Unraveling the Adipose Tissue Proteome of Transition Cows through Severe Negative Energy Balance
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
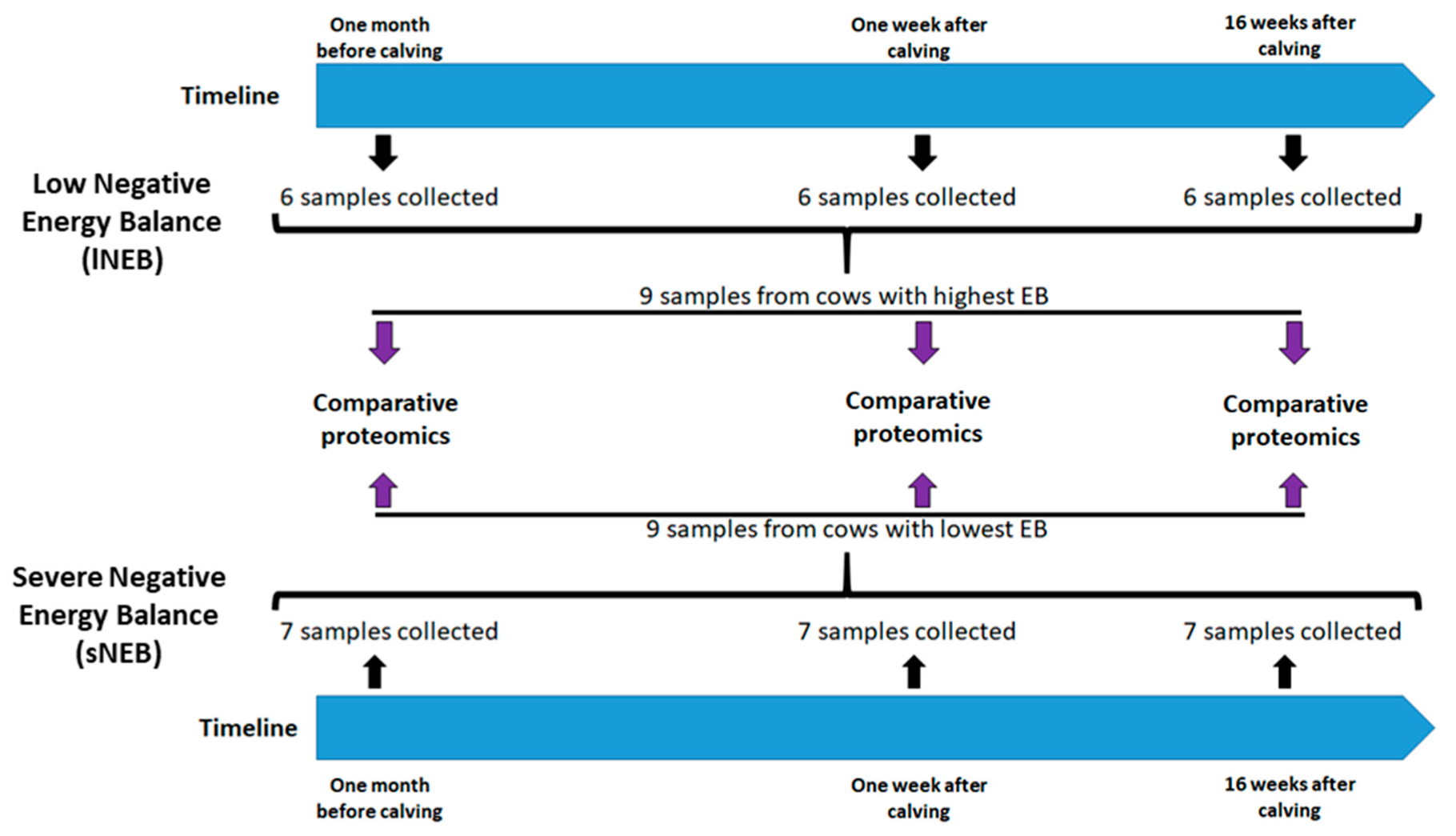
2.1. Animals and Experimental Design
2.2. Body Weight, Milk Yield, Feeding, and Energy Balance
2.3. Subcutaneous Adipose Tissue Thickness
2.4. Biopsies of Adipose Tissue
2.5. Plasma Non-Esterified Fatty Acids (NEFA) Assays
2.6. Proteomics
2.7. Protein Samples Preparation for 2DE
2.8. 2D SDS-PAGE
2.9. MALDI-TOF MS Analysis
2.10. Statistical Analyses
2.11. Bioinformatics Analysis
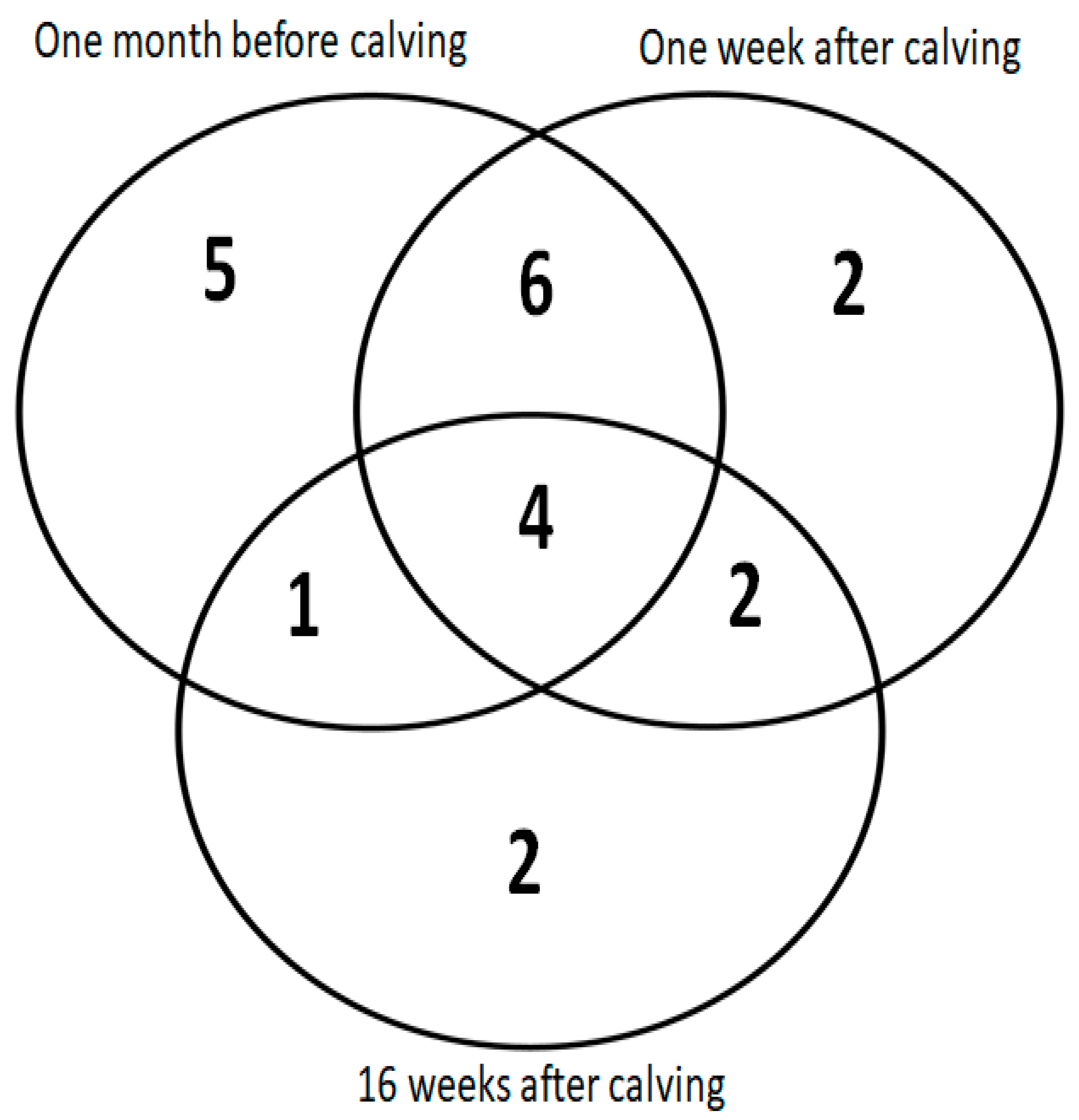
3. Results
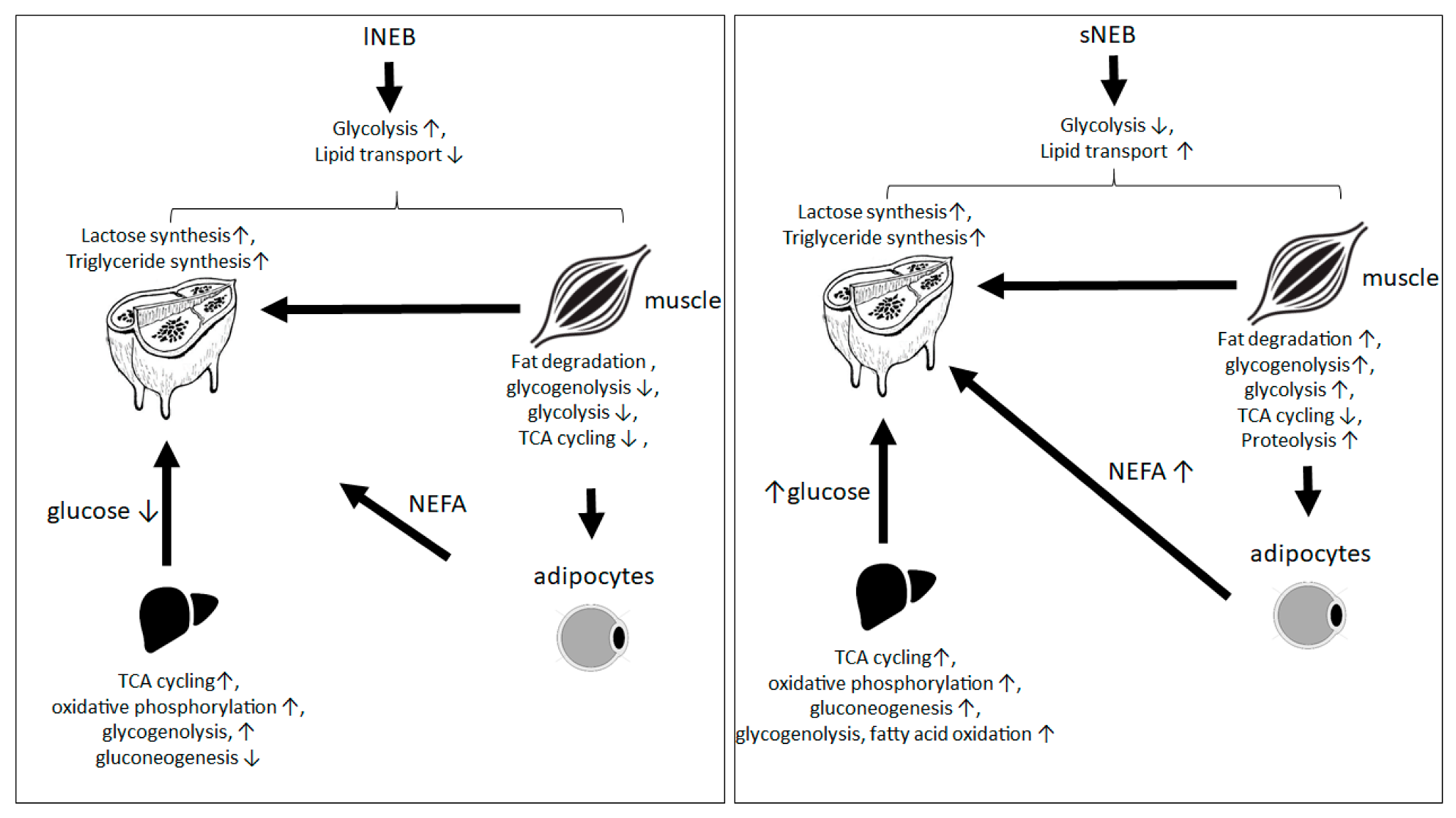
4. Discussion
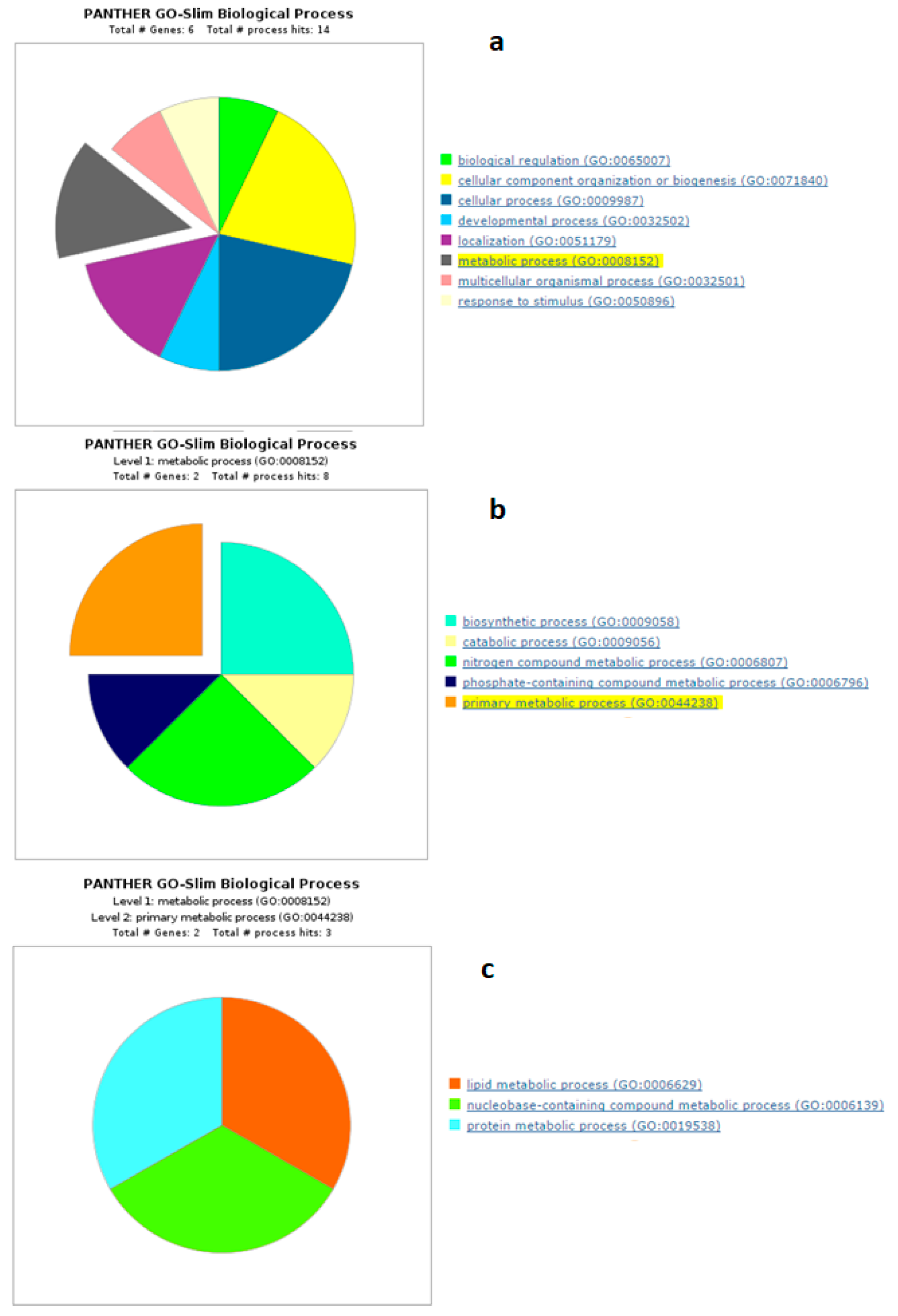
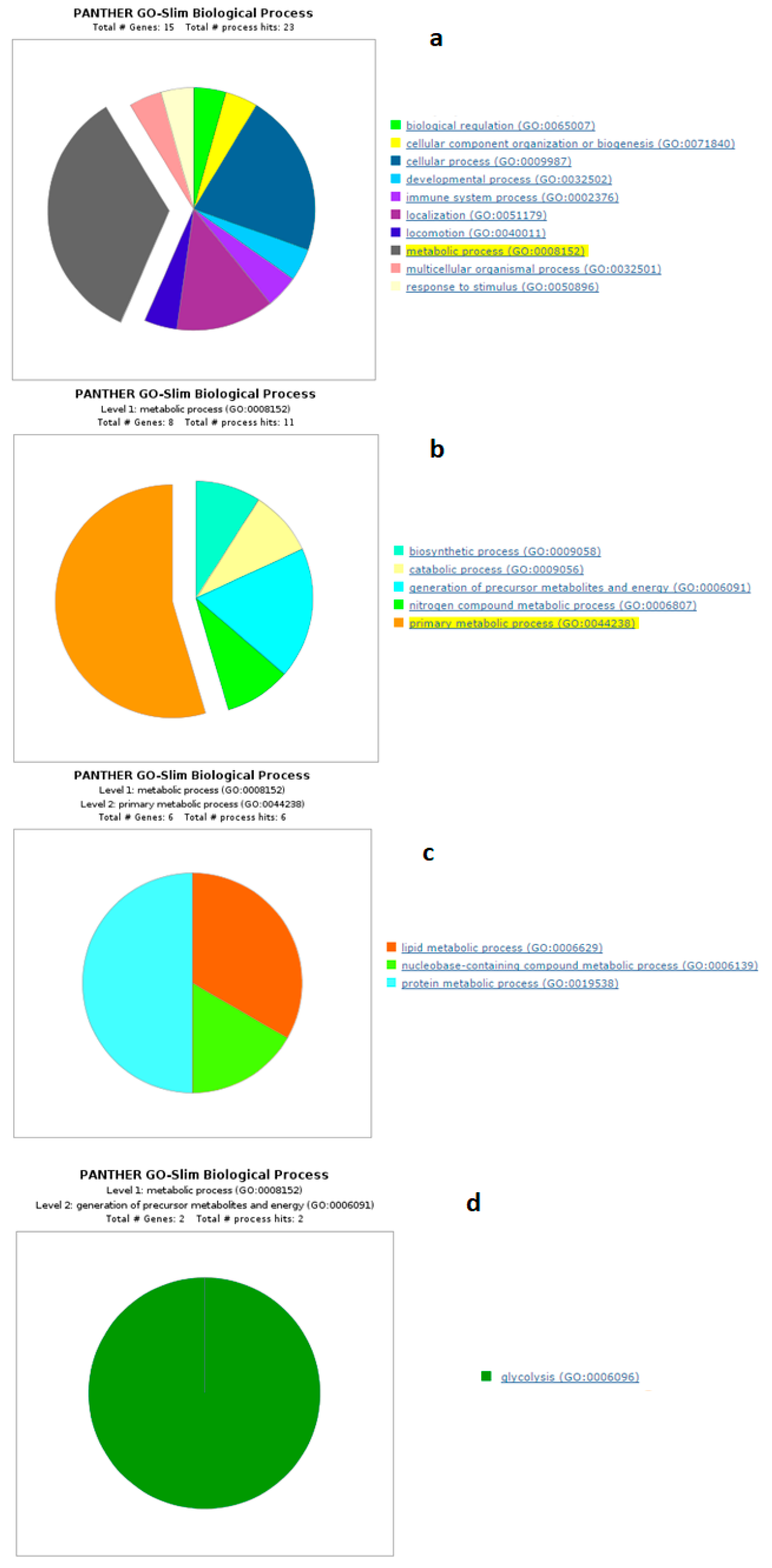
4.1. Biological Processes of Highly Abundant Proteins
4.2. Biological Processes of Less Abundant Proteins
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schröder, U.J.; Staufenbiel, R. Invited review: Methods to determine body fat reserves in the dairy cow with special regard to ultrasonographic measurement of backfat thickness. J. Dairy Sci. 2006, 89, 1–14. [Google Scholar] [CrossRef]
- Gröhn, Y.; Rajala-Schultz, P.; Allore, H.; DeLorenzo, M.; Hertla, J.; Galliganc, D. Optimizing replacement of dairy cows: Modeling the effects of diseases. Prev. Vet. Med. 2003, 61, 27–43. [Google Scholar] [CrossRef]
- Patton, J.; Kenny, D.; McNamara, S.; Mee, J.; O’Mara, F.; Diskin, M.; Murphy, J. Relationships among milk production, energy balance, plasma analytes, and reproduction in Holstein-Friesian cows. J. Dairy Sci. 2007, 90, 649–658. [Google Scholar] [CrossRef]
- Veerkamp, R.; Oldenbroek, J.; Van Der Gaast, H.; Van Der Werf, J. Genetic correlation between days until start of luteal activity and milk yield, energy balance, and live weights. J. Dairy Sci. 2000, 83, 577–583. [Google Scholar] [CrossRef]
- Grummer, R.R.; Mashek, D.G.; Hayirli, A. Dry matter intake and energy balance in the transition period. Vet. Clin. Food Anim. Pract. 2004, 20, 447–470. [Google Scholar] [CrossRef]
- McNamara, J. Regulation of Adipose Tissue Metabolism in Support of Lactation. J. Dairy Sci. 1991, 74, 706–719. [Google Scholar] [CrossRef]
- Mohamed-Ali, V.; Pinkney, J.; Coppack, S. Adipose tissue as an endocrine and paracrine organ. Int. J. Obes. Relat. Metab. Disord. 1998, 22, 1145–1158. [Google Scholar] [CrossRef]
- Trayhurn, P.; Beattie, J.H. Physiological role of adipose tissue: White adipose tissue as an endocrine and secretory organ. Proc. Nutr. Soc. 2001, 60, 329–339. [Google Scholar] [CrossRef]
- Guerre-Millo, M. Adipose tissue hormones. J. Endocrinol. Investig. 2002, 25, 855–861. [Google Scholar] [CrossRef]
- Zachut, M. Defining the adipose tissue proteome of dairy cows to reveal biomarkers related to peripartum insulin resistance and metabolic status. J. Proteome Res. 2015, 14, 2863–2871. [Google Scholar] [CrossRef]
- Zachut, M.; Kra, G.; Livshitz, L.; Portnick, Y.; Yakoby, S.; Friedlander, G.; Levin, Y. Proteome dataset of subcutaneous adipose tissue obtained from late pregnant dairy cows during summer heat stress and winter seasons. Data Brief 2017, 12, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Coyral-Castel, S.; Faverdin, P.; Rame, C.; Freret, S.; Guillaume, D.; Fritz, S.; Dupont, J. Significant differences in fertility between dairy cows selected for one QTL located on bovine chromosome 3 are not attributable to energy balance, although eating behaviour is affected. Animal 2013, 7, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Wessel, D.; Flügge, U. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984, 138, 141–143. [Google Scholar] [CrossRef]
- Piras, C.; Soggiu, A.; Bonizzi, L.; Greco, V.; Ricchi, M.; Arrigoni, N.; Bassols, A.; Urbani, A.; Roncada, P. Identification of immunoreactive proteins of Mycobacterium avium subsp. paratuberculosis. Proteomics 2015, 15, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Piras, C.; Guo, Y.; Soggiu, A.; Chanrot, M.; Greco, V.; Urbani, A.; Charpigny, G.; Bonizzi, L.; Roncada, P.; Humblotb, P. Changes in protein expression profiles in bovine endometrial epithelial cells exposed to E. coli LPS challenge. Mol. Biosyst. 2017, 13, 392–405. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014, 43, 447–452. [Google Scholar] [CrossRef]
- Leonardini, A.; Laviola, L.; Perrini, S.; Natalicchio, A.; Giorgino, F. Cross-Talk between PPAR and Insulin Signaling and Modulation of Insulin Sensitivity. PPAR Res. 2010. [Google Scholar] [CrossRef]
- Ruan, X.; Li, Z.; Zhang, Y.; Yang, L.; Pan, Y.; Wang, Z.; Feng, G.-S.; Chen, Y. Apolipoprotein A-I possesses an anti-obesity effect associated with increase of energy expenditure and up-regulation of UCP1 in brown fat. J. Cell. Mol. Med. 2011, 15, 763–772. [Google Scholar] [CrossRef]
- Folnožić, I.; Turk, R.; Duričić, D.; Vince, S.; Pleadin, J.; Flegar-Meštrić, Z.; Valpotić, H.; Dobranić, T.; Gračner, D.; Samardžija, M. Influence of Body Condition on Serum Metabolic Indicators of Lipid Mobilization and Oxidative Stress in Dairy Cows During the Transition Period. Reprod. Domest. Anim. 2015, 50, 910–917. [Google Scholar] [CrossRef]
- Karavia, E.A.; Papachristou, D.J.; Liopeta, K.; Triantaphyllidou, I.-E.; Dimitrakopoulos, O.; Kypreos, K.E. Apolipoprotein AI modulates processes associated with diet-induced nonalcoholic fatty liver disease in mice. Mol. Med. 2012, 18, 901–912. [Google Scholar] [CrossRef] [PubMed]
- Turk, R.; Juretić, D.; Gereš, D.; Svetina, A.; Turk, N.; Flegar-Meštrić, Z. Influence of oxidative stress and metabolic adaptation on PON1 activity and MDA level in transition dairy cows. Anim. Reprod. Sci. 2008, 108, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Turk, R.; Podpečan, O.; Mrkun, J.; Kosec, M.; Flegar-Meštrić, Z.; Perkov, S.; Starič, J.; Robić, M.; Belić, M.; Zrimšek, P. Lipid mobilisation and oxidative stress as metabolic adaptation processes in dairy heifers during transition period. Anim. Reprod. Sci. 2013, 141, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Hertzel, A.V.; Bernlohr, D.A. The mammalian fatty acid-binding protein multigene family: Molecular and genetic insights into function. Trends Endocrinol. Metab. 2000, 11, 175–180. [Google Scholar] [CrossRef]
- Furuhashi, M.; Saitoh, S.; Shimamoto, K.; Miura, T. Fatty acid-binding protein 4 (FABP4): Pathophysiological insights and potent clinical biomarker of metabolic and cardiovascular diseases. Clin. Med. Insights. Cardiol. 2014. [Google Scholar] [CrossRef]
- Furuhashi, M.; Hotamisligil, G.S. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008, 7, 489–503. [Google Scholar] [CrossRef]
- Veerkamp, J.; Van Moerkerk, H. Cellular Fatty Acid-Binding Proteins II; Springer: New York, NY, USA, 1993; pp. 101–106. [Google Scholar]
- De Vries, M.; Veerkamp, R. Energy balance of dairy cattle in relation to milk production variables and fertility. J. Dairy Sci. 2000, 83, 62–69. [Google Scholar] [CrossRef]
- Zhao, Y.M.; Basu, U.; Dodson, M.V.; Basarb, J.A. Proteome differences associated with fat accumulation in bovine subcutaneous adipose tissues. Proteome Sci. 2010, 8, 14. [Google Scholar] [CrossRef]
- Salameh, A.; Daquinag, A.C.; Staquicini, D.I.; An, Z.; Hajjar, K.A.; Pasqualini, R.; Arap, W.; Kolonin, M.G. Prohibitin/annexin 2 interaction regulates fatty acid transport in adipose tissue. JCI Insight 2016, 1, e86351. [Google Scholar] [CrossRef]
- Gerke, V.; Creutz, C.E.; Moss, S.E. Annexins: Linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 2005, 6, 449–461. [Google Scholar] [CrossRef]
- Kuhla, B.; Metges, C. Energy and Protein Metabolism and Nutrition in Sustainable Animal Production; Wageningen Academic Publishers: Wageningen, The Netherlands, 2013; pp. 213–221. [Google Scholar]
- Xu, W.; Vervoort, J.; Saccenti, E.; van Hoeij, R.; Kemp, B.; van Knegsel, A. Milk Metabolomics Data Reveal the Energy Balance of Individual Dairy Cows in Early Lactation. Sci. Rep. 2018, 8, 15828. [Google Scholar] [CrossRef] [PubMed]


| Group | lNEB | sNEB | p-Values | ||||
|---|---|---|---|---|---|---|---|
| Lsmeans | SEM | Lsmeans | SEM | NEB | Week | NEB × Week | |
| Live body weight, kg/day | 560.56 | 0.66 | 540.26 | 0.78 | 0.649 | <0.0001 | 0.822 |
| Variation of empty body weight, kg/day | −12.19 | 0.10 | −18.66 | 0.11 | 0.041 | <0.0001 | 0.774 |
| Milk yield, kg/day | 22.92 | 0.11 | 24.29 | 0.07 | 0.364 | <0.0001 | 0.004 |
| Dry matter intake, kg/day | 16.10 | 0.07 | 12.74 | 0.12 | 0.002 | <0.0001 | 0.004 |
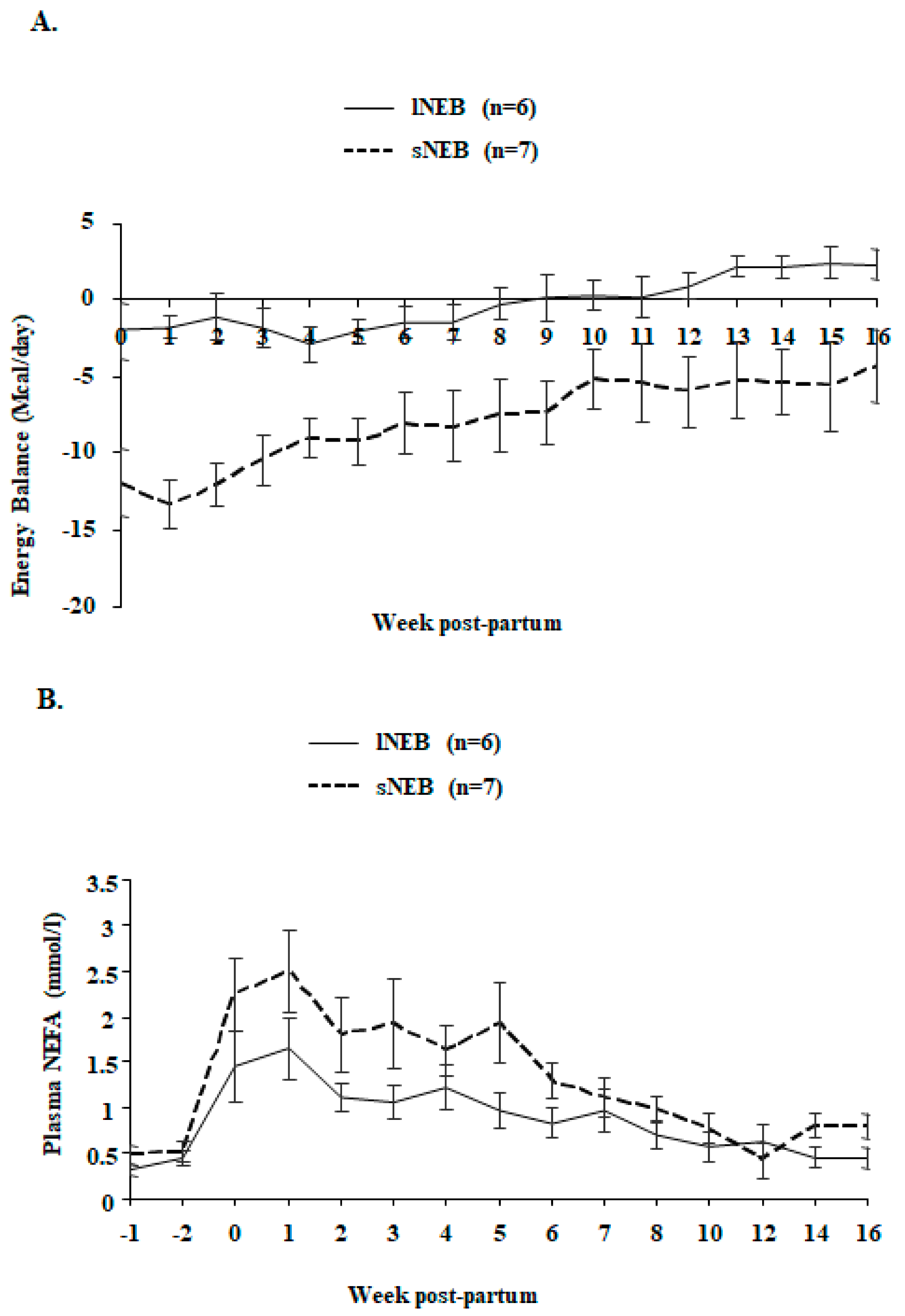
| Energy balance, Mcal/day | −0.26 | 0.08 | −7.92 | 0.11 | <0.0001 | <0.0001 | 0.189 |
| Variation of backfat thickness (%/week 4 before calving) | −26.75 | 1.04 | −45.26 | 0.85 | <0.001 | <0.0001 | 0.494 |
| Non Esterified Fatty Acids, mmol/L | 0.86 | 0.02 | 1.28 | 0.04 | 0.042 | <0.0001 | 0.485 |
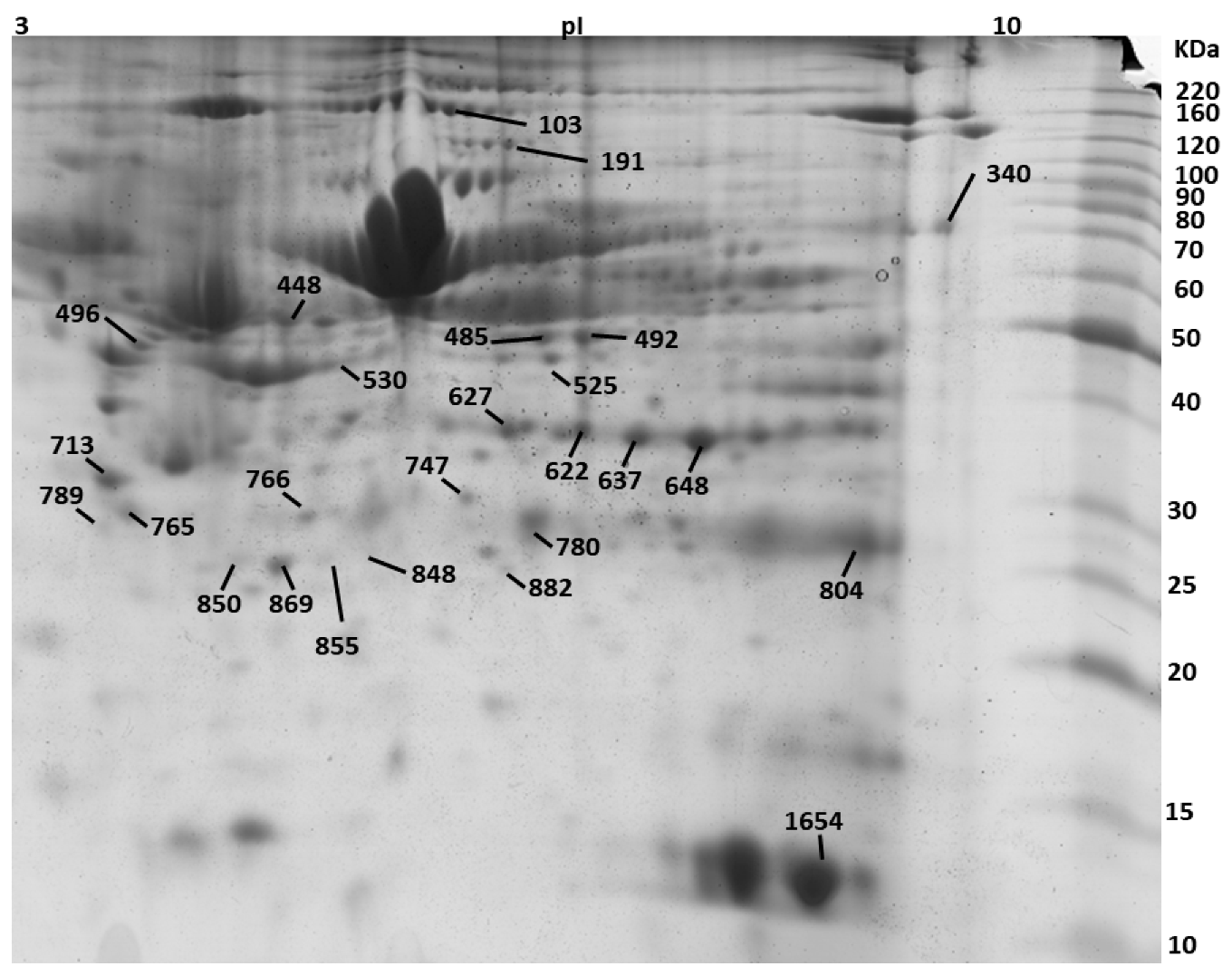
| Spot No. | Protein Description a | Accession No. b | Theoretical Mr(kDa)/pI | PMF | Tandem Mass Spectrometry | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mascot Score c | % Seq. Coverage d | Matched Peptides e | Peptide Sequence f | Mascot Score g | m/zh | ||||
| 103 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 OS = Bos taurus | E1B7X9 | 118/5.5 | 74 | 14 | Nov-81 | 801-EMSQLMLK-808 | 86 | 78 |
| 763-SINNMEKNTEMCNVMMQLR-781 | 979.49 | 2302.007 | |||||||
| 191 | Phospholipase D2 OS = Bos taurus GN = PLD2 PE = 2 SV = 1 | Q0V8L6 | 106.6/7.8 | 72 | 12 | 14/41 | 558-HFIQRWNFTK-567 | 76 | 1376.73 |
| 419-ALMLLHPNIKVMR-431 | 82 | 1551.96 | |||||||
| 340 | 3-ketoacyl-CoA thiolase, mitochondrial OS = Bos taurus | Q3T0R7 | 42.6/8.06 | 64 | 29 | 26-Oct | 333-SLNLDPSK-340 | 96 | 873.43 |
| 77-VGIPKETPAITINR-90 | 98 | 1508.8 | |||||||
| 448 | Endonuclease/exonuclease/phosphatase family domain-containing protein 1 OS = Bos taurus | Q3MHJ7 | 63.2/8.6 | 71 | 24 | Oct-61 | 154-GLTEKMAVSIVDYR-167 | 102 | 1597.82 |
| 40-LNINTATEEELMTLPGVTR-58 | 86 | 2118.81 | |||||||
| 485 | WD repeat and FYVE domain-containing protein 1 OS = Bos taurus GN = WDFY1 PE = 2 SV = 1 | Q2KIY3 | 47.29/6.9 | 76 | 35 | 24-Sep | 109-TYPAHQNR-116 | 92 | 986.5 |
| 104-MNFIKTYPAHQNR-116 | 106 | 1619.7 | |||||||
| 492 | Alpha-enolase OS = Bos taurus GN = ENO1 PE = 1 SV = 4 | Q9XSJ4 | 47.64/6.37 | 132 | 35 | 13/39 | 240-VVIGMDVAASEFYR-253 | 126 | 1556.8 |
| 163-LAMQEFMILPVGAENFR-179 | 104 | 1965.98 | |||||||
| 530 | Actin, cytoplasmic 1 OS = Bos taurus GN = ACTB PE = 1 SV = 1 | P60712 | 42.05/5.29 | 94 | 28 | Oct-47 | 197-GYSFTTTAER-206 | 86 | 1132.96 |
| 360-QEYDESGPSIVHR-372 | 74 | 1516.66 | |||||||
| 622 | Annexin A1 OS = Bos taurus GN = ANXA1 PE = 2 SV = 2 | P46193 | 63.2/8.6 | 98 | 46 | 15/74 | 154-GLTEKMAVSIVDYR-167 | 102 | 1597.82 |
| 40-LNINTATEEELMTLPGVTR-58 | 86 | 2118.81 | |||||||
| 627 | Annexin A1 OS = Bos taurus GN = ANXA1 PE = 2 SV = 2 | P46193 | 63.2/8.6 | 66 | 30 | 13/74 | 119-DAEELRAAMK-129 | 76 | 1071,55 |
| 154-GLTEKMAVSIVDYR-167 | 78 | 1597.82 | |||||||
| 637 | Annexin A2 OS = Bos taurus GN = ANXA2 PE = 1 SV = 2 | P04272 | 38.8/6.2 | 106 | 49 | Nov-81 | 48-TKGVDEVTIVNILTNR-63 | 122 | 1771.86 |
| 314-SLYYYIQQDTKGDYQK-329 | 102 | 2012.91 | |||||||
| 648 | Actin-related protein 10 OS = Bos taurus GN = ACTR10 PE = 2 SV = 1 | Q3ZBD2 | 46.84/7.06 | 86 | 404-NQPPLMKR-411 | 82 | 999.61 | ||
| 36 | 26/109 | 279-SVATLILDSLMQCPIDTR-296 | 70 | 2012.91 | |||||
| 713 | Coiled-coil domain-containing protein 71 OS = Bos taurus GN = CCDC71 PE = 2 SV = 1 | Q2HJ91 | 48.08/5.13 | 76 | 31 | Nov-87 | 220-LGNAQLKAPR-229 | 78 | 1067.68 |
| 198-AQSLQLSLGDSPLKVR-213 | 96 | 1712.08 | |||||||
| 747 | Purine nucleoside phosphorylase OS = Bos taurus GN = PNP PE = 1 SV = 3 | P55859 | 32.24/5.92 | 90 | 28 | 20-Aug | 68-LVFGILNGR-76 | 92 | 988.59 |
| 40-DHINLPGFSGENPLR-58 | 86 | 2118.81 | |||||||
| 765 | Elongation factor 1-delta OS = Bos taurus | A5D989 | 30.97/5.42 | 82 | 36 | Jul-57 | 1-MATNFLVHEK-10 | 82 | 1189.691 |
| 259-SHQVEEHVQSVDIAAFNKI-277 | 80 | 2151.12 | |||||||
| 766 | Glyceraldehyde-3-phosphate dehydrogenase OS = Bos taurus GN = GAPDH PE = 1 SV = 4 | P10096 | 36.07/8.5 | 66 | 32 | May-57 | 131-MGVNHEKYN-140 | 66 | 929.27 |
| 171-GLMTTVHAIT ATQKTVDGPS-190 | 88 | 1615.88 | |||||||
| 780 | E3 ubiquitin-protein ligase RNF114 OS = Bos taurus | Q4U5R4 | 26.65/6.5 | 96 | 36 | Oct-64 | 141-YTFPCPYCPEK-151 | 76 | 1463.61 |
| 11-DGGAQLAGPAAEADPLGR-28 | 98 | 1665.74 | |||||||
| 789 | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 OS = Bos taurus GN = DDAH1 PE = 1 SV = 3 | P56965 | 31.6/5.7 | 84 | 58 | 12-Jul | 150-GAEILADTFK-159 | 78 | 1063.55 |
| 231-GHVLLHRTPEEYPESAK-247 | 84 | 1963.08 | |||||||
| 848 | Succinate dehydrogenase cytochrome b560 subunit, mitochondrial OS = Bos taurus GN = SDHC PE = 1 SV = 2 | P35720 | 18.7/9.8 | 76 | 44 | 14-Jun | 8-HVGRHCLR-15 | 67 | 1034.52 |
| 18-NLGSNRPLSPHITIYR-36 | 97 | 1836.99 | |||||||
| 850 | General transcription factor IIF subunit 2 OS = Bos taurus GN = GTF2F2 PE = 2 SV = 1 | Q2T9L9 | 28.49/9.24 | 83 | 29 | 30-Aug | 26-YLSQQWAK-33 | 86 | 1023.57 |
| 208-QPVSYLKDILK-218 | 92 | 1303.8 | |||||||
| 855 | Fatty acid-binding protein, adipocyte OS = Bos taurus GN = FABP4 PE = 1 SV = 4 | P48035 | 15.35/7.57 | 96 | 39 | Dec-60 | 11-WRLVESK-17 | 76 | 917.5 |
| 111-LEDGKLVVVCVMNNVTCTR-129 | 98 | 2225.11 | |||||||
| 869 | Apolipoprotein A-I OS = Bos taurus GN = APOA1 PE = 1 SV = 3 | P15497 | 30.25/5.71 | 118 | 39 | 14/54 | 142-VAPLGEEFR-150 | 102 | 1017.53 |
| 131-WHEEVEIYR-159 | 98 | 1260.57 | |||||||
| 882 | Heat shock protein beta-1 OS = Bos taurus GN = HSPB1 PE = 2 SV = 1 | Q3T149 | 22.43/5.98 | 90 | 36 | 23-Jul | 29-LFDQAFGLPR-38 | 96 | 1163.97 |
| 13-GPSWDPFRDWYPAHSR-28 | 92 | 1973.91 | |||||||
| Uniprot Name/Accession Number | String Name | Same Spots Coding Number | Protein Name | One Month before Calving: Trend/Fold Change/p-Value | One Week after Calving: Trend/Fold Change/p-Value | 16 Weeks after Calving: Trend/Fold Change/p-Value |
|---|---|---|---|---|---|---|
| E1B7X9 | SMARCAD1 | 103 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1 OS = Bos taurus | ↓/1.9/0.0193 | ||
| Q0V8L6 | PLD2 | 191 | Phospholipase D2 OS = Bos taurus GN = PLD2 PE = 2 SV = 1 | ↓/2.1/0.004 | ↓/2.3/0.030 | |
| Q3T0R7 | ACAA2 | 340 | 3-ketoacyl-CoA thiolase, mitochondrial OS = Bos taurus | ↓/3.3/0.028 | ||
| Q3MHJ7 | EEPD1 | 448 | Endonuclease/exonuclease/phosphatase family domain-containing protein 1 OS = Bos taurus | ↑/1.6/0.0086 | ↑/1.5/0.0072 | |
| Q2KIY3 | WDFY1 | 485 | WD repeat and FYVE domain-containing protein 1 OS = Bos taurus GN = WDFY1 PE = 2 SV = 1 | ↓/1.6/0.0207 | ||
| Q9XSJ4 | ENO1 | 492 | Alpha-enolase OS = Bos taurus GN = ENO1 PE = 1 SV = 4 | ↓/1.8/0.0004 | ↓/1.4/0.050 | |
| P60712 | NA | 530 | Actin, cytoplasmic 1 OS = Bos taurus GN = ACTB PE = 1 SV = 1 | ↑/1.7/0.0297 | ↑/2/0.0080 | |
| P46193 | ANXA1 | 622 | Annexin A1 OS = Bos taurus GN = ANXA1 PE = 2 SV = 2 | ↓/2/0.00186 | ||
| P46193 | ANXA1 | 627 | Annexin A1 OS = Bos taurus GN = ANXA1 PE = 2 SV = 2 | ↓/2.9/0.0429 | ↓/1.8/0.0502 | |
| P04272 | ANXA2 | 637 | Annexin A2 OS = Bos taurus GN = ANXA2 PE = 1 SV = 2 | ↓/2.7/0.0118 | ↓/1.7/0.0085 | ↓/1.8/0.0098 |
| Q3ZBD2 | ACTR10 | 648 | Actin-related protein 10 OS = Bos taurus GN = ACTR10 PE = 2 SV = 1 | ↓/2.1/0.0050 | ↓/1.7/0.0085 | ↓/1.4/0.038 |
| Uniprot Name/Accession Number | String Name | SameSpots Coding Number | Protein Name | One Month before Calving: Trend/Fold Change/p-Value | One Week after Calving: Trend/Fold Change/p-Value | 16 Weeks after Calving: Trend/Fold Change/p-Value |
| Q2HJ91 | CCDC71 | 713 | Coiled-coil domain-containing protein 71 OS = Bos taurus GN = CCDC71 PE = 2 SV = 1 | ↓/1.2/0.0026 | ||
| P55859 | PNP | 747 | Purine nucleoside phosphorylase OS = Bos taurus GN = PNP PE = 1 SV = 3 | ↓/1.4/0.0439 | ↓/1.3/0.0165 | |
| A5D989 | EEF1D | 765 | Elongation factor 1-delta | ↓/1.6/0.00141 | ||
| P10096 | G3PDH | 766 | Glyceraldehyde-3-phosphate dehydrogenase OS = Bos taurus GN = GAPDH PE = 1 SV = 4 | ↓/1.8/0.0201 | ↓/1.6/0.0260 | ↓/1.2/0.040 |
| Q4U5R4 | RNF114 | 780 | E3 ubiquitin-protein ligase RNF114 OS = Bos taurus | ↓/1.3/0.024 | ||
| P56965 | DDAH1 | 789 | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 OS = Bos taurus GN = DDAH1 PE = 1 SV = 3 | ↓/2.2/ | ↓/1.8/0.0011 | |
| 8.84 × 10−5 | ||||||
| P35720 | SDHC | 848 | Succinate dehydrogenase cytochrome b560 subunit, mitochondrial OS = Bos taurus GN = SDHC PE = 1 SV = 2 | ↑/1.8/0.0059 | ||
| Q2T9L9 | GTF2F2 | 850 | General transcription factor IIF subunit 2 OS = Bos taurus GN = GTF2F2 PE = 2 SV = 1 | ↑/3/0.0104 | ||
| P48035 | NA | 855 | Fatty acid-binding protein, adipocyte OS = Bos taurus GN = FABP4 PE = 1 SV = 4 | ↑/2.2/0.0053 | ↑/2.7/0.0231 | ↑/3.3/0.0127 |
| P15497 | APOA1 | 869 | Apolipoprotein A-I OS = Bos taurus GN = APOA1 PE = 1 SV = 3 | ↑/3/0.0028 | ↑/2.6/0.0343 | |
| Q3T149 | HSPB1 | 882 | Heat shock protein beta-1 OS = Bos taurus GN = HSPB1 PE = 2 SV = 1 | ↓/2.8/0.0122 | ↓/2.7/0.005 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piras, C.; Morittu, V.M.; Spina, A.A.; Soggiu, A.; Greco, V.; Ramé, C.; Briant, E.; Mellouk, N.; Tilocca, B.; Bonizzi, L.; et al. Unraveling the Adipose Tissue Proteome of Transition Cows through Severe Negative Energy Balance. Animals 2019, 9, 1013. https://doi.org/10.3390/ani9121013
Piras C, Morittu VM, Spina AA, Soggiu A, Greco V, Ramé C, Briant E, Mellouk N, Tilocca B, Bonizzi L, et al. Unraveling the Adipose Tissue Proteome of Transition Cows through Severe Negative Energy Balance. Animals. 2019; 9(12):1013. https://doi.org/10.3390/ani9121013
Chicago/Turabian StylePiras, Cristian, Valeria Maria Morittu, Anna Antonella Spina, Alessio Soggiu, Viviana Greco, Christelle Ramé, Eric Briant, Namya Mellouk, Bruno Tilocca, Luigi Bonizzi, and et al. 2019. "Unraveling the Adipose Tissue Proteome of Transition Cows through Severe Negative Energy Balance" Animals 9, no. 12: 1013. https://doi.org/10.3390/ani9121013
APA StylePiras, C., Morittu, V. M., Spina, A. A., Soggiu, A., Greco, V., Ramé, C., Briant, E., Mellouk, N., Tilocca, B., Bonizzi, L., Roncada, P., & Dupont, J. (2019). Unraveling the Adipose Tissue Proteome of Transition Cows through Severe Negative Energy Balance. Animals, 9(12), 1013. https://doi.org/10.3390/ani9121013







