Genetic Parameter Estimation and Genomic Prediction of Duroc Boars’ Sperm Morphology Abnormalities
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statements
2.2. Phenotypes, Genotypes, and Pedigree
- PD: Proximal droplet—a droplet of cytoplasm attached to the base of the head where the midpiece emerges.
- DD: Distal droplet—a cytoplasmic droplet attached further down the midpiece/tail from the base of the head. If the droplet is located at more than 4 µm from the base of the head, the droplet is defined as distal. It may indicate an immature sperm and is considered a defect.
- BT: Bent tail—the bending rate of the tail exceeds 20°/µm.
- CT: Coiled tail—the tail bends 180° or more over its length.
- DMR: Distal midpiece reflex—the tail is wrapped around a distal cytoplasmic droplet, typically at the end of the midpiece (close to 4 µm from the end base of the sperm head), and the tail returns to the sperm head, usually becoming visible at the top of the head.
2.3. Statistical Model
2.4. Estimation of Genetic Parameters
2.5. Assessment of Genomic Predictive Ability
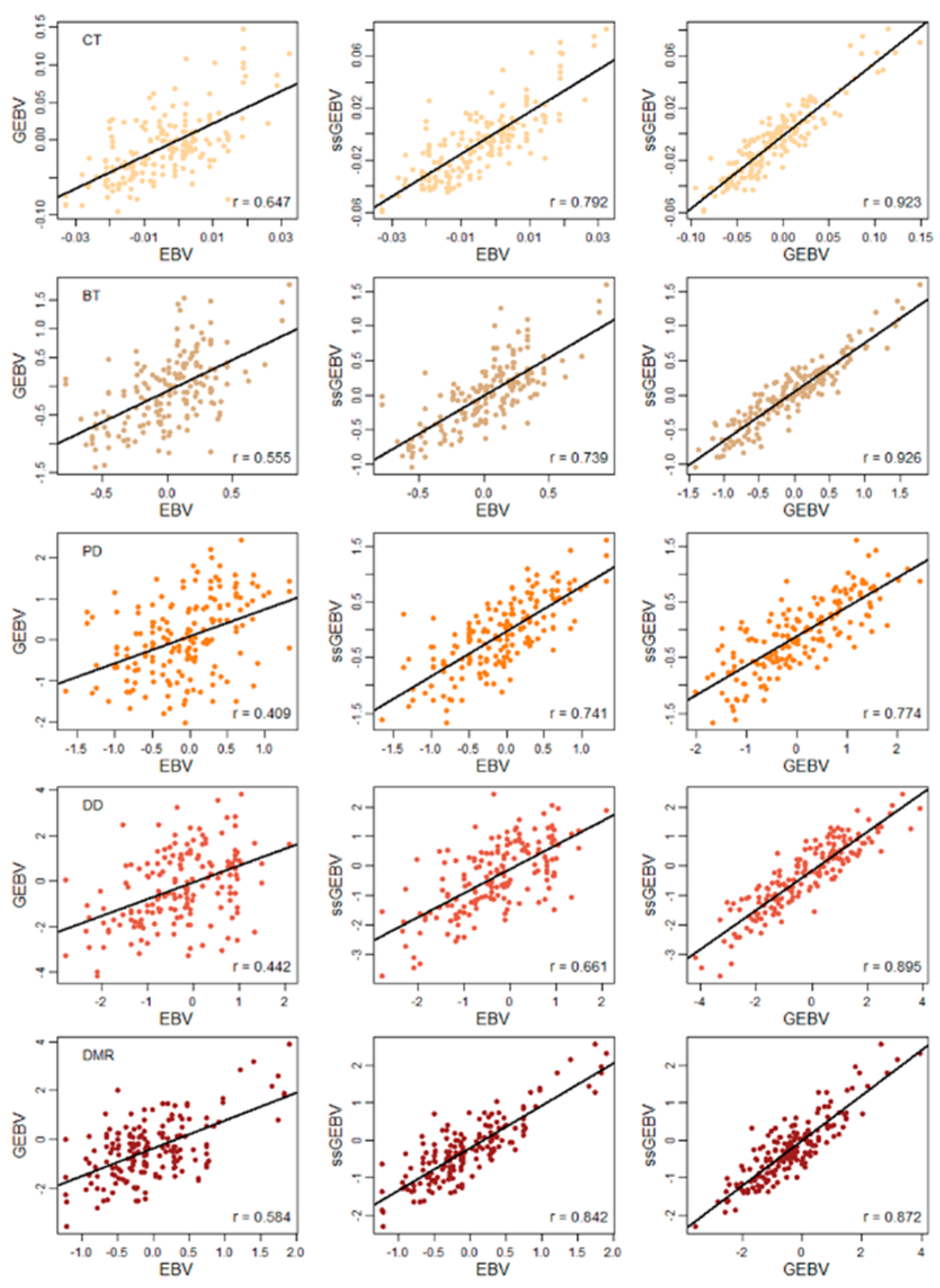
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [PubMed]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Habier, D.; Fernando, R.L.; Kizilkaya, K.; Garrick, D.J. Extension of the bayesian alphabet for genomic selection. BMC Bioinf. 2011, 12, 186. [Google Scholar] [CrossRef] [PubMed]
- Gianola, D. Priors in whole-genome regression: The Bayesian alphabet returns. Genetics 2013, 194, 573–596. [Google Scholar] [CrossRef] [PubMed]
- Legarra, A.; Aguilar, I.; Misztal, I. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 2009, 92, 4656–4663. [Google Scholar] [CrossRef] [PubMed]
- Christensen, O.F.; Lund, M.S. Genomic prediction when some animals are not genotyped. Genet. Sel. Evol. 2010, 42, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ober, U.; Erbe, M.; Zhang, H.; Gao, N.; He, J.; Li, J.; Simianer, H. Improving the accuracy of whole genome prediction for complex traits using the results of genome wide association studies. PLoS ONE 2014, 9, e93017. [Google Scholar] [CrossRef] [PubMed]
- Fangmann, A.; Sharifi, R.A.; Heinkel, J.; Danowski, K.; Schrade, H.; Erbe, M.; Simianer, H. Empirical comparison between different methods for genomic prediction of number of piglets born alive in moderate sized breeding populations. J. Anim. Sci. 2017, 95, 1434–1443. [Google Scholar] [CrossRef] [PubMed]
- Christensen, O.F.; Madsen, P.; Nielsen, B.; Ostersen, T.; Su, G. Single-step methods for genomic evaluation in pigs. Animal 2012, 6, 1565–1571. [Google Scholar] [CrossRef]
- Kang, H.; Ning, C.; Zhou, L.; Zhang, S.; Yan, Q.; Liu, J. Short communication: Single-step genomic evaluation of milk production traits using multiple-trait random regression model in Chinese Holsteins. J. Dairy Sci. 2018, 101. [Google Scholar] [CrossRef]
- Koivula, M.; Strandén, I.; Pösö, J.; Aamand, G.P.; Mäntysaari, E.A. Single-step genomic evaluation using multitrait random regression model and test-day data. J. Dairy Sci. 2015, 98, 2775–2784. [Google Scholar] [CrossRef] [PubMed]
- Teissier, M.; Larroque, H.; Granié, C.R. Weighted single—Step genomic BLUP improves accuracy of genomic breeding values for protein content in French dairy goats: A quantitative trait influenced by a major gene. Genet. Sel. Evol. 2018, 31, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, H.; Garrick, D.; Golden, B.; Dekkers, J.; Park, K.; Lee, D.; Fernando, R. Comparison of alternative approaches to single-trait genomic prediction using genotyped and non-genotyped Hanwoo beef cattle. Genet. Sel. Evol. 2017, 49, 2. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J. Heritabilities and genetic correlations for litter size and semen traits in Czech Large White and Landrace pigs. J. Anim. Sci. 2010, 9, 2893–2903. [Google Scholar] [CrossRef] [PubMed]
- Marques, D.B.D.; Lopes, M.S.; Broekhuijse, M.L.W.J.; Guimarães, S.E.F.; Knol, E.F.; Bastiaansen, J.W.M.; Silva, F.F.; Lopes, P.S. Genetic parameters for semen quality and quantity traits in five pig lines. J. Anim. Sci. 2017, 10, 4251–4259. [Google Scholar] [CrossRef]
- Sutovsky, P. New Approaches to Boar Semen Evaluation, Processing and Improvement. Reprod. Domest. Anim. 2015, 50 (Suppl. 2), 11–19. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 813, 559–575. [Google Scholar] [CrossRef]
- Henderson, C.R. Best Linear Unbiased Estimation and Prediction under a Selection Model. Biometrics 1975, 31, 423. [Google Scholar] [CrossRef]
- Jensen, J.; Mantysaari, E.A.; Madsen, P.; Thompson, R. Residual Maximum Likelihood Estimation of (Co) Variance Components in Multivariate Mixed Linear Models using Average Information. J. Indian Soc. Agric. Stat. 1997, 49, 215–236. [Google Scholar]
- Misztal, I.; Tsuruta, S.; Strabel, T.; Auvray, B.; Druet, T.; Lee, D.H. BLUPF90 and related programs (BGF90). In Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, 19–23 August 2002; pp. 21–22. [Google Scholar]
- Garrick, D.J.; Taylor, J.F.; Fernando, R.L. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet. Sel. Evol. 2009, 41, 1–8. [Google Scholar] [CrossRef]
- Hayes, B.J.; Bowman, P.J.; Chamberlain, A.C.; Verbyla, K.; Goddard, M.E. Accuracy of genomic breeding values in multi-breed dairy cattle populations. Genet. Sel. Evol. 2009, 41, 51. [Google Scholar] [CrossRef] [PubMed]
- Tier, B.B.; Meyer, K. Approximating prediction error covariances among additive genetic effects within animals in multiple-trait and random regression models. J. Anim. Breed. Genet. 2004, 121, 77–89. [Google Scholar] [CrossRef]
- Rodriguez, A.L.; Van Soom, A.; Arsenakis, I.; Maes, D. Boar management and semen handling factors affect the quality of boar extended semen. Porc. Heal. Manag. 2017, 15, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Pena, D.; Knox, R.V.; Rodriguez-Zas, S.L. Contribution of semen trait selection, artificial insemination technique, and semen dose to the profitability of pig production systems: A simulation study. Theriogenology 2016, 2, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Broekhuijse, M.; Gaustad, A.H.; Guill, A.B.; Knol, E. Efficient Boar Semen Production and Genetic Contribution: The Impact of Low-Dose Artificial Insemination on Fertility. Reprod. Domest. Anim. 2015, 50, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Zak, L.J.; Gaustad, A.H.; Bolarin, A.; Broekhuijse, M.L.W.J.; Walling, G.A.; Knol, E.F. Genetic control of complex traits, with a focus on reproduction in pigs. Mol. Reprod. Dev. 2017, 84, 1004–1011. [Google Scholar] [CrossRef] [PubMed]
- Martini, J.W.R.; Schrauf, M.F.; Garcia-Baccino, C.A.; Pimentel, E.C.G.; Munilla, S.; Rogberg-Munoz, A.; Cantet, R.J.C.; Reimer, C.; Gao, N.; Wimmer, V.; et al. The effect of the H(-1) scaling factors tau and omega on the structure of H in the single-step procedure. Genet. Sel. Evol. 2018, 50, 16. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Muir, W.M. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet. Res. 2012, 94, 73–83. [Google Scholar] [CrossRef]
- Zhang, X.; Lourenco, D.; Aguilar, I.; Legarra, A. Weighting Strategies for Single-Step Genomic BLUP: An Iterative Approach for Accurate Calculation of GEBV and GWAS. Front. Genet. 2016, 7, 1–14. [Google Scholar] [CrossRef]
- Teissier, M.; Larroque, H.; Robert-Granie, C. Accuracy of genomic evaluation with weighted single-step genomic best linear unbiased prediction for milk production traits, udder type traits, and somatic cell scores in French dairy goats. J. Dairy Sci. 2019, 102, 3142–3154. [Google Scholar] [CrossRef]
| Traits 1 | #obs. 2 | #indiv 3 | Min. | Mean | Max. | SD | CV (%) |
|---|---|---|---|---|---|---|---|
| CT (%) | 29,526 | 1304 | 0 | 0.15 | 5.3 | 0.24 | 162.22 |
| BT (%) | 29,526 | 1304 | 0 | 2.41 | 48 | 2.12 | 88.17 |
| PD (%) | 29,526 | 1304 | 0 | 4.94 | 41.7 | 3.07 | 62.12 |
| DD (%) | 29,526 | 1304 | 0 | 7.25 | 50.4 | 4.77 | 65.73 |
| DMR (%) | 28,973 | 1232 | 0.10 | 2.82 | 38.1 | 2.58 | 91.22 |
| Dataset | #. Pedigree 1 | #. Genotypes 2 | #. Phenotypes 3 | #. Pheno & Geno 4 |
|---|---|---|---|---|
| Training | 4256 | 1247 | 1012 | 657 |
| Test | 1028 | 376 | 292 | 172 |
| Traits 1 | σa 2 | σp 2 | σe 2 | h 2 (SE) | r (SE) |
|---|---|---|---|---|---|
| CT | 0.0016 | 0.0047 | 0.0494 | 0.0287 (0.0106) | 0.1133 (0.0068) |
| BT | 0.5960 | 0.6929 | 3.0432 | 0.1376 (0.0299) | 0.2975 (0.0123) |
| PD | 2.5734 | 3.2141 | 4.7412 | 0.2444 (0.0453) | 0.5497 (0.0131) |
| DD | 7.2729 | 4.2530 | 13.1157 | 0.2951 (0.0453) | 0.4677 (0.0147) |
| DMR | 2.2786 | 2.8257 | 3.4195 | 0.2673 (0.0481) | 0.5988 (0.0127) |
| Traits 1 | CT | BT | PD | DD | DMR |
|---|---|---|---|---|---|
| CT | 0.036 (0.011) | 0.955 (0.027) | 0.329 (0.158) | 0.207 (0.161) | 0.403 (0.159) |
| BT | 0.033 (0.009) | 0.134 (0.029) | 0.325 (0.134) | 0.305 (0.131) | 0.378 (0.134) |
| PD | 0.023 (0.014) | 0.394 (0.199) | 0.239 (0.045) | 0.607 (0.091) | 0.052 (0.142) |
| DD | 0.025 (0.020) | 0.631 (0.301) | 2.609 (0.639) | 0.297 (0.045) | 0.207 (0.123) |
| DMR | 0.027 (0.012) | 0.434 (0.181) | 0.124 (0.345) | 0.845 (0.550) | 0.264 (0.048) |
| Traits 1 | Cross-Validation | Forward Prediction | ||
|---|---|---|---|---|
| GBLUP | ssGBLUP | GBLUP | ssGBLUP | |
| CT | 0.212 ± 0.004 | 0.249 ± 0.004 | 0.069 | 0.085 |
| BT | 0.373 ± 0.003 | 0.439 ± 0.003 | 0.311 | 0.311 |
| PD | 0.330 ± 0.003 | 0.565 ± 0.002 | 0.250 | 0.311 |
| DD | 0.386 ± 0.003 | 0.507 ± 0.002 | 0.389 | 0.442 |
| DMR | 0.417 ± 0.003 | 0.561 ± 0.002 | 0.344 | 0.483 |
| Average | 0.344 | 0.464 | 0.273 | 0.326 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Gao, N.; Cheng, J.; El-Ashram, S.; Zhu, L.; Zhang, C.; Li, Z. Genetic Parameter Estimation and Genomic Prediction of Duroc Boars’ Sperm Morphology Abnormalities. Animals 2019, 9, 710. https://doi.org/10.3390/ani9100710
Zhao Y, Gao N, Cheng J, El-Ashram S, Zhu L, Zhang C, Li Z. Genetic Parameter Estimation and Genomic Prediction of Duroc Boars’ Sperm Morphology Abnormalities. Animals. 2019; 9(10):710. https://doi.org/10.3390/ani9100710
Chicago/Turabian StyleZhao, Yunxiang, Ning Gao, Jian Cheng, Saeed El-Ashram, Lin Zhu, Conglin Zhang, and Zhili Li. 2019. "Genetic Parameter Estimation and Genomic Prediction of Duroc Boars’ Sperm Morphology Abnormalities" Animals 9, no. 10: 710. https://doi.org/10.3390/ani9100710
APA StyleZhao, Y., Gao, N., Cheng, J., El-Ashram, S., Zhu, L., Zhang, C., & Li, Z. (2019). Genetic Parameter Estimation and Genomic Prediction of Duroc Boars’ Sperm Morphology Abnormalities. Animals, 9(10), 710. https://doi.org/10.3390/ani9100710





