Liver Transcriptome Changes of Hyla Rabbit in Response to Chronic Heat Stress
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statements
2.2. Experiment Design and Sample Collection
2.3. RNA Extraction, cDNA Library Construction and Sequencing
2.4. Quality Control and Mapping of Reads
2.5. Analysis of Differentially Expressed Genes
2.6. Functional Enrichment Analysis
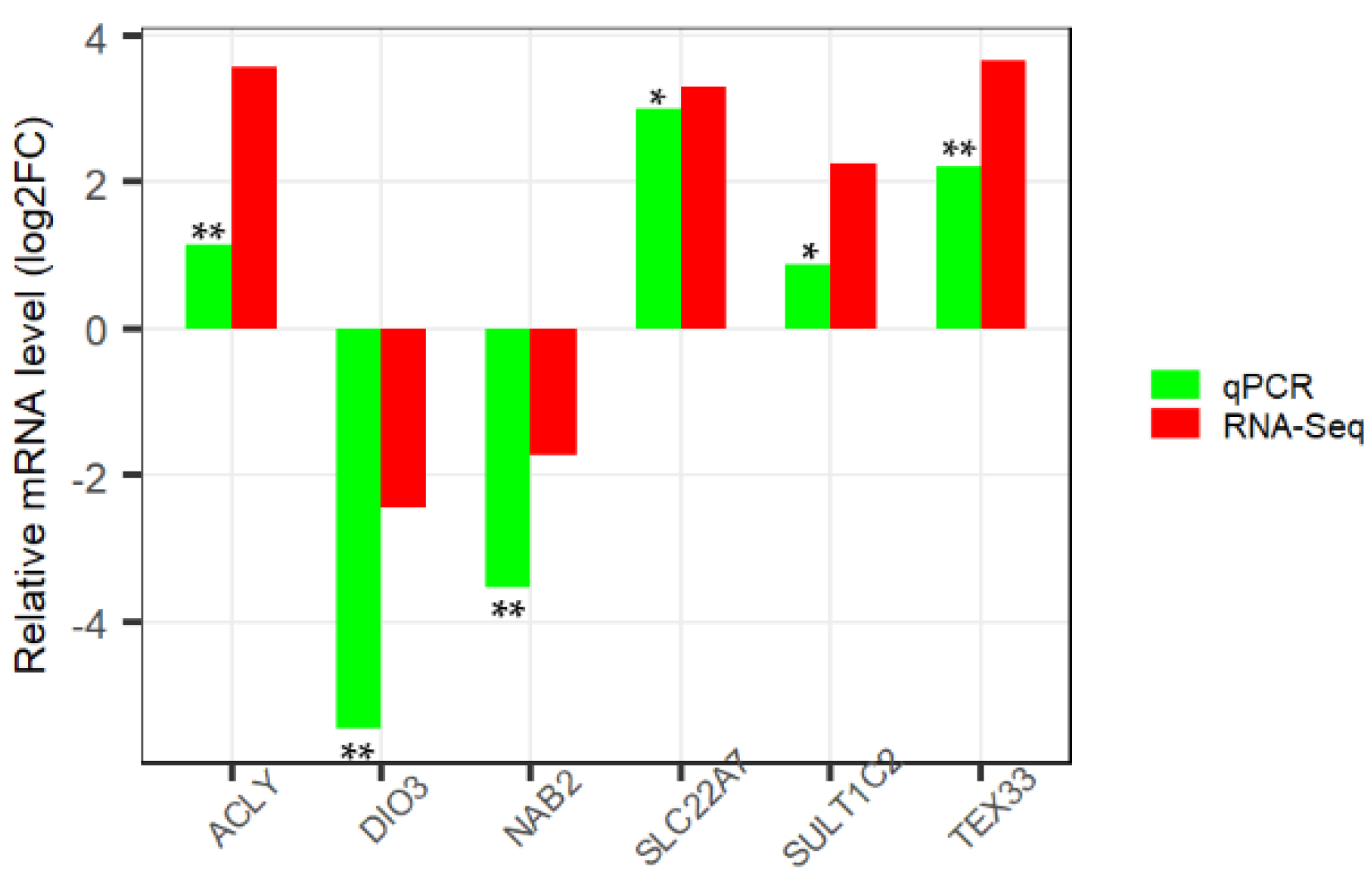
2.7. Validation of RNA-Seq Data by qPCR
3. Results
3.1. Influence of Chronic Heat Stress on Physiological Characteristics
3.2. Sequencing and Mapping of Reads
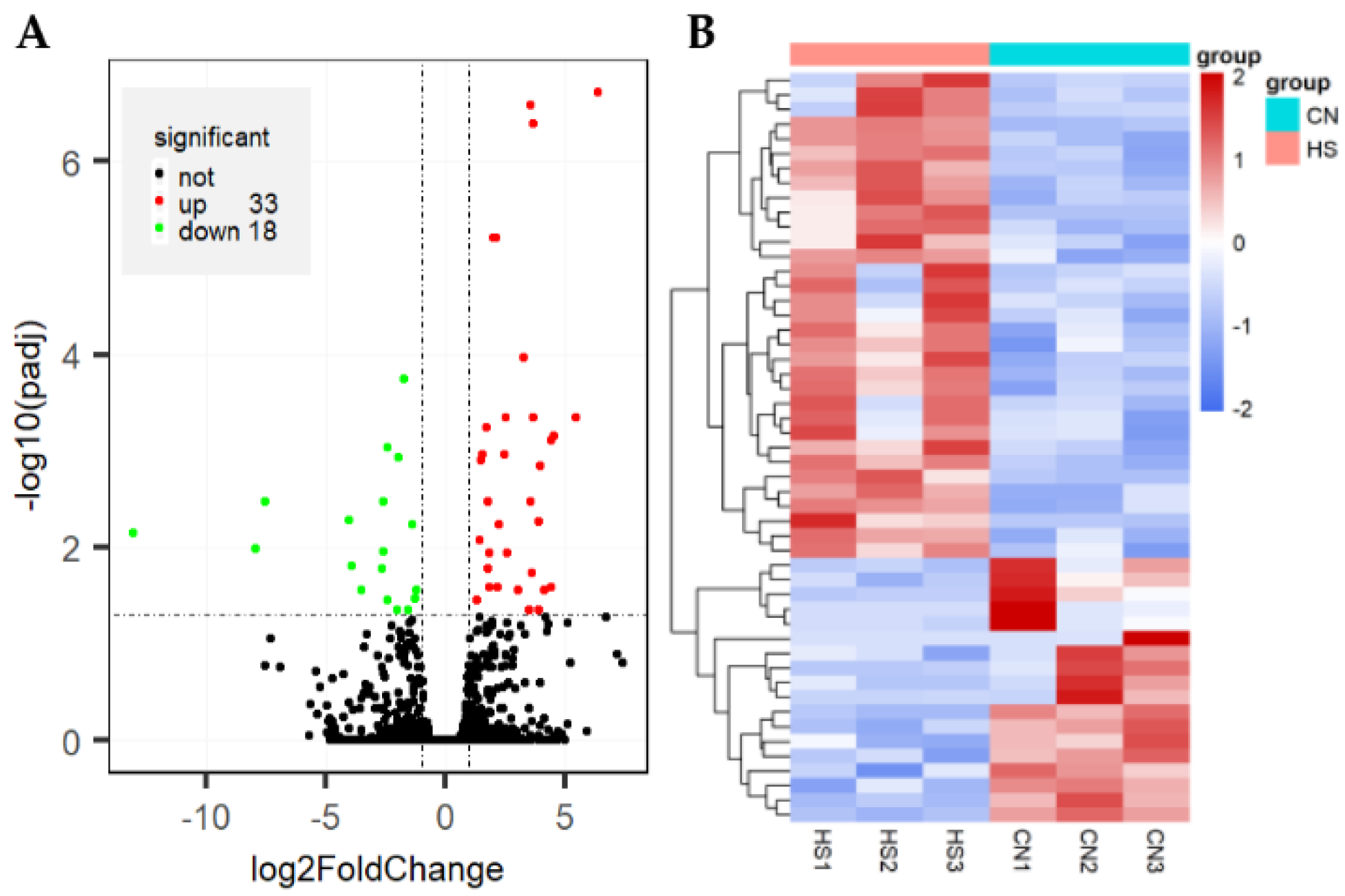
3.3. Analysis of Gene Expression
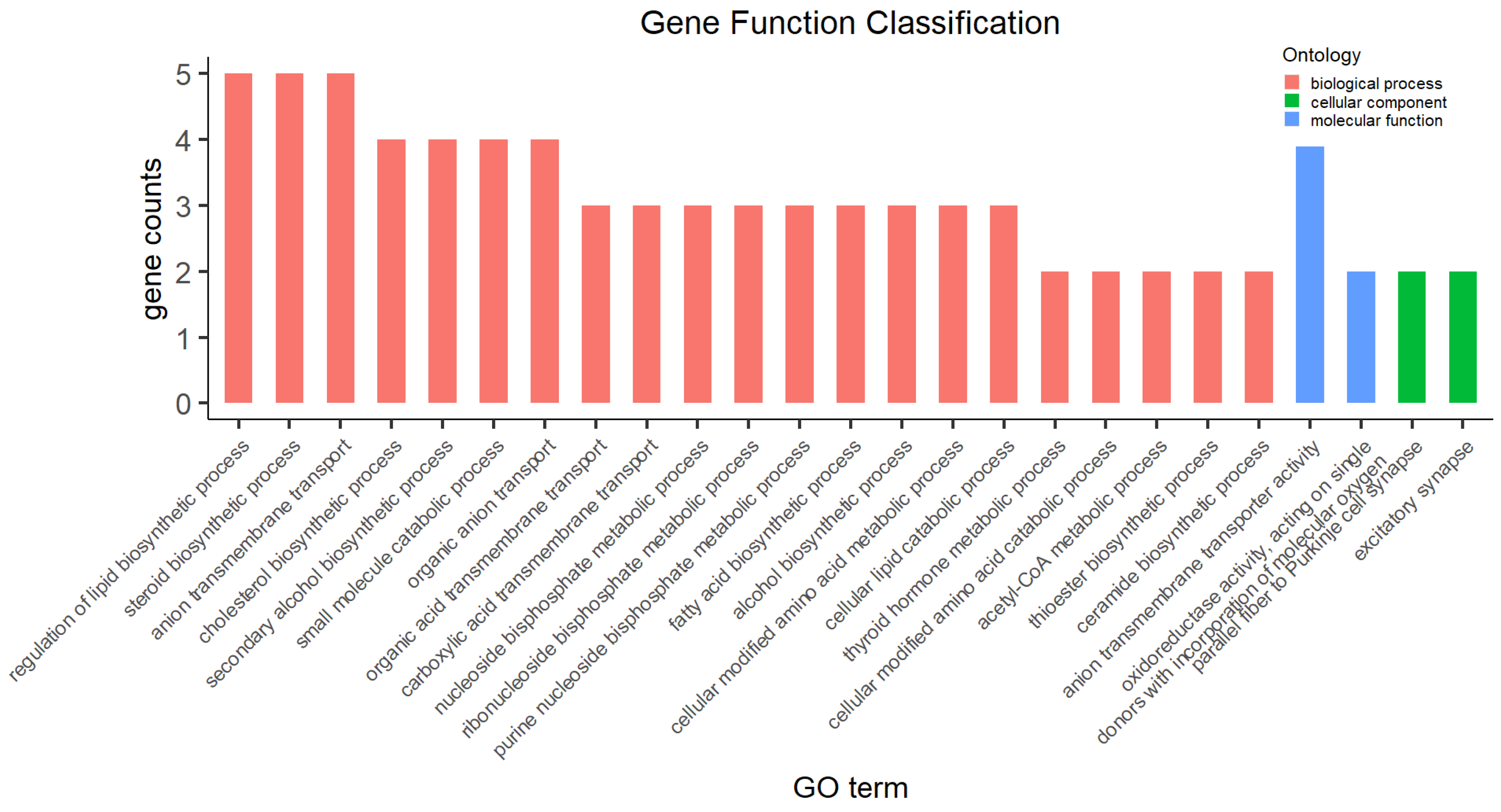
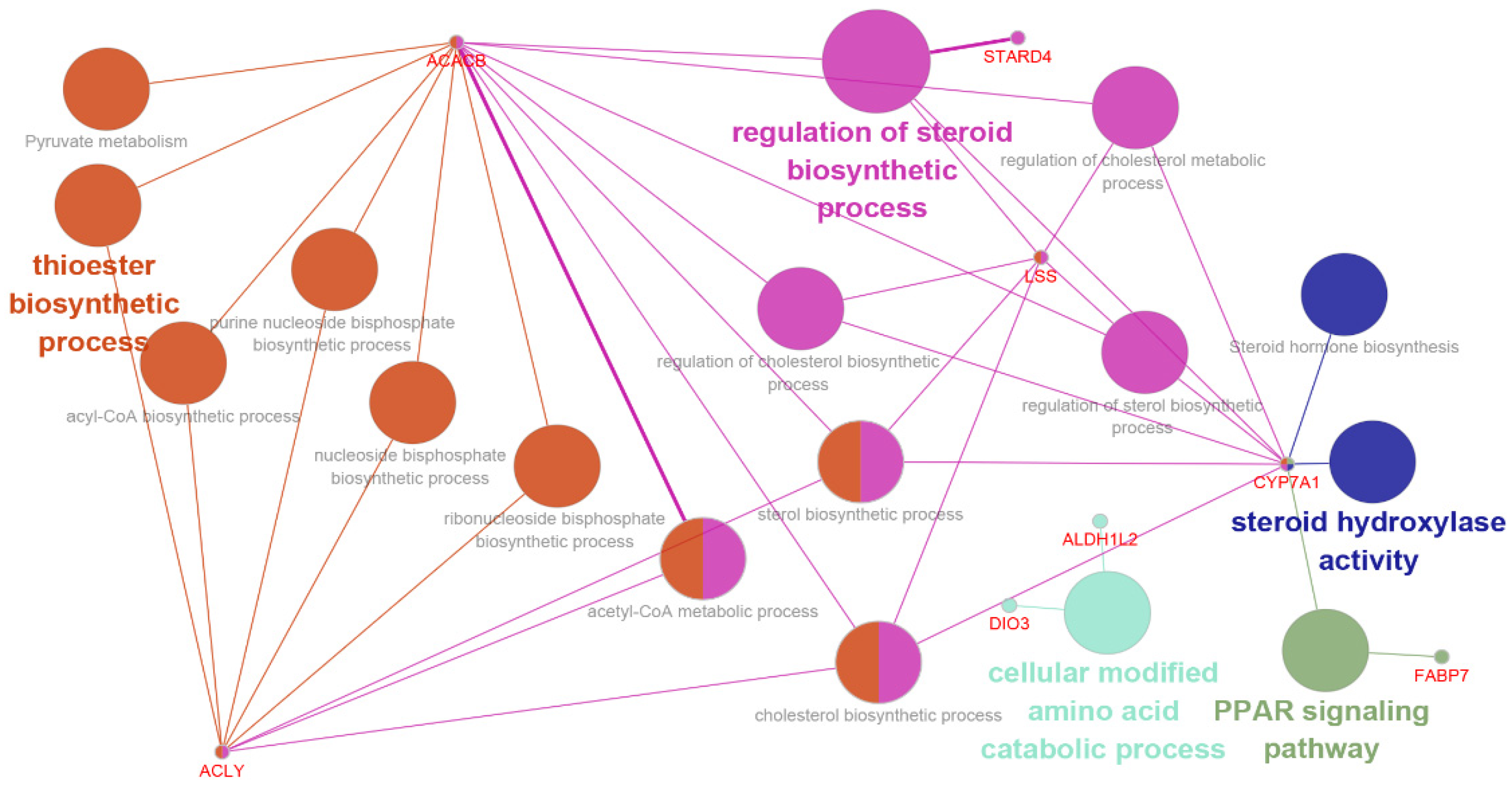
3.4. Functional Categories of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Min, L.; Zheng, N.; Zhao, S.; Cheng, J.; Yang, Y.; Zhang, Y.; Yang, H.; Wang, J. Long-Term Heat Stress Induces the Inflammatory Response in Dairy Cows Revealed by Plasma Proteome Analysis. Biochem. Biophys. Res. Commun. 2016, 471, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Do Amaral, B.C.; Connor, E.E.; Tao, S.; Hayen, M.J.; Bubolz, J.W.; Dahl, G.E. Heat Stress Abatement During the Dry Period Influences Metabolic Gene Expression and Improves Immune Status In the Transition Period of Dairy Cows. J. Dairy Sci. 2011, 94, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Coble, D.J.; Fleming, D.; Persia, M.E.; Ashwell, C.M.; Rothschild, M.F.; Schmidt, C.J.; Lamont, S.J. RNA-Seq Analysis of Broiler Liver Transcriptome Reveals Novel Responses to High Ambient Temperature. BMC Genom. 2014, 15, 1084. [Google Scholar] [CrossRef] [PubMed]
- Jimoh, O.A.; Ayedun, E.S.; Oyelade, W.A.; Oloruntola, O.D.; Daramola, O.T.; Ayodele, S.O.; Omoniyi, I.S. Protective Effect of Soursop (Annona Muricata Linn.) Juice on Oxidative Stress in Heat Stressed Rabbits. J. Anim. Sci. Technol. 2018, 60, 28. [Google Scholar] [CrossRef] [PubMed]
- Jastrebski, S.F.; Lamont, S.J.; Schmidt, C.J. Chicken Hepatic Response to Chronic Heat Stress Using Integrated Transcriptome and Metabolome Analysis. PLoS ONE 2017, 12, e0181900. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Lim, K.S.; Byun, M.; Lee, K.T.; Yang, Y.R.; Park, M.; Lim, D.; Chai, H.H.; Bang, H.T.; Hwangbo, J.; et al. Identification of the Acclimation Genes in Transcriptomic Responses to Heat Stress of White Pekin duck. Cell Stress Chaperones 2017, 22, 787–797. [Google Scholar] [CrossRef]
- Li, Y.; Kong, L.; Deng, M.; Lian, Z.; Han, Y.; Sun, B.; Guo, Y.; Liu, G.; Liu, D. Heat Stress-Responsive Transcriptome Analysis in the Liver Tissue of Hu Sheep. Genes 2019, 10, 395. [Google Scholar] [CrossRef]
- Lu, Z.; Chu, M.; Li, Q.; Jin, M.; Fei, X.; Ma, L.; Zhang, L.; Wei, C. Transcriptomic Analysis Provides Novel Insights into Heat Stress Responses in Sheep. Animals 2019, 9, 387. [Google Scholar] [CrossRef]
- Fan, J.; Chen, Y.; Yan, H.; Liu, B.; Wang, Y.; Zhang, J.; Chen, Y.E.; Liu, E.; Liang, J. Genomic and Transcriptomic Analysis of Hypercholesterolemic Rabbits: Progress and Perspectives. Int. J. Mol. Sci. 2018, 19, 3512. [Google Scholar] [CrossRef]
- Li, T.; Wan, B.; Huang, J.; Zhang, X. Comparison of Gene Expression in Hepatocellular Carcinoma, Liver Development, and Liver Regeneration. Mol. Genet. Genom. 2010, 283, 485–492. [Google Scholar] [CrossRef]
- Marai, I.F.; Ayyat, M.S.; Abd El-Monem, U.M. Growth Performance and Reproductive Traits at First Parity of New Zealand White Female Rabbits as Affected by Heat Stress and Its Alleviation under Egyptian Conditions. Trop. Anim. Health Prod. 2001, 33, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ File Format for Sequences with Quality Scores, and the Solexa/Illumina FASTQ Variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods. 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. Feature Counts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. Cluster Profiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape Plug-In to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zeferino, C.P.; Moura, A.S.A.M.T.; Fernandes, S.; Kanayama, J.S.; Scapinello, C.; Sartori, J.R. Genetic Group × Ambient Temperature Interaction Effects on Physiological Responses and Growth Performance of Rabbits. Livest. Sci. 2011, 140, 177–183. [Google Scholar] [CrossRef]
- Ondruska, L.; Rafay, J.; Okab, A.B.; Ayoub, M.A.; Al-Haidary, A.A.; Samara, E.M.; Parkanyi, V.; Chrastinova, L.; Jurcik, R.; Massanyi, P. Influence of Elevated Ambient Temperature upon Some Physiological Measurements of New Zealand White Rabbits. Vet. Med. (Praha) 2011, 56, 180–186. [Google Scholar] [CrossRef]
- Baker, T.A.; Romero, J.; Bach Iv, H.H.; Strom, J.A.; Gamelli, R.L.; Majetschak, M. Systemic Release of Cytokines and Heat Shock Proteins in Porcine Models of Polytrauma and Hemorrhage. Crit. Care Med. 2012, 40, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.J.; Yoon, K.Y.; Hong, H.D.; Lee, B.Y. Role of the Red Ginseng in Defense Against the Environmental Heat Stress in Sprague Dawley Rats. Molecules 2015, 20, 20240–20253. [Google Scholar] [CrossRef] [PubMed]
- Wheelock, J.B.; Rhoads, R.P.; Vanbaale, M.J.; Sanders, S.R.; Baumgard, L.H. Effects of Heat Stress on Energetic Metabolism in Lactating Holstein Cows. J. Dairy Sci. 2010, 93, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Wakil, S.J.; Abu-Elheiga, L.A. Fatty Acid Metabolism: Target for Metabolic Syndrome. J. Lipid Res. 2009, 50, S138–S143. [Google Scholar] [CrossRef]
- Han, B.; Liang, W.; Liu, L.; Li, Y.; Sun, D. Genetic Association of the ACACB Gene with Milk Yield and Composition Traits in Dairy Cattle. Anim. Genet. 2018, 49, 169–177. [Google Scholar] [CrossRef]
- Chypre, M.; Zaidi, N.; Smans, K. ATP-Citrate Lyase: A Mini-Review. Biochem. Biophys. Res. Commun. 2012, 422, 1–4. [Google Scholar] [CrossRef]
- Flees, J.; Rajaei-Sharifabadi, H.; Greene, E.; Beer, L.; Hargis, B.M.; Ellestad, L.; Porter, T.; Donoghue, A.; Bottje, W.G.; Dridi, S. Effect of Morinda Citrifolia (noni)-Enriched Diet on Hepatic Heat Shock Protein and Lipid Metabolism-Related Genes in Heat Stressed Broiler Chickens. Front. Physiol. 2017, 8, 919. [Google Scholar] [CrossRef]
- Maione, F.; Oliaro-Bosso, S.; Meda, C.; Di Nicolantonio, F.; Bussolino, F.; Balliano, G.; Viola, F.; Giraudo, E. The Cholesterol Biosynthesis Enzyme Oxidosqualene Cyclase Is a New Target to Impair Tumour Angiogenesis and Metastasis Dissemination. Sci. Rep. 2015, 5, 9054. [Google Scholar] [CrossRef]
- Björkhem, I.; Araya, Z.; Rudling, M.; Angelin, B.; Einarsson, C.; Wikvall, K. Differences in the Regulation of the Classical and the Alternative Pathway for Bile Acid Synthesis in Human Liver-No Coordinate Regulation of CYP7A1 and CYP27A1. J. Biol. Chem. 2002, 277, 26804–26807. [Google Scholar] [CrossRef]
- Hylemon, P.B.; Zhou, H.; Pandak, W.M.; Ren, S.; Gil, G.; Dent, P. Bile Acids as Regulatory Molecules. J. Lipid Res. 2009, 50, 1509–1520. [Google Scholar] [CrossRef]
- Xiao, Y.; Yan, W.; Zhou, K.; Cao, Y.; Cai, W. Glucocorticoid Treatment Alters Systemic Bile Acid Homeostasis by Regulating the Biosynthesis and Transport of Bile Salts. Dig. Liver Dis. 2016, 48, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.J.; Hong, H.D.; Lee, O.H.; Lee, B.Y. The Effects of Acanthopanax Senticosus on Global Hepatic Gene Expression in Rats Subjected to Heat Environmental Stress. Toxicology 2010, 278, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Hsieh, J.C.; Schmidt, C.J.; Zhu, Q.; Lamont, S.J. Liver Transcriptome Response to Hyperthermic Stress in Three Distinct Chicken Lines. BMC Genom. 2016, 17, 955. [Google Scholar] [CrossRef] [PubMed]
- Aengwanich, W.; Simaraks, S. Pathology of Heart, Lung, Liver and Kidney in Broilers under Chronic Heat Stress. Pathology 2004, 26, 418. [Google Scholar]
- Azad, M.a.K.; Kikusato, M.; Maekawa, T.; Shirakawa, H.; Toyomizu, M. Metabolic Characteristics and Oxidative Damage to Skeletal Muscle in Broiler Chickens Exposed to Chronic Heat Stress. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2010, 155, 401–406. [Google Scholar] [CrossRef]
| Item | Pretreatment | Treatment | ||
|---|---|---|---|---|
| HS | CN | HS | CN | |
| respiration rate (/min) | 120 ± 11 | 126 ± 17 | 126 ± 14 | 124 ± 17 |
| heart rate (/min) | 210 ± 27 B | 223 ± 30 AB | 245 ± 35 A | 213 ± 29 B |
| rectal temperature (°C) | 39.5 ± 0.3 B | 39.6 ± 0.2 B | 40.6 ± 0.4 A | 39.5 ± 0.3 B |
| Items | HS1 | HS2 | HS3 | CN1 | CN2 | CN3 |
|---|---|---|---|---|---|---|
| Raw reads | 64,568,316 | 64,568,316 | 64,568,316 | 64,568,316 | 64,568,316 | 64,568,316 |
| Clean reads | 63,191,812 | 63,262,266 | 63,292,936 | 63,065,116 | 63,170,720 | 63,559,320 |
| Clean bases | 9.48G | 9.49G | 9.49G | 9.46G | 9.48G | 9.53G |
| Error rate | 0.03% | 0.03% | 0.03% | 0.03% | 0.03% | 0.03% |
| Q20 | 96.50% | 95.95% | 96.52% | 96.38% | 97.38% | 97.70% |
| Q30 | 91.13% | 90.05% | 91.09% | 90.82% | 92.87% | 93.58% |
| GC content | 55.51% | 55.76% | 54.95% | 55.30% | 55.83% | 55.52% |
| Total map | 53,694,163 (84.97%) | 52,919,164 (83.65%) | 54,192,938 (85.62%) | 53,806,908 (85.32%) | 53,743,538 (85.08%) | 54,514,687 (85.77%) |
| Unique map | 51,618,791 (81.69%) | 51,024,408 (80.66%) | 51,954,752 (82.09%) | 51,853,433 (82.22%) | 51,701,154 (81.84%) | 52,454,172 (82.53%) |
| Multi map | 2,075,372 (3.28%) | 1,894,756 (3.0%) | 2,238,186 (3.54%) | 1,953,475 (3.1%) | 2,042,384 (3.23%) | 2,060,515 (3.24%) |
| Proper map | 48,422,878 (76.63%) | 47,632,360 (75.29%) | 48,881,796 (77.23%) | 48,638,926 (77.12%) | 48,810,104 (77.27%) | 49,723,966 (78.23%) |
| FPKM Interval | HS1 | HS2 | HS3 | CN1 | CN2 | CN3 |
|---|---|---|---|---|---|---|
| 0–1 | 4319 (28.65%) | 4315 (28.56%) | 4373 (28.79%) | 4349 (29.08%) | 4497 (29.08%) | 4582 (29.47%) |
| 1–3 | 2379 (15.78%) | 2413 (15.97%) | 2399 (15.79%) | 2315 (15.48%) | 2426 (15.69%) | 2397 (15.42%) |
| 3–15 | 4657 (30.89%) | 4635 (30.68%) | 4748 (31.26%) | 4523 (30.24%) | 4829 (31.23%) | 4916 (31.62%) |
| 15–60 | 2448 (16.24%) | 2492 (16.49%) | 2411 (15.87%) | 2468 (16.50%) | 2545 (16.46%) | 2542 (16.35%) |
| >60 | 1272 (8.44%) | 1254 (8.30%) | 1259 (8.29%) | 1300 (8.69%) | 1167 (7.55%) | 1110 (7.14%) |
| Total | 15,075 | 15,109 | 15,190 | 14,955 | 15,464 | 15,547 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Z.-L.; Yang, X.; Chen, S.-Y.; Deng, F.-L.; Jia, X.-B.; Hu, S.-Q.; Wang, J.; Lai, S.-J. Liver Transcriptome Changes of Hyla Rabbit in Response to Chronic Heat Stress. Animals 2019, 9, 1141. https://doi.org/10.3390/ani9121141
Wu Z-L, Yang X, Chen S-Y, Deng F-L, Jia X-B, Hu S-Q, Wang J, Lai S-J. Liver Transcriptome Changes of Hyla Rabbit in Response to Chronic Heat Stress. Animals. 2019; 9(12):1141. https://doi.org/10.3390/ani9121141
Chicago/Turabian StyleWu, Zhou-Lin, Xue Yang, Shi-Yi Chen, Fei-Long Deng, Xian-Bo Jia, Shen-Qiang Hu, Jie Wang, and Song-Jia Lai. 2019. "Liver Transcriptome Changes of Hyla Rabbit in Response to Chronic Heat Stress" Animals 9, no. 12: 1141. https://doi.org/10.3390/ani9121141
APA StyleWu, Z.-L., Yang, X., Chen, S.-Y., Deng, F.-L., Jia, X.-B., Hu, S.-Q., Wang, J., & Lai, S.-J. (2019). Liver Transcriptome Changes of Hyla Rabbit in Response to Chronic Heat Stress. Animals, 9(12), 1141. https://doi.org/10.3390/ani9121141





