and activity through a permafrost/ground ice core profile from the Canadian high Arctic. Environ. Microbiol. 2008, 10, 3388–3403. [Google Scholar] [CrossRef]
- Singh, P.; Singh, S.M.; Singh, R.N.; Naik, S.; Roy, U.; Srivastava, A.; Bolter, M. Bacterial communities in ancient permafrost profiles of Svalbard, Arctic. J. Basic Microbiol. 2017, 57, 1018–1036. [Google Scholar] [CrossRef]
- Balan, B.M.; Binish, S.; Sruthy, K.; Kottekkattu, P.; Mohan, M. Mercury tolerance and biosorption in bacteria isolated from Ny-Alesund, Svalbard, Arctic. J. Basic Microbiol. 2018, 58, 286–295. [Google Scholar] [CrossRef]
- Liu, Y.; Yao, T.; Zhu, L.; Jiao, N.; Liu, X.; Zeng, Y.; Jiang, H. Bacterial diversity of freshwater alpine lake Puma Yumco on the Tibetan Plateau. Geomicrobiol. J. 2009, 26, 131–145. [Google Scholar] [CrossRef]
- Raymond-Bouchard, I.; Goordial, J.; Zolotarov, Y.; Ronholm, J.; Stromvik, M.; Bakermans, C.; Whyte, L.G. Conserved genomic and amino acid traits of cold adaptation in subzero-growing Arctic permafrost bacteria. FEMS Microbiol. Ecol. 2018, 94, fiy023. [Google Scholar] [CrossRef] [PubMed]
- Steven, B.; Leveille, R.; Pollard, W.H.; Whyte, L.G. Microbial ecology and biodiversity in permafrost. Extremophiles 2006, 10, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Zhao, Q.; Zhang, G.; Jiang, Z.; Sheng, H.; Feng, H.; An, L. Paracoccus tibetensis sp. nov.; isolated from Qinghai-Tibet Plateau permafrost. Int. J. Syst. Evol. Microbiol. 2013, 63, 1902–1905. [Google Scholar] [CrossRef]
- Romanenko, L.A.; Tanaka, N.; Frolova, G.M.; Mikhailov, V.V. Pseudochrobactrum glaciei sp. nov.; isolated from sea ice collected from Peter the Great Bay of the Sea of Japan. Int. J. Syst. Evol. Microbiol. 2008, 58, 2454–2458. [Google Scholar] [CrossRef][Green Version]
- Zhang, D.C.; Busse, H.J.; Liu, H.C.; Zhou, Y.G.; Schinner, F.; Margesin, R. Sphingomonas glacialis sp. nov.; a psychrophilic bacterium isolated from alpine glacier cryoconite. Int. J. Syst. Evol. Microbiol. 2011, 61, 587–591. [Google Scholar] [CrossRef]
- D’Amico, S.; Collins, T.; Marx, J.C.; Feller, G.; Gerday, C. Psychrophilic microorganisms: Challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef]
- Yi Pan, S.; Tan, G.Y.; Convey, P.; Pearce, D.A.; Tan, I.K. Diversity and bioactivity of actinomycetes from Signy Island terrestrial soils, maritime Antarctic. Adv. Polar Sci. 2013, 24, 208–212. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Sonjak, S.; Zalar, P.; Frisvad, J.C.; Diderichsen, B.; Plemenitas, A. Extremophilic fungi in arctic ice: A relationship between adaptation to low temperature and water activity. Phys. Chem. Earth. 2003, 28, 1273–1278. [Google Scholar] [CrossRef]
- Rivkina, E.; Laurinavichius, K.; McGrath, J.; Tiedje, J.; Shcherbakova, V.; Gilichinsky, D. Microbial life in permafrost. Adv. Space Res. 2004, 33, 1215–1221. [Google Scholar] [CrossRef]
- Pedros-Alio, C. The rare bacterial biosphere. Annu. Rev. Mar. Sci. 2012, 4, 449–466. [Google Scholar] [CrossRef] [PubMed]
- Mindlin, S.Z.; Soina, V.S.; Petrova, M.A.; Gorlenko, Z.M. Isolation of antibiotic resistance bacterial strains from Eastern Siberia permafrost sediments. Russ. J. Genet. 2008, 44, 27–34. [Google Scholar] [CrossRef]
- Bjedov, I.; Tenaillon, O.; Gerard, B.; Souza, V.; Denamur, E.; Radman, M.; Taddei, F.; Matic, I. Stress-induced mutagenesis in bacteria. Science 2003, 300, 1404–1409. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).