Transcriptome of Chicken Liver Tissues Reveals the Candidate Genes and Pathways Responsible for Adaptation into Two Different Climatic Conditions
Abstract
Simple summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement and Sample Collection
2.2. RNA Extraction, Library Construction, and Sequencing
2.3. Quality Analysis and Mapping of Reads
2.4. Differential Gene Expression Analysis
2.5. GO, KEGG Pathway Analysis, and Network Representation
3. Results
3.1. Raw Reads Quality, Sequencing and Mapping
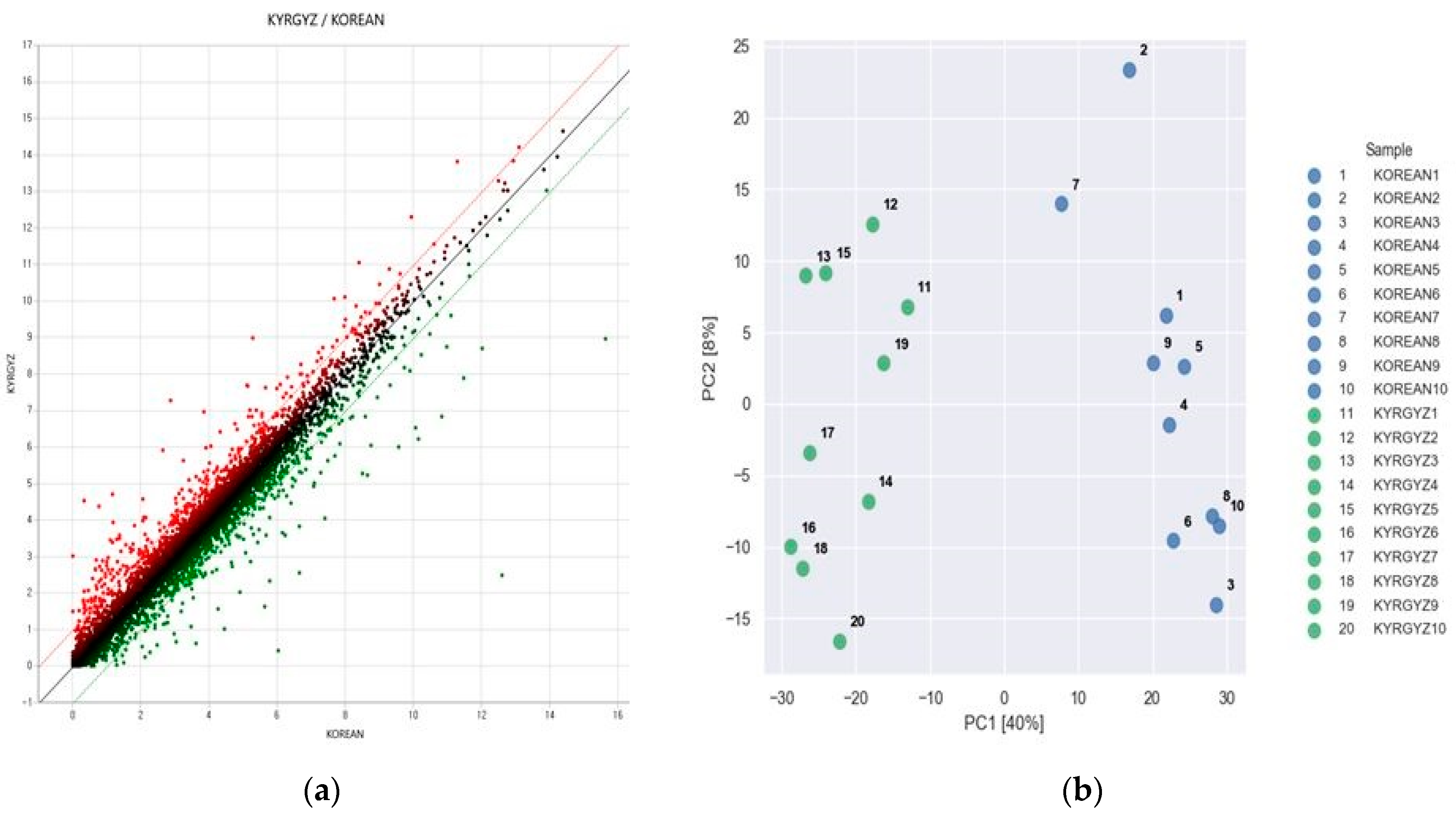
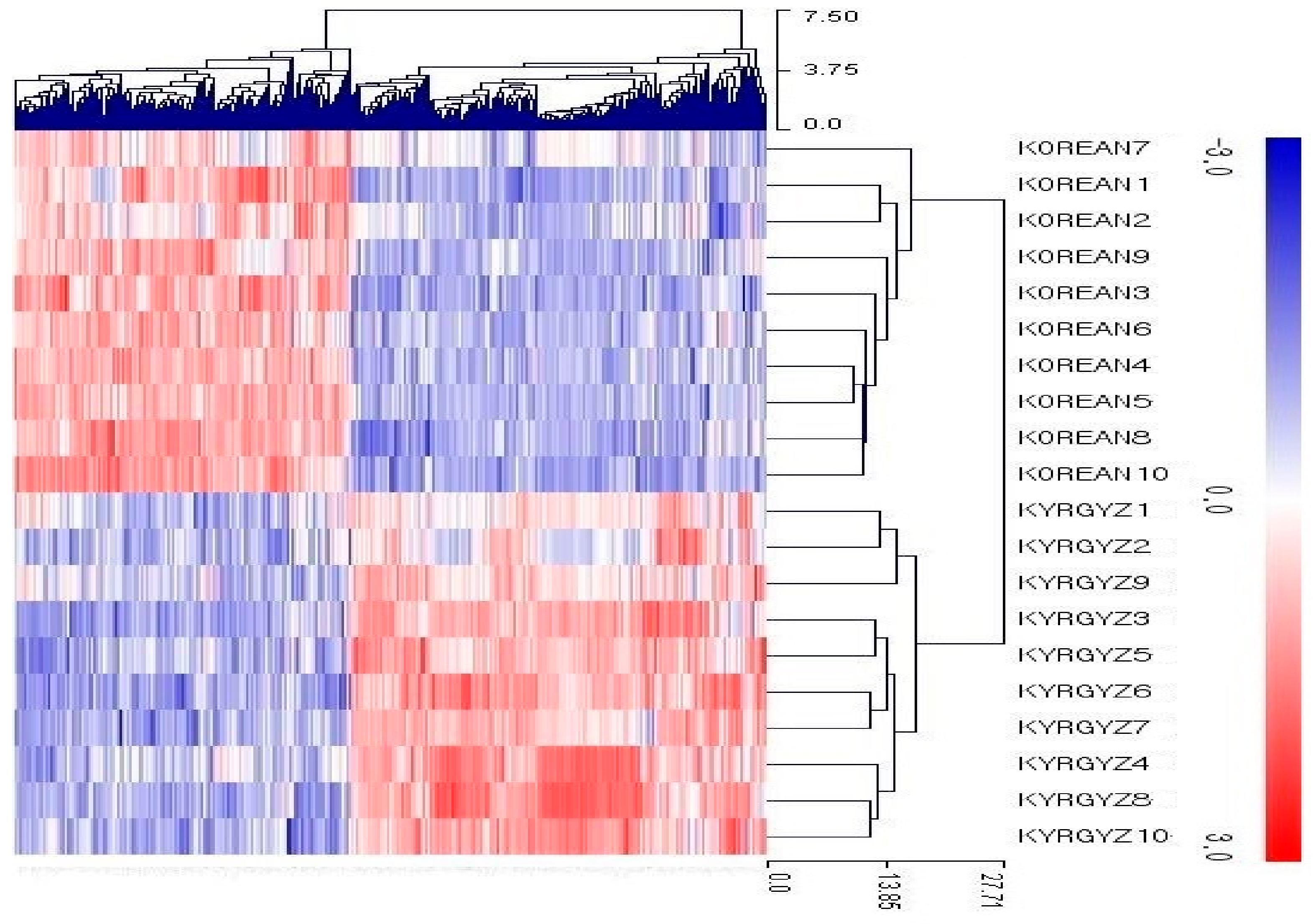
3.2. RNA-Seq of Liver Tissue
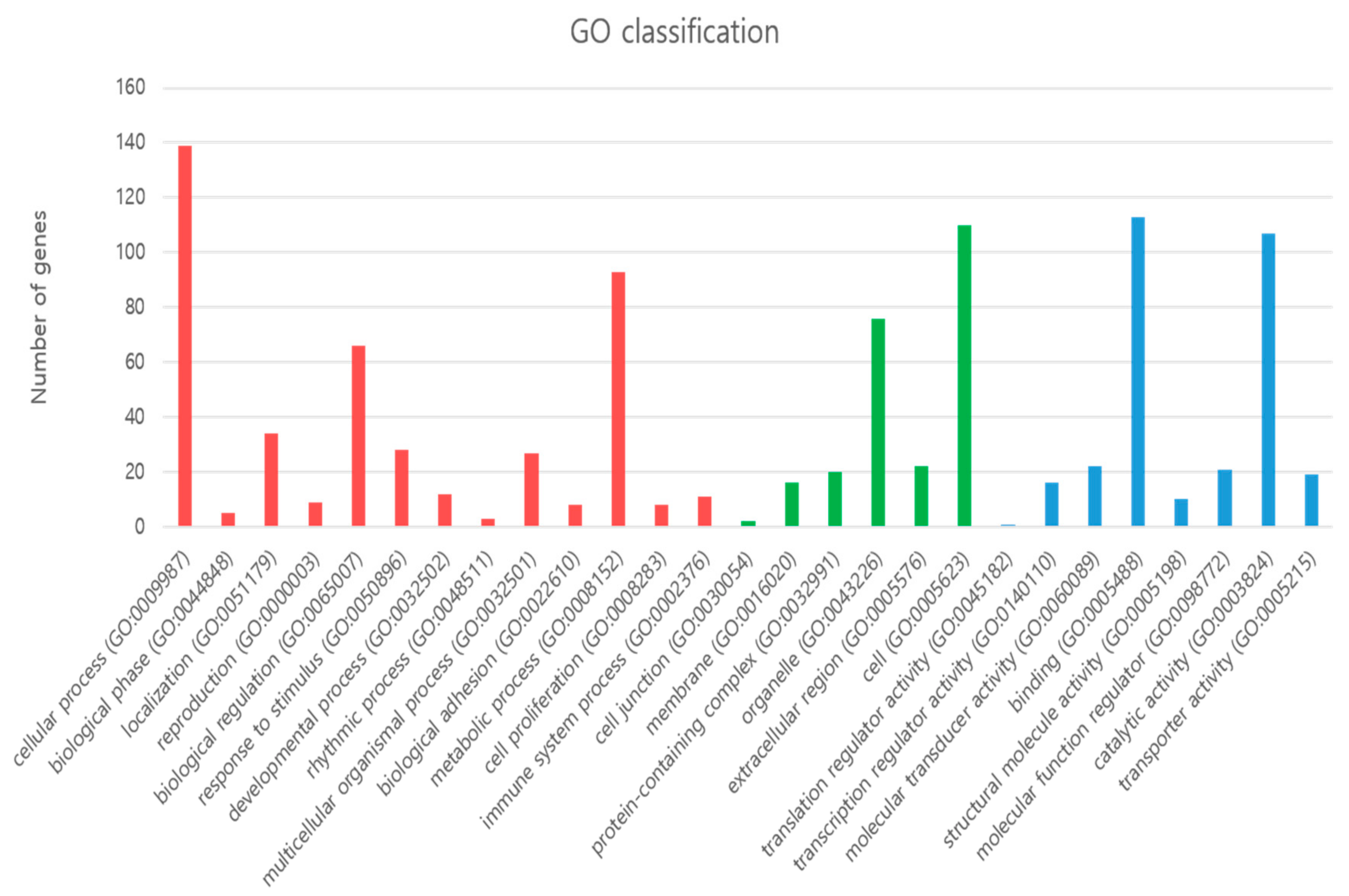
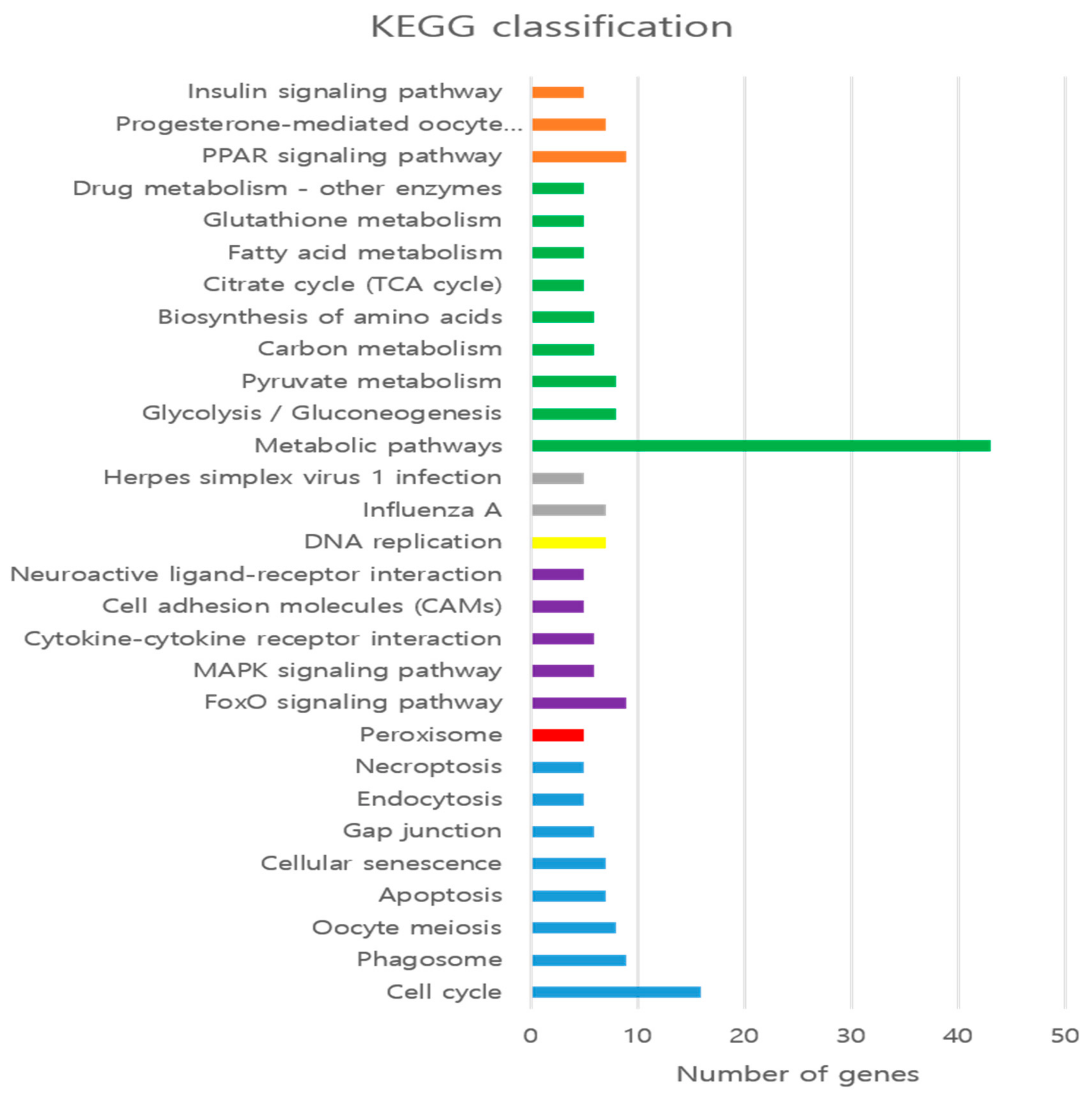
3.3. GO Enrichment and KEGG Pathways Analysis
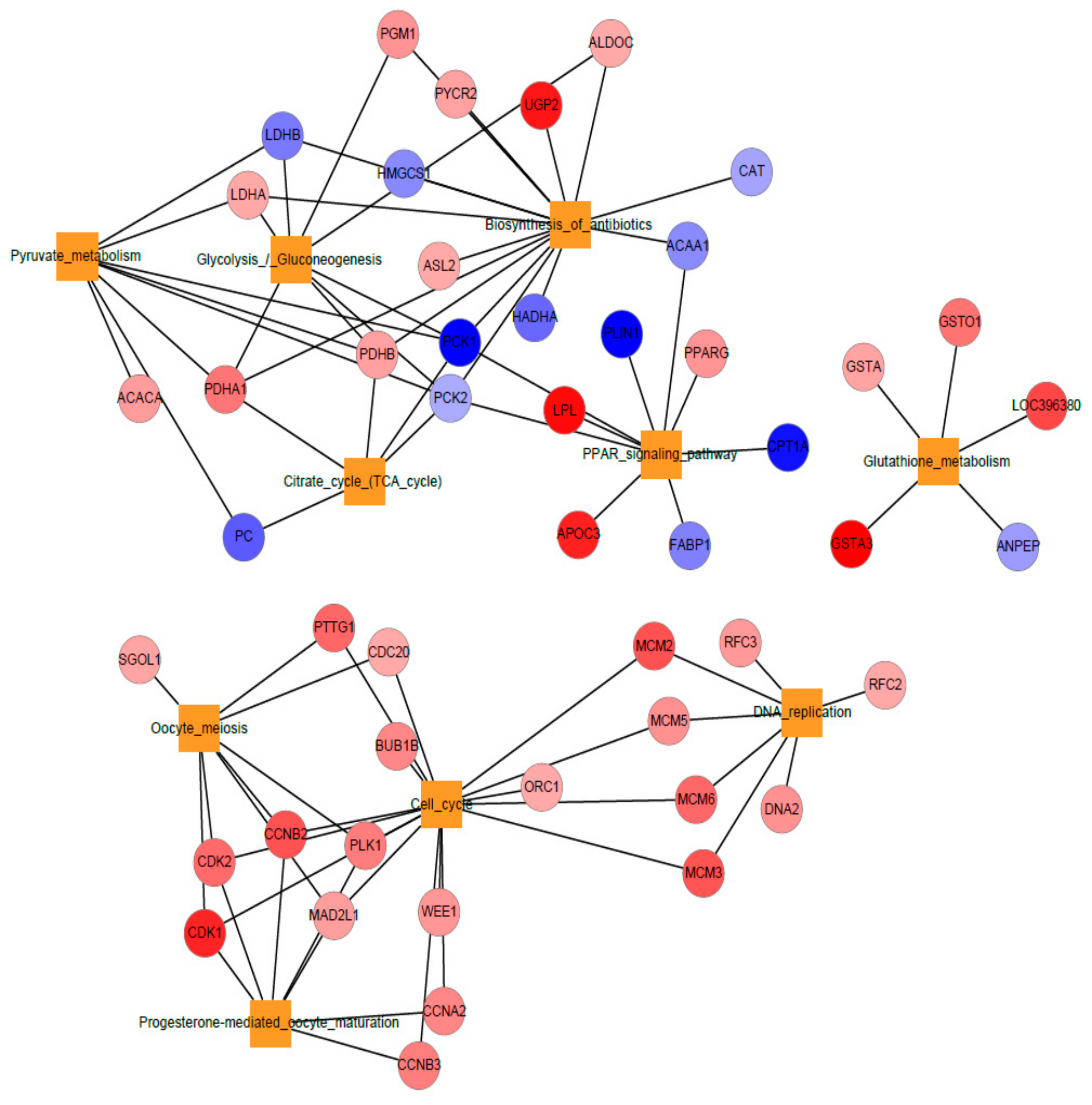
3.4. Network Representation of Top Ten KEGG Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Le Bihan-Duval, E.; Debut, M.; Berri, C.M.; Sellier, N.; Santé-Lhoutellier, V.; Jégo, Y.; Beaumont, C. Chicken meat quality: Genetic variability and relationship with growth and muscle characteristics. BMC genetics 2008, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, D. Poultry meat quality. World’s Poul. Sci. J. 2002, 58, 131–145. [Google Scholar] [CrossRef]
- Ahn, D.-H.; Park, S.-Y. Studies on components related to taste such as free amino acids and nucleotides in Korean native chicken meat. J. Korean Societ.Food Sci.Nutr. 2002, 31, 547–552. [Google Scholar]
- Choo, Y.K.; Chung, S.H. Brief Review on Local Chicken Breeds in Korea with Respect to Growth Performance and Meat Quality. Inter. J. Poult. Sci. 2014, 13, 662. [Google Scholar]
- Désert, C.; Duclos, M.J.; Blavy, P.; Lecerf, F.; Moreews, F.; Klopp, C.; Aubry, M.; Herault, F.; Le Roy, P.; Berri, C. Transcriptome profiling of the feeding-to-fasting transition in chicken liver. BMC Genom. 2008, 9, 611. [Google Scholar] [CrossRef] [PubMed]
- Kassahn, K.S.; Crozier, R.H.; Pörtner, H.O.; Caley, M.J. Animal performance and stress: Responses and tolerance limits at different levels of biological organisation. Biol. Rev. 2009, 84, 277–292. [Google Scholar] [CrossRef]
- Aimee, Y.Y.; Shimoda, L.A.; Iyer, N.V.; Huso, D.L.; Sun, X.; McWilliams, R.; Beaty, T.; Sham, J.S.; Wiener, C.M.; Sylvester, J. Impaired physiological responses to chronic hypoxia in mice partially deficient for hypoxia-inducible factor 1α. J. Clinic. Investig. 1999, 103, 691–696. [Google Scholar]
- Li, H.; Wang, T.; Xu, C.; Wang, D.; Ren, J.; Li, Y.; Tian, Y.; Wang, Y.; Jiao, Y.; Kang, X. Transcriptome profile of liver at different physiological stages reveals potential mode for lipid metabolism in laying hens. BMC Genom. 2015, 16, 763. [Google Scholar] [CrossRef]
- Srikanth, K.; Kumar, H.; Park, W.; Byun, M.-J.; Lim, D.; Kemp, S.; Te Pas, M.; Kim, J.-M.; Park, J.-E. Cardiac and skeletal muscle transcriptome response to heat stress in Kenyan chicken ecotypes adapted to low and high altitudes reveal differences in thermal tolerance and stress response. Front. Genet. 2019, 10, 993. [Google Scholar] [CrossRef]
- Park, W.; Srikanth, K.; Lim, D.; Park, M.; Hur, T.; Kemp, S.; Dessie, T.; Kim, M.; Lee, S.R.; Te Pas, M. Comparative transcriptome analysis of Ethiopian indigenous chickens from low and high altitudes under heat stress condition reveals differential immune response. Anim. Genetic. 2019, 50, 42–53. [Google Scholar] [CrossRef]
- Si, Y.; Wen, H.; Li, Y.; He, F.; Li, J.; Li, S.; He, H. Liver transcriptome analysis reveals extensive transcriptional plasticity during acclimation to low salinity in Cynoglossus semilaevis. BMC Genom. 2018, 19, 464. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, M.M.; Emmenegger, Y.; Hubbard, J.; Franken, P. Cold-inducible RNA-binding protein (CIRBP) adjusts clock-gene expression and REM-sleep recovery following sleep deprivation. Elife 2019, 8, e43400. [Google Scholar] [CrossRef] [PubMed]
- Gracey, A.Y.; Fraser, E.J.; Li, W.; Fang, Y.; Taylor, R.R.; Rogers, J.; Brass, A.; Cossins, A.R. Coping with cold: An integrative, multitissue analysis of the transcriptome of a poikilothermic vertebrate. Proc. Nation. Acad. Sci. 2004, 101, 16970–16975. [Google Scholar] [CrossRef] [PubMed]
- Baze, M.M.; Schlauch, K.; Hayes, J.P. Gene expression of the liver in response to chronic hypoxia. Physiol. Genom. 2010, 41, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Gao, G.; Palti, Y.; Cleveland, B.M.; Weber, G.M.; Rexroad III, C.E. RNA-seq analysis of early hepatic response to handling and confinement stress in rainbow trout. PLoS ONE 2014, 9, e88492. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Xu, G.; Nie, Z.; Xu, P.; Gu, R. Transcriptome analysis gene expression in the liver of Coilia nasus during the stress response. BMC Genom. 2014, 15, 558. [Google Scholar] [CrossRef]
- Coble, D.J.; Fleming, D.; Persia, M.E.; Ashwell, C.M.; Rothschild, M.F.; Schmidt, C.J.; Lamont, S.J. RNA-seq analysis of broiler liver transcriptome reveals novel responses to high ambient temperature. BMC Genom. 2014, 15, 1084. [Google Scholar] [CrossRef]
- Singh, D.; Swarup, V.; Le, H.; Kumar, V. Transcriptional signatures in liver reveal metabolic adaptations to seasons in migratory blackheaded buntings. Front. Physiol. 2018, 9, 1568. [Google Scholar] [CrossRef]
- Trefts, E.; Williams, A.S.; Wasserman, D.H. Exercise and the regulation of hepatic metabolism. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2015; Volume 135, pp. 203–225. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genom. Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Trapnell, C. Cufflinks: Transcriptome assembly and differential expression analysis for RNA-Seq. 2014. [Google Scholar]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genom. Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Roberts, A.; Trapnell, C.; Donaghey, J.; Rinn, J.L.; Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genom. Biol. 2011, 12, R22. [Google Scholar] [CrossRef]
- Team, R.C. R development Core team. R: A Language and Environment for Statistical Computing; R Foundation forStatistical Computing: Vienna, Austria, 2017; Volume 55, pp. 275–286. [Google Scholar]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-Seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef]
- Takada, I.; Kobayashi, M. Structural features and transcriptional activity of chicken PPARs (, and). PPAR Res. 2013, 2013, 186312. [Google Scholar] [CrossRef] [PubMed]
- Willson, N.-L.; Forder, R.E.; Tearle, R.; Williams, J.L.; Hughes, R.J.; Nattrass, G.S.; Hynd, P.I. Transcriptional analysis of liver from chickens with fast (meat bird), moderate (F1 layer x meat bird cross) and low (layer bird) growth potential. BMC Genom. 2018, 19, 309. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kong, L.; Deng, M.; Lian, Z.; Han, Y.; Sun, B.; Guo, Y.; Liu, G.; Liu, D. Heat Stress-Responsive Transcriptome Analysis in the Liver Tissue of Hu Sheep. Genes 2019, 10, 395. [Google Scholar] [CrossRef]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef]
- Jastrebski, S.F.; Lamont, S.J.; Schmidt, C.J. Chicken hepatic response to chronic heat stress using integrated transcriptome and metabolome analysis. PLoS ONE 2017, 12, e0181900. [Google Scholar] [CrossRef]
- Guo, M.; Li, C.; Lei, Y.; Xu, S.; Zhao, D.; Lu, X.-Y. Role of the adipose PPARγ-adiponectin axis in susceptibility to stress and depression/anxiety-related behaviors. Mol. Psych. 2017, 22, 1056. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xian, X.; Huang, W.; Chen, L.; Wu, L.; Zhu, Y.; Fan, J.; Ross, C.; Hayden, M.R.; Liu, G. Expression of LPL in endothelial-intact artery results in lipid deposition and vascular cell adhesion molecule-1 upregulation in both LPL and ApoE-deficient mice. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 197–203. [Google Scholar] [CrossRef]
- Foretz, M.; Ancellin, N.; Andreelli, F.; Saintillan, Y.; Grondin, P.; Kahn, A.; Thorens, B.; Vaulont, S.; Viollet, B. Short-term overexpression of a constitutively active form of AMP-activated protein kinase in the liver leads to mild hypoglycemia and fatty liver. Diabetes 2005, 54, 1331–1339. [Google Scholar] [CrossRef]
- Huang, P.; Zhang, Y.; Xiao, K.; Jiang, F.; Wang, H.; Tang, D.; Liu, D.; Liu, B.; Liu, Y.; He, X. The chicken gut metagenome and the modulatory effects of plant-derived benzylisoquinoline alkaloids. Microbiome 2018, 6, 211. [Google Scholar] [CrossRef]
- Führing, J.; Damerow, S.; Fedorov, R.; Schneider, J.; Münster-Kühnel, A.-K.; Gerardy-Schahn, R. Octamerization is essential for enzymatic function of human UDP-glucose pyrophosphorylase. Glycobiology 2012, 23, 426–437. [Google Scholar] [CrossRef]
- Owen, O.E.; Kalhan, S.C.; Hanson, R.W. The key role of anaplerosis and cataplerosis for citric acid cycle function. J. Biol. Chem. 2002, 277, 30409–30412. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, H.; Iskender, A.U.; Srikanth, K.; Kim, H.; Zhunushov, A.T.; Chooq, H.; Jang, G.W.; Lim, Y.; Song, K.D.; Park, J.E. Transcriptome of Chicken Liver Tissues Reveals the Candidate Genes and Pathways Responsible for Adaptation into Two Different Climatic Conditions. Animals 2019, 9, 1076. https://doi.org/10.3390/ani9121076
Kumar H, Iskender AU, Srikanth K, Kim H, Zhunushov AT, Chooq H, Jang GW, Lim Y, Song KD, Park JE. Transcriptome of Chicken Liver Tissues Reveals the Candidate Genes and Pathways Responsible for Adaptation into Two Different Climatic Conditions. Animals. 2019; 9(12):1076. https://doi.org/10.3390/ani9121076
Chicago/Turabian StyleKumar, Himansu, Asankadyr U. Iskender, Krishnamoorthy Srikanth, Hana Kim, Asankadyr T. Zhunushov, Hyojun Chooq, Gul Won Jang, Youngjo Lim, Ki Duk Song, and Jong Eun Park. 2019. "Transcriptome of Chicken Liver Tissues Reveals the Candidate Genes and Pathways Responsible for Adaptation into Two Different Climatic Conditions" Animals 9, no. 12: 1076. https://doi.org/10.3390/ani9121076
APA StyleKumar, H., Iskender, A. U., Srikanth, K., Kim, H., Zhunushov, A. T., Chooq, H., Jang, G. W., Lim, Y., Song, K. D., & Park, J. E. (2019). Transcriptome of Chicken Liver Tissues Reveals the Candidate Genes and Pathways Responsible for Adaptation into Two Different Climatic Conditions. Animals, 9(12), 1076. https://doi.org/10.3390/ani9121076




