Transcriptome Analysis of the Effects of Fasting Caecotrophy on Hepatic Lipid Metabolism in New Zealand Rabbits
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Design
2.1.1. Ethics Statement
2.1.2. Animals
2.1.3. Sample Collection
2.1.4. Histology
2.1.5. Serum Biochemical Indices
2.2. Transcriptome Analysis
2.2.1. Liver Transcriptome Library Construction and Illumina Sequencing
2.2.2. Data Analysis
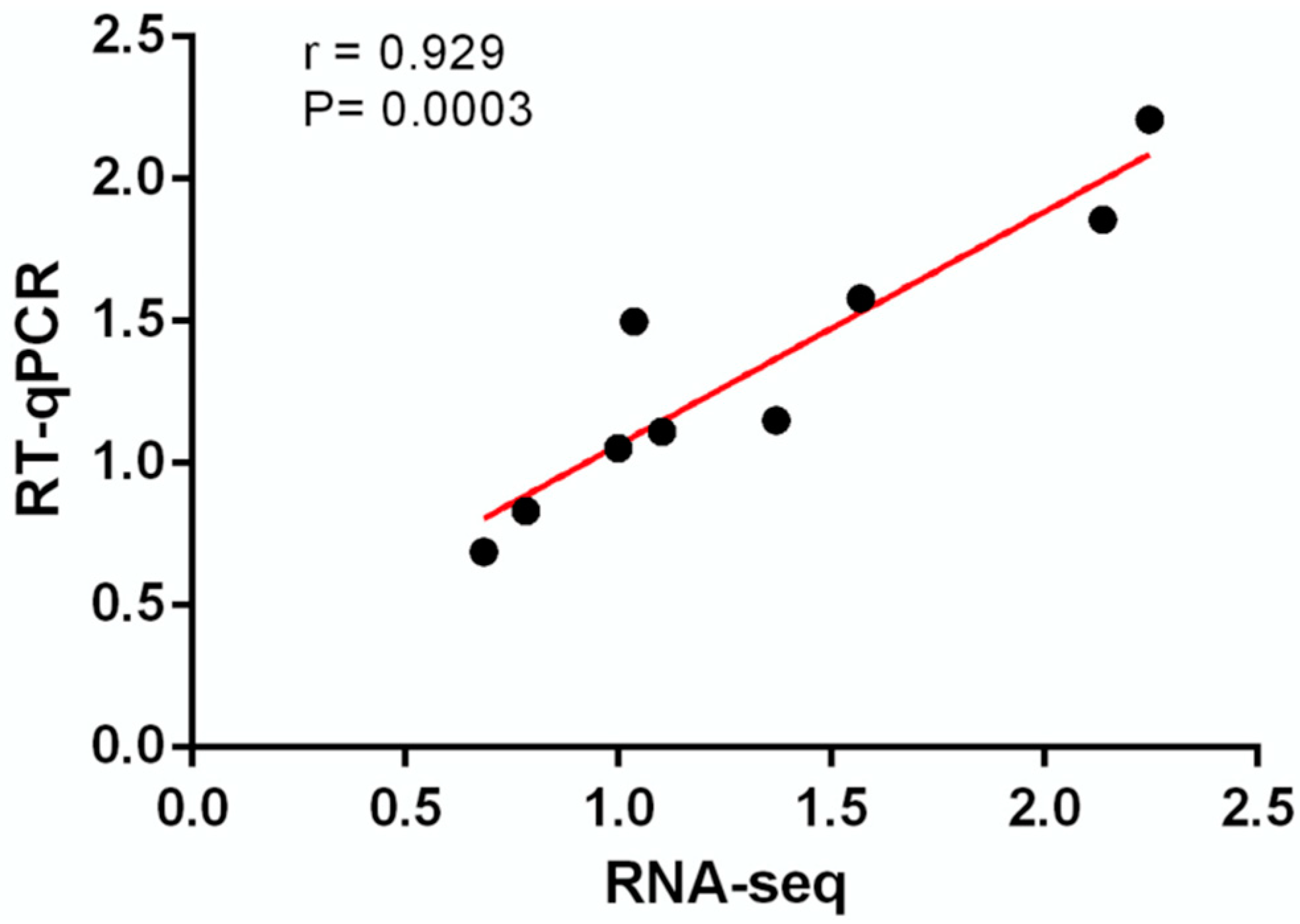
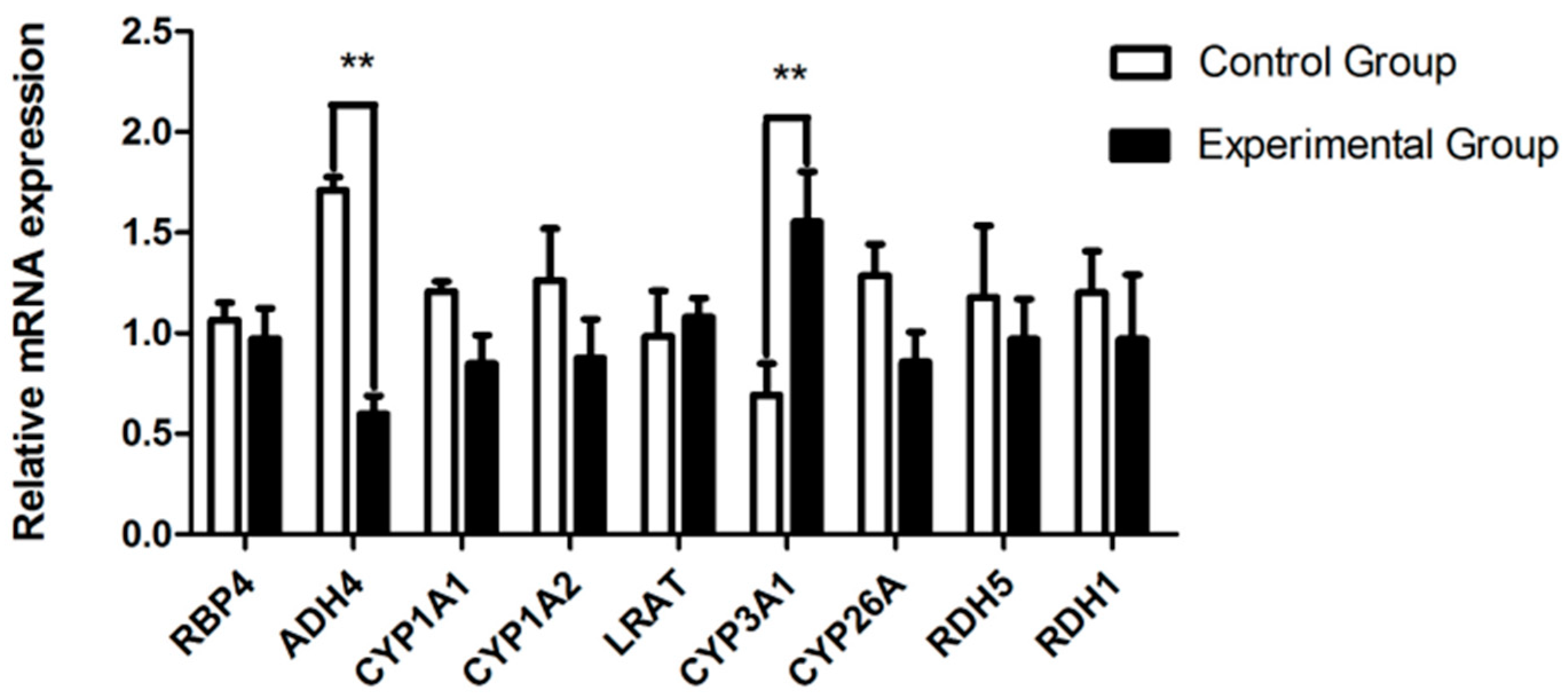
2.3. RT-qPCR
2.4. Data Analysis
3. Results
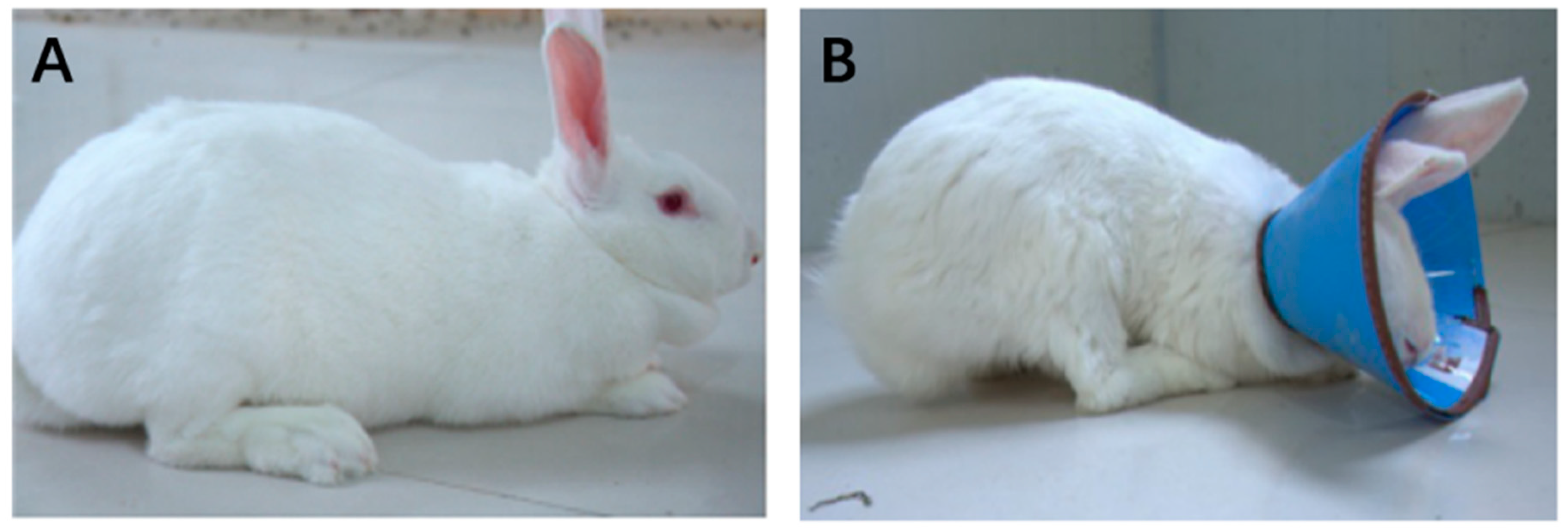
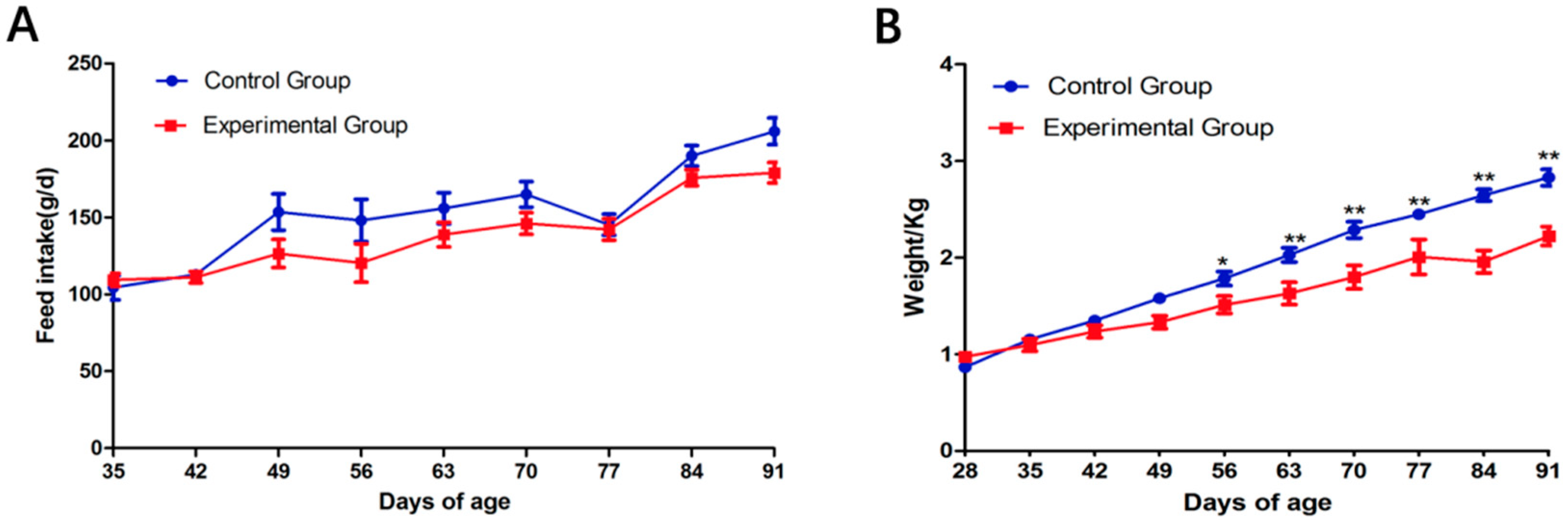
3.1. Effects of Wearing a Collar on Rabbit Growth
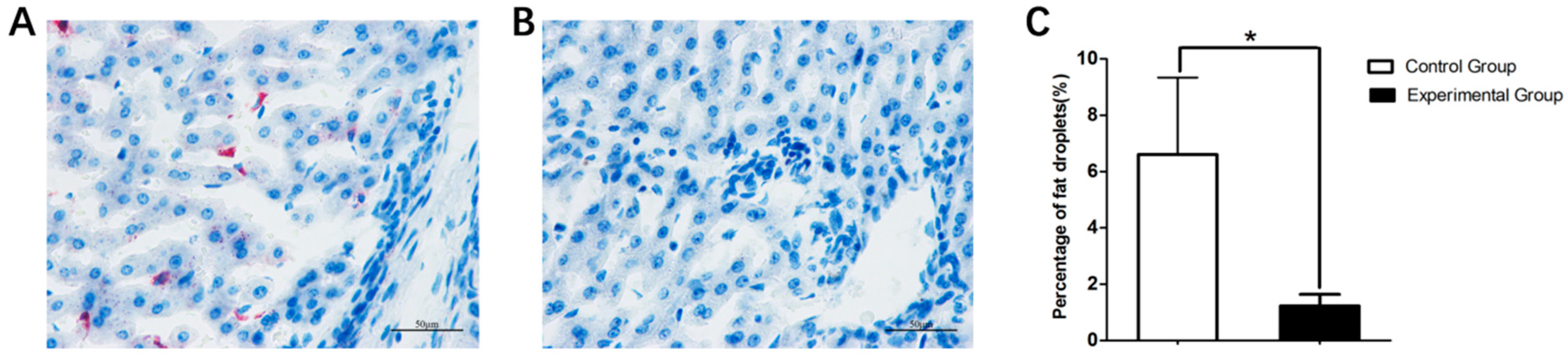
3.2. Body Weight and Liver Histology Changes
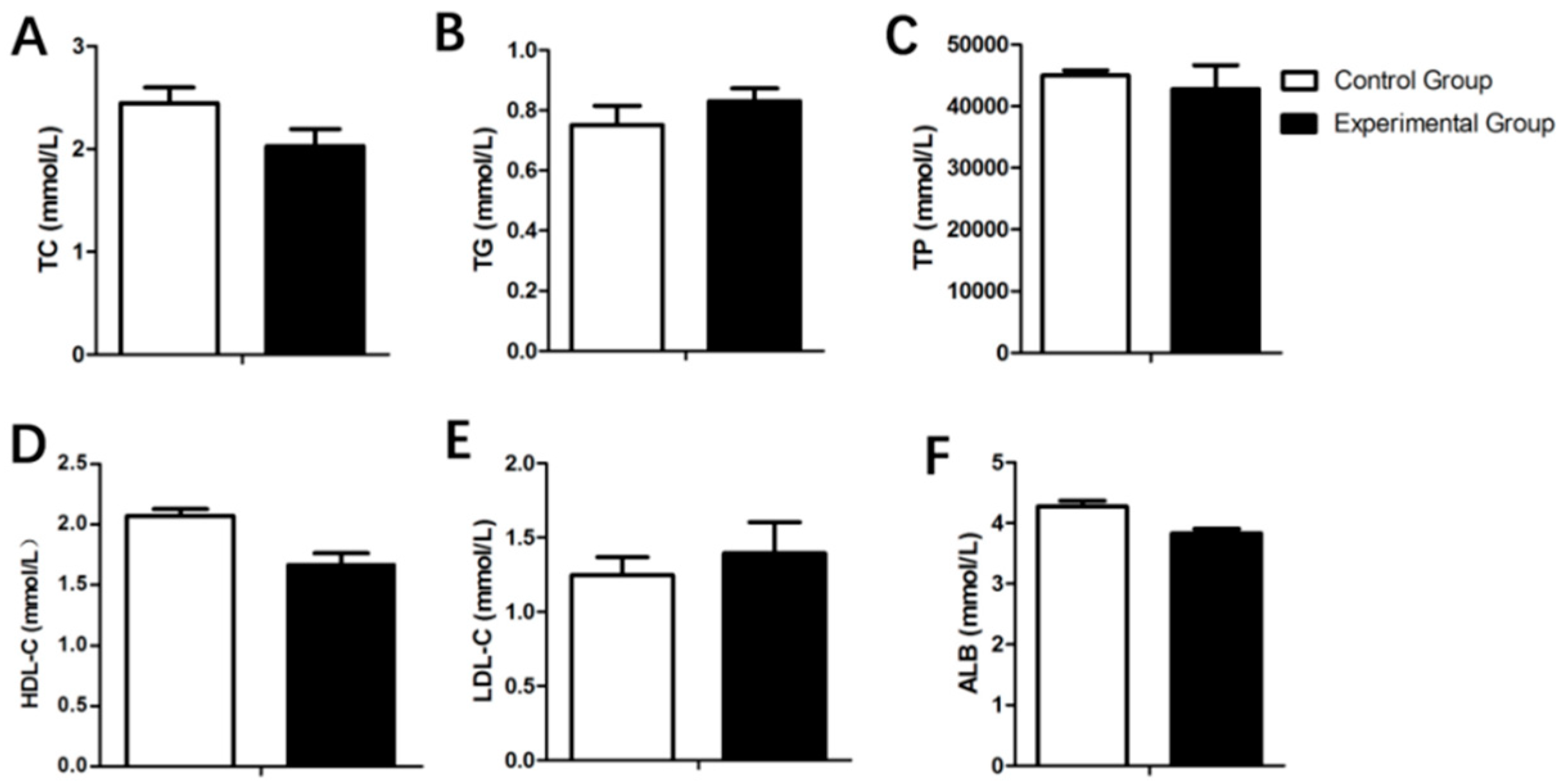
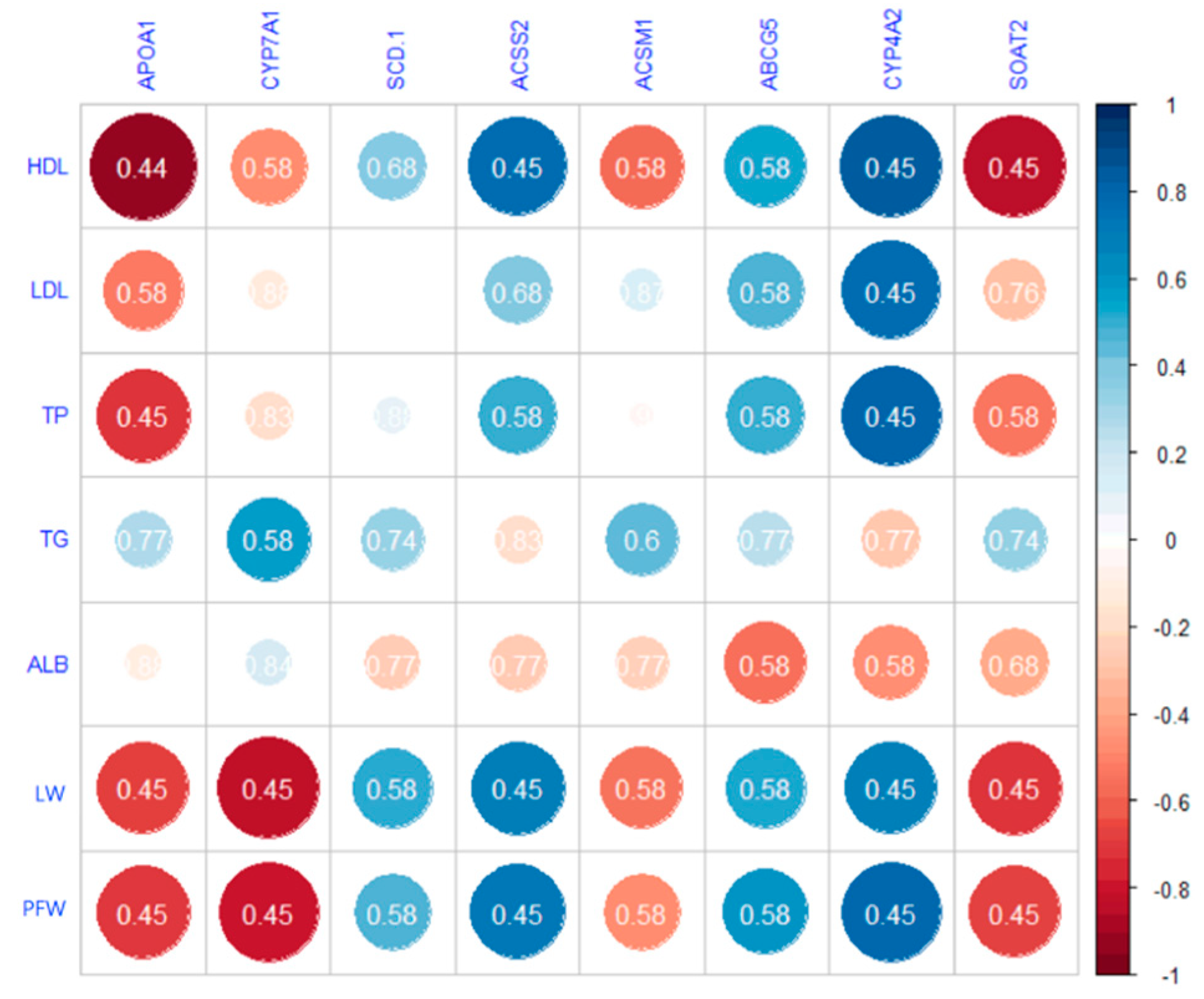
3.3. Serum Biochemistry
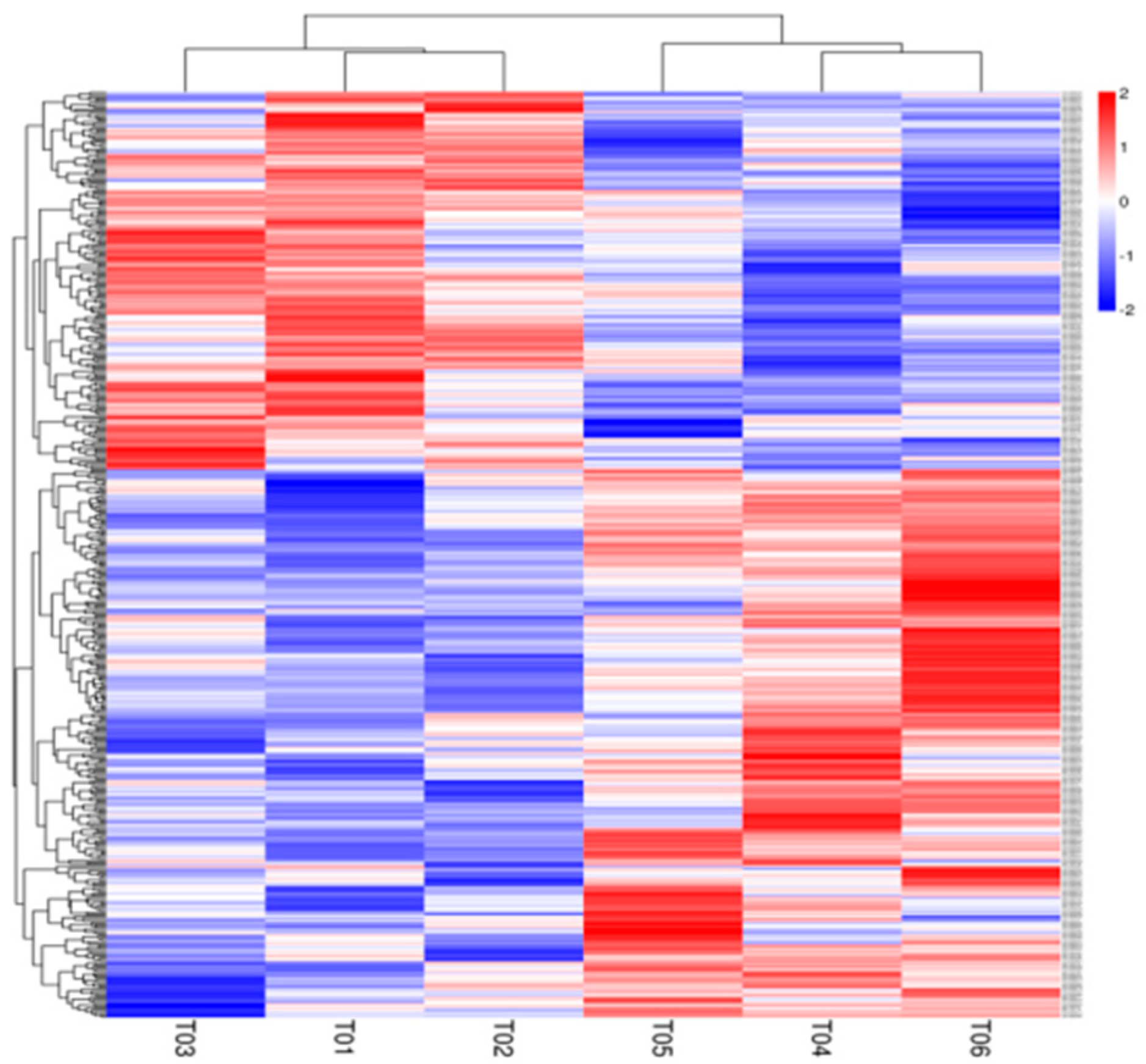
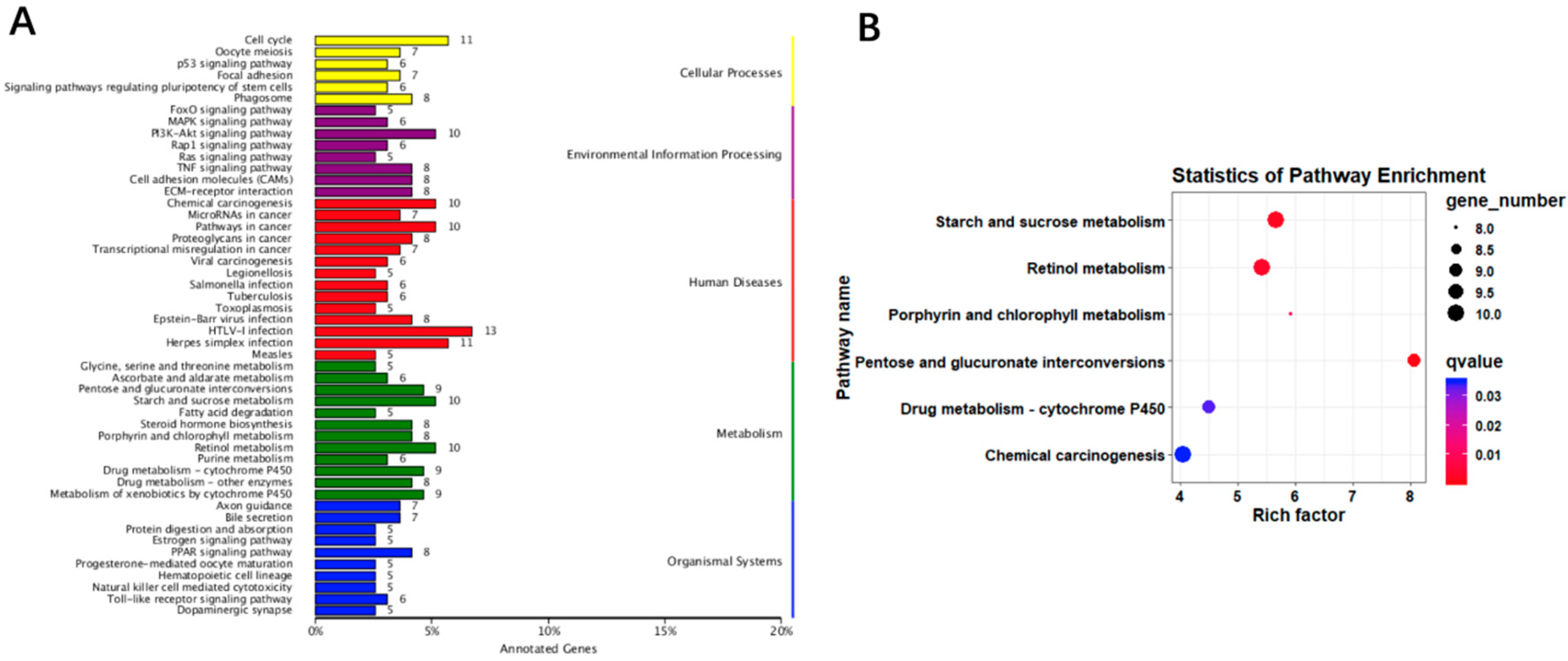
3.4. Liver Transcriptomic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability Statement
References
- Piñero, F.S.; López, F.J.P. Coprophagy in Lepidoptera: Observational and experimental evidence in the pyralid moth Aglossa pinguinalis. Proc. Zool. Soc. Lond. 1998, 244, 357–362. [Google Scholar] [CrossRef]
- Müller, D.W.; Caton, J.; Codron, D.; Schwarm, A.; Lentle, R.; Streich, W.J.; Hummel, J.; Clauss, M. Phylogenetic constraints on digesta separation: Variation in fluid throughput in the digestive tract in mammalian herbivores. Comp. Biochem. Physiol. Part A 2011, 160, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.P.O.; Cargninferreira, E. Morphology and Histology of the Pirarucu (Arapaima gigas) Digestive Tract. Int. J. Morphol. 2017, 35, 950–957. [Google Scholar] [CrossRef]
- Van Bokhorst–de van der Schueren, M.A.E.; Roosemalen, M.M.; Weijs, P.J.M.; Langius, J.A.E. High Waste Contributes to Low Food Intake in Hospitalized Patients. Nutr. Clin. Pract. 2012, 27, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Camp, M.J.; Shipley, L.A.; Milling, C.R.; Rachlow, J.L.; Forbey, J.S. Interacting effects of ambient temperature and food quality on the foraging ecology of small mammalian herbivores. J. Therm. Biol. 2018, 71, 83. [Google Scholar] [CrossRef]
- Feng, Y.; Duan, C.J.; Pang, H.; Mo, X.C.; Wu, C.F.; Yu, Y.; Hu, Y.L.; Wei, J.; Tang, J.L.; Feng, J.X. Cloning and identification of novel cellulase genes from uncultured microorganisms in rabbit cecum and characterization of the expressed cellulases. Appl. Microbiol. Biotechnol. 2007, 75, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Partanen, K.; Jalava, T.; Valaja, J. Effects of a dietary organic acid mixture and of dietary fibre levels on ileal and faecal nutrient apparent digestibility, bacterial nitrogen flow, microbial metabolite concentrations and rate of passage in the digestive tract of pigs. Animal 2007, 1, 389–401. [Google Scholar] [CrossRef]
- Rehman, H.; Böhm, J.; Zentek, J. Effects of differentially fermentable carbohydrates on the microbial fermentation profile of the gastrointestinal tract of broilers. J. Anim. Physiol. Anim. Nutr. 2008, 92, 471. [Google Scholar] [CrossRef]
- Tham, H.T.; Udén, P. Effect of water hyacinth (eichhornia crassipes) silage on intake and nutrient digestibility in cattle fed rice straw and cottonseed cake. Asian Australas. J. Anim. Sci. 2013, 26, 646. [Google Scholar] [CrossRef]
- Yoshida, T.; Kandatsu, M. Studies on cecum digestion. 6. Free amino acids in the cecal contents, hard feces, and soft feces of the rabbit. Nihon Chikusan Gakkaiho 1964, 35, 64–69. [Google Scholar] [CrossRef]
- Yoshida, T.; Kandatsu, M. Studies on Cecum Digestion:III. Nitrogen Distribution in Cecal Contents, Hard Feces, and Soft Feces of the Rabbit. Nihon Chikusan Gakkaiho 1959, 30. Available online: http://xueshu.baidu.com/usercenter/paper/show?paperid=3bfd82708702e12b45d7a50596422f73&site=xueshu_se (accessed on 24 June 2019). [CrossRef]
- Clauss, W. Resorption and Secretion of Water and Electrolytes in the Colon of Rabbit in Relation to Soft and Hard Feces Formation. 1978. Available online: http://agris.fao.org/agris-search/search.do?recordID=US201300572521 (accessed on 24 June 2019).
- Janječić, Z. Impact of Feeding on the Performance and Meat Quality of Fattening Rabbit. Krmiva Časopis O Hranidbi Životinja Proizv. I Tehnol. Krme 2016, 58, 25–32. [Google Scholar]
- Kujawinski, D. Gross Me Out: Why Rabbits Eat Poop and Other Gross Facts about Pets; Nsta Recommends Capstone Press: Arlington, VA, USA, 2012. [Google Scholar]
- Khayyal, A.A.M. Nutritional Effect of Rabbit Manure on the Performance of Growing Rabbits. 1997. Available online: http://agris.fao.org/agris-search/search.do?recordID=EG1999001114 (accessed on 24 June 2019).
- Yun-Fei, L.I.; Qin, C.L.; Wang, Z.Z.; Yang, R.L.; Yang, L.I. Establishment and Analysis of ERIC-PCR DNA Fingerprint of Bacteria in Rabbit Manure. Chin. J. Anim. Vet. Sci. 2016, 47, 716–722. [Google Scholar]
- Liu, T.; Yang, Z.; Zhang, X.; Han, N.; Yuan, J.; Cheng, Y. 16S rDNA analysis of the effect of fecal microbiota transplantation on pulmonary and intestinal flora. Biotech 2017, 7, 370. [Google Scholar] [CrossRef] [PubMed]
- Zou, F.; Zeng, D.; Wen, B.; Sun, H.; Zhou, Y.; Yang, M.; Peng, Z.; Xu, S.; Wang, H.; Fu, X. Illumina Miseq platform analysis caecum bacterial communities of rex rabbits fed with different antibiotics. AMB Express 2016, 6, 100. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Zeng, B.; Wang, P.; Wang, L.; Wen, B.; Li, Y.; Liu, H.; Bai, S.; Jia, G. Microbiome of Total Versus Live Bacteria in the Gut of Rex Rabbits. Front. Microbiol. 2018, 9, 733. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Han, S.; Wang, P.; Wen, B.; Jian, W.; Guo, W.; Yu, Z.; Du, D.; Fu, X.; Kong, F.; et al. The bacterial communities associated with fecal types and body weight of rex rabbits. Sci. Rep. 2015, 5, 9342. [Google Scholar] [CrossRef]
- Goto, Y.; Kurashima, Y.; Kiyono, H. Roles of the gut mucosal immune system in symbiosis and immunity. [Rinshō Ketsueki] Jpn. J. Clin. Hematol. 2015, 56, 2205. [Google Scholar]
- Heinken, A.; Sahoo, S.; Fleming, R.M.T.; Thiele, I. Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut. Gut Microbes 2013, 4, 28. [Google Scholar] [CrossRef]
- Mouzaki, M.; Wang, A.Y.; Bandsma, R.; Comelli, E.M.; Arendt, B.M.; Zhang, L.; Fung, S.; Fischer, S.E.; Mcgilvray, I.G.; Allard, J.P. Bile Acids and Dysbiosis in Non-Alcoholic Fatty Liver Disease. PLoS ONE 2016, 11, e0151829. [Google Scholar] [CrossRef]
- Alexander, K.S.; Zakai, N.A.; Lidofsky, S.D.; Callas, P.W.; Judd, S.E.; Tracy, R.P.; Cushman, M. Non-alcoholic fatty liver disease, liver biomarkers and stroke risk: The Reasons for Geographic and Racial Differences in Stroke cohort. PLoS ONE 2018, 13, e0194153. [Google Scholar] [CrossRef]
- Chen, S.Q.; Zhang, J.; Fang, Q.C. Recent Progress of the Role of Bile Acids in Regulating Glucose, Lipid and Energy Metabolism. Med. Recapitul. 2016, 5, 947–950. [Google Scholar]
- Brody, H. Fatty liver disease. Nature 2017, 551, 7681. [Google Scholar]
- Do, A.; Lim, J.K. Epidemiology of nonalcoholic fatty liver disease: A primer. Clin. Liver Dis. 2016, 7, 106–108. [Google Scholar] [CrossRef]
- Lee, S.; Xu, H. The crystal structure and Gibbs free energy of formation of chukanovite as an oxidation product of carbon steel in human liver. Chem. Geol. 2018, 488, 180–188. [Google Scholar] [CrossRef]
- Xie, X.; Tao, Z.; Shuang, Z.; Wei, L.; Ma, L.; Ming, D.; Yuan, L. Effects of n-3 polyunsaturated fatty acids high fat diet intervention on the synthesis of hepatic high-density lipoprotein cholesterol in obesity-insulin resistance rats. Lipids Health Dis. 2016, 15, 81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, C.; Zhang, Y.; Tang, F.; Li, H.; Zhang, Q.; Lin, H.; Wu, S.; Liu, Y.; Xue, F. Metabolic syndrome and its components as predictors of nonalcoholic fatty liver disease in a northern urban Han Chinese population: A prospective cohort study. Atherosclerosis 2015, 240, 144–148. [Google Scholar] [CrossRef]
- Kächele, M.; Hennige, A.M.; Machann, J.; Hieronimus, A.; Lamprinou, A.; Machicao, F.; Schick, F.; Fritsche, A.; Stefan, N.; Nürnberg, B. Variation in the Phosphoinositide 3-Kinase Gamma Gene Affects Plasma HDL-Cholesterol without Modification of Metabolic or Inflammatory Markers. PLoS ONE 2015, 10, e0144494. [Google Scholar] [CrossRef]
- Gusev, A.; Ko, A.; Shi, H.; Bhatia, G.; Chung, W.; Penninx, B.W.; Jansen, R.; de Geus, E.J.; Boomsma, D.I.; Wright, F.A. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016, 48, 245. [Google Scholar] [CrossRef]
- Lappalainen, T.; Al, E. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013, 501, 506–511. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.R.; Neff, N.F.; Kalisky, T.; Dalerba, P.; Treutlein, B.; Rothenberg, M.E.; Mburu, F.M.; Mantalas, G.L.; Sim, S.; Clarke, M.F. Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods 2014, 11, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Demaux, G.; Gallouin, F.; Guemon, L.; Papantonakis, C. Effects in the rabbit of preventing caecotrophy for a prolonged period. Reprod. Nutr. Dev. 1980, 1651–1659. [Google Scholar] [CrossRef][Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Olsen, H.M. Investigations on pseudo-rumination in rabbits. Vidensk. Medd. Dansk. Naturh. Fohren. Kbh. 1944, 107, 37. [Google Scholar]
- Yoshida, T.; Pleasants, J.R.; Reddy, B.S.; Wostmann, B.S. Efficiency of digestion in germ-free and conventional rabbits. Br. J. Nutr. 1968, 22, 723–737. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fraga, M.J.; Pérezd, A.P.; Carabaño, R.; de Blas, J.C. Effect of type of fiber on the rate of passage and on the contribution of soft feces to nutrient intake of finishing rabbits. J. Anim. Sci. 1991, 69, 1566–1574. [Google Scholar] [CrossRef] [PubMed]
- Michelland, R.; Combes, S.; Cauquil, L.; Gidenne, T.; Monteils, V.; Fortunlamothe, L.; Xicato, G.; Trocino, A.; Lukefahr, S.D. Characterization of bacterial communities in caeum, hard and soft feces of rabbit using 16S rRNA genes capillary electrophoresis single-strand conformation polymorphism (CE-SSCP). In Proceedings of the 9th World Rabbit Congress, Verona, Italy, 10–13 June 2008; pp. 36–40. [Google Scholar]
- Zhang, G. A study on the Behavior of Pseudo-rumination of the Rabbit. Ecol. Domest. Anim. 1994, 14, 10–14. [Google Scholar]
- Taylor, E.L. Pseudo-rumination in the Rabbit. J. Zool. 2010, A110, 159–163. [Google Scholar] [CrossRef]
- Summers, R.W.; Kent, T.H.; Osborne, J.W. Effects of drugs, ileal obstruction, and irradiation on rat gastrointestinal propulsion. Gastroenterology 1970, 59, 731. [Google Scholar] [CrossRef]
- Chilcott, M.J.; Hume, I.D. Coprophagy and Selective Retention of Fluid Digesta: Their Role in the Nutrition of the Common Ringtail Possum, Pseudocheirus Peregrinus. Aust. J. Zool. 1985, 33, 1–15. [Google Scholar] [CrossRef]
- Leiber, F.; Meier, J.S.; Burger, B.; Wettstein, H.R.; Kreuzer, M.; Hatt, J.M.; Clauss, M. Significance of coprophagy for the fatty acid profile in body tissues of rabbits fed different diets. Lipids 2008, 43, 853–865. [Google Scholar] [CrossRef] [PubMed]
- Slade, L.M.; Hintz, H.F. Comparison of digestion in horses, ponies, rabbits and guinea pigs. J. Anim. Sci. 1969, 28, 842–843. [Google Scholar] [CrossRef]
- Sakaguchi, E.; Nabata, A. Comparison of fibre digestion and digesta retention time between nutrias (Myocaster coypus) and guinea-pigs (Cavia porcellus). Comp. Biochem. Physiol. Part A Physiol. 1992, 103, 601–604. [Google Scholar] [CrossRef]
- Miśta, D.; Króliczewska, B.; Marounek, M.; Pecka, E.; Zawadzki, W.; Nicpoń, J. In Vitro Study and Comparison of Caecal Methanogenesis and Fermentation Pattern in the Brown Hare (Lepus europaeus) and Domestic Rabbit (Oryctolagus cuniculus). PLoS ONE 2015, 10, e0117117. [Google Scholar] [CrossRef][Green Version]
- Sánchez, D.H.; Peralta, M.A.C.; Muñoz, S.S.G.; Gama, R.B.; García, J.L.A.; López, F.G. Microbial populations and fermentation in the cecum of growing rabbits fed cecal bacteria inocula. Interciencia 2004, 29, 442–446. [Google Scholar]
- Combes, S.; Gidenne, T.; Cauquil, L.; Bouchez, O.; Fortun-Lamothe, L. Coprophagous behavior of rabbit pups affects implantation of cecal microbiota and health status. J. Anim. Sci. 2015, 92, 652–665. [Google Scholar] [CrossRef]
- Jacquier, V.; Combes, S.; Oswald, I.P.; Rogelgaillard, C.; Gidenne, T. Early modulation of the cecal microbial activity in the young rabbit with rapidly fermentable fiber: Impact on health and growth. J. Anim. Sci. 2014, 92, 5551–5559. [Google Scholar] [CrossRef]
- Dulal, S.; Keku, T.O. Gut microbiome and colorectal adenomas. Cancer J. 2014, 20, 225. [Google Scholar] [CrossRef] [PubMed]
- Carabaño, R.; Fraga, M.J.; Santoma, G.; de Blas, J.C. Effect of diet on composition of cecal contents and on excretion and composition of soft and hard feces of rabbits. J. Anim. Sci. 1988, 66, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Heimesaat, M.M.; Boelke, S.; Fischer, A.; Haag, L.M.; Loddenkemper, C.; Kühl, A.A.; Göbel, U.B.; Bereswill, S. Comprehensive postmortem analyses of intestinal microbiota changes and bacterial translocation in human flora associated mice. PLoS ONE 2012, 7, e40758. [Google Scholar] [CrossRef]
- Belyaeva, O.V.; Wu, L.; Shmarakov, I.; Nelson, P.S.; Kedishvili, N.Y. Retinol dehydrogenase 11 is essential for the maintenance of retinol homeostasis in liver and testis in mice. J. Biol. Chem. 2018, 293, 6996–7007. [Google Scholar] [CrossRef] [PubMed]
- Stuetz, W.; Weber, D.; Dollé, M.E.; Jansen, E.; Grubeck-Loebenstein, B.; Fiegl, S.; Toussaint, O.; Bernhardt, J.; Gonos, E.S.; Franceschi, C.; et al. Plasma Carotenoids, Tocopherols, and Retinol in the Age-Stratified (35–74 Years) General Population: A Cross-Sectional Study in Six European Countries. Nutrients 2016, 8, 614. [Google Scholar] [CrossRef] [PubMed]
- Barochia, A.V.; Gordon, E.M.; Kaler, M.; Cuento, R.A.; Theard, P.; Figueroa, D.M.; Yao, X.; Weir, N.A.; Sampson, M.L.; Stylianou, M. High Density Lipoproteins and Type 2 Inflammatory Biomarkers Are Negatively Correlated in Atopic Asthmatics. J. Lipid Res. 2017, 58, 1713–1721. [Google Scholar] [CrossRef] [PubMed]
- Marschner, R.A.; Pinto, G.; Borges, J.; Markoski, M.M.; Schaan, B.D.; Lehnen, A.M. Short-Term Detraining does not Change Insulin Sensitivity and RBP4 in Rodents Previously Submitted to Aerobic Exercise. Horm. Metab. Res. 2017, 49, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Isvoran, A.; Louet, M.; Vladoiu, D.L.; Craciun, D.; Loriot, M.A.; Villoutreix, B.O.; Miteva, M.A. Pharmacogenomics of the cytochrome P450 2C family: Impacts of amino acid variations on drug metabolism. Drug Discov. Today 2017, 22, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Wen, N.; Liu, W.; Hou, Y.; Zhao, Z. The kinetics behavior of the reduction of formaldehyde catalyzed by Alcohol Dehydrogenase (ADH) and partial uncompetitive substrate inhibition by NADH. Appl. Biochem. Biotechnol. 2013, 170, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Neis, M.M.; Ladd, P.A.; Keeney, D.S. Differentiation-specific factors modulate epidermal CYP1-4 gene expression in human skin in response to retinoic acid and classic aryl hydrocarbon receptor ligands. J. Pharmacol. Exp. Ther. 2006, 319, 1162–1171. [Google Scholar] [CrossRef]
- Zolfaghari, R.; Cifelli, C.J.; Lieu, S.O.; Chen, Q.; Li, N.Q.; Ross, A.C. Lipopolysaccharide opposes the induction of CYP26A1 and CYP26B1 gene expression by retinoic acid in the rat liver in vivo. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G1029–G1036. [Google Scholar] [CrossRef][Green Version]
- Klempt, M.; Vobach, M.; Wiegand, H.; Preuss, T.G.; Schäffer, A.; Kammann, U. 353-nonylphenol induces expression of the T-box6 gene in zebrafish embryos—linking transcriptional information with deformities. J. Fish. Com. 2011, 7, 30–42. [Google Scholar] [CrossRef]
- Wu, C.; Cheng, J.; Hu, S.; Deng, R.; Muangu, Y.W.; Shi, L.; Wu, K.; Zhang, P.; Chang, W.; Wang, G.; et al. Reduced proliferation and increased apoptosis of the SGC-7901 gastric cancer cell line on exposure to GDC-0449. Mol. Med. Rep. 2015, 13, 1434–1440. [Google Scholar] [CrossRef][Green Version]
- Jang, Y.; Park, Y.E.; Yun, C.W.; Kim, D.H.; Chung, H. The vest-collar as a rodent collar to prevent licking and scratching during experiments. Lab. Anim. 2016, 50, 296–304. [Google Scholar] [CrossRef] [PubMed]


| Items | Groups | |||
|---|---|---|---|---|
| CON | CA | CP | ||
| IBW (g) | 1 300.42 ± 128.02 | 1 288.75 ± 129.91 | 1 273.54 ± 116.05 | |
| ADFI (g) | Total Feed intake/g | 143.98 ± 10.32 | 140.48 ± 10.01 | 138.60 ± 7.87 |
| Feed intake at daylight/g | 48.12 ± 8.41 | 43.21 ± 3.99 | 47.29 ± 4.64 | |
| Feed intake at night/g | 95.86 ± 8.49 | 97.27 ± 10.93 | 91.40 ± 5.76 | |
| ADG (g) | 21.60 ± 3.52 a | 21.69 ± 2.11 a | 19.64 ± 1.98 b | |
| Final weight (g) | 2 736.36 ± 271.13 a | 2 722.00 ± 165.15 a | 2 524.76 ± 211.59 b | |
| F/G | 6.80 ± 1.08 | 6.52 ± 0.61 | 7.12 ± 0.75 | |
| Items | Treatment | |
|---|---|---|
| Control Group | Experimental Group | |
| Initial weight (kg) | 0.913 ± 0.118 | 0.922 ± 0.163 |
| Final weight (kg) | 2.935 ± 1.188 a | 2.292 ± 0.170 b |
| Weight gain (%) | 2.021 ± 0.159 a | 1.370 ± 0.262 b |
| SGR (%day-1) | 0.033 ± 0.002 a | 0.022 ± 0.004 b |
| FCR | 3.555 ± 0.341 a | 4.795 ± 1.238 b |
| HIS (%) | 44.702 ± 4.918 | 45.971 ± 3.971 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Xu, H.; Sun, G.; Xue, M.; Sun, S.; Huang, T.; Zhou, J.; Loor, J.J.; Li, M. Transcriptome Analysis of the Effects of Fasting Caecotrophy on Hepatic Lipid Metabolism in New Zealand Rabbits. Animals 2019, 9, 648. https://doi.org/10.3390/ani9090648
Wang Y, Xu H, Sun G, Xue M, Sun S, Huang T, Zhou J, Loor JJ, Li M. Transcriptome Analysis of the Effects of Fasting Caecotrophy on Hepatic Lipid Metabolism in New Zealand Rabbits. Animals. 2019; 9(9):648. https://doi.org/10.3390/ani9090648
Chicago/Turabian StyleWang, Yadong, Huifen Xu, Guirong Sun, Mingming Xue, Shuaijie Sun, Tao Huang, Jianshe Zhou, Juan J. Loor, and Ming Li. 2019. "Transcriptome Analysis of the Effects of Fasting Caecotrophy on Hepatic Lipid Metabolism in New Zealand Rabbits" Animals 9, no. 9: 648. https://doi.org/10.3390/ani9090648
APA StyleWang, Y., Xu, H., Sun, G., Xue, M., Sun, S., Huang, T., Zhou, J., Loor, J. J., & Li, M. (2019). Transcriptome Analysis of the Effects of Fasting Caecotrophy on Hepatic Lipid Metabolism in New Zealand Rabbits. Animals, 9(9), 648. https://doi.org/10.3390/ani9090648






