Microbial Distribution and Biofilm-Forming Capacity in the Reproductive Tract of Farm Ruminants
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Sample Collection and Bacterial Isolation
2.3. DNA Extraction of Vaginal Microorganisms
2.4. Polymerase Chain Reaction (PCR)
2.5. Biofilm Formation Assay
2.6. Statistical Analysis
2.7. Limitations Section
3. Results
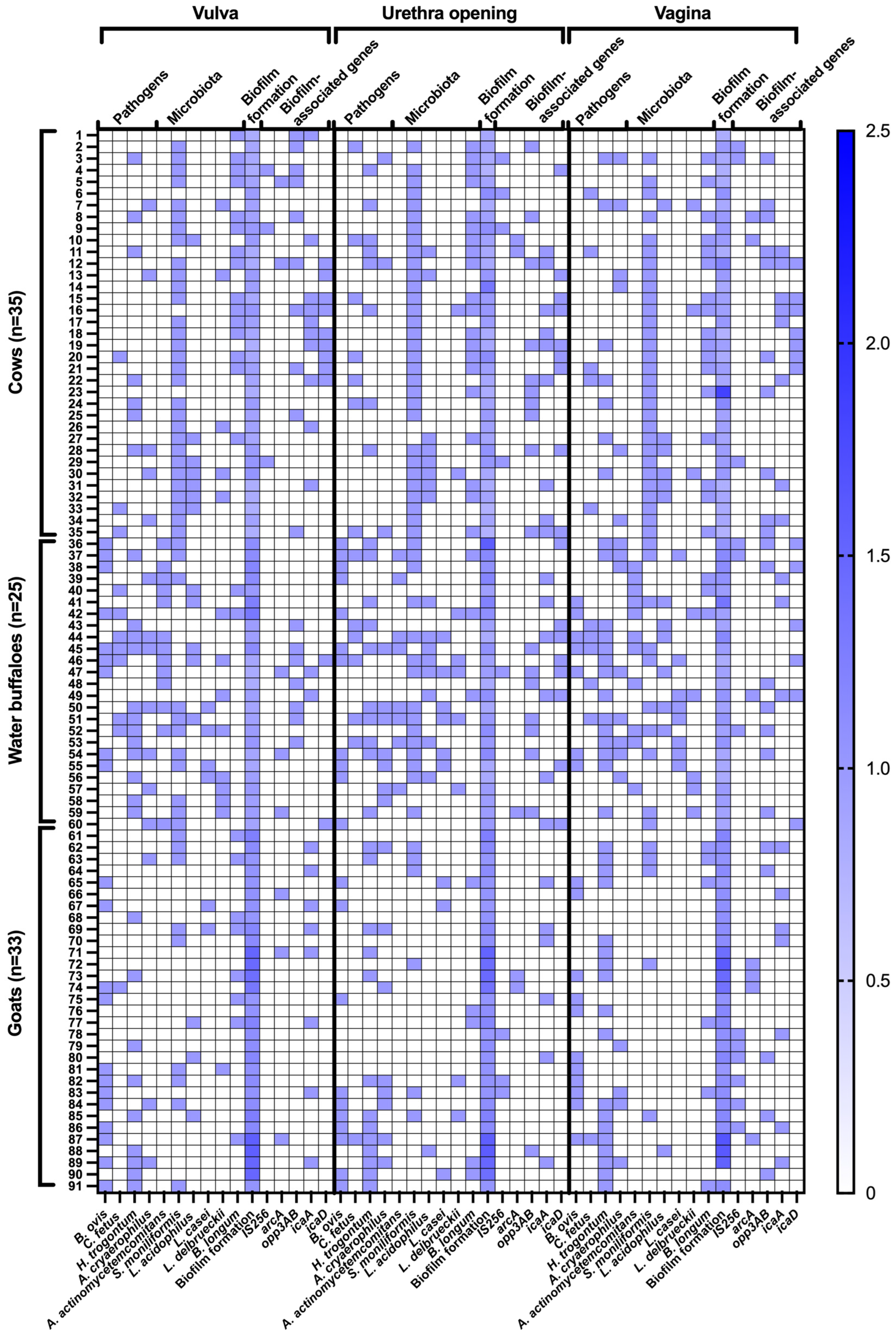
3.1. Distribution of Pathogens and Microbiota
3.2. Biofilm Formation Patterns
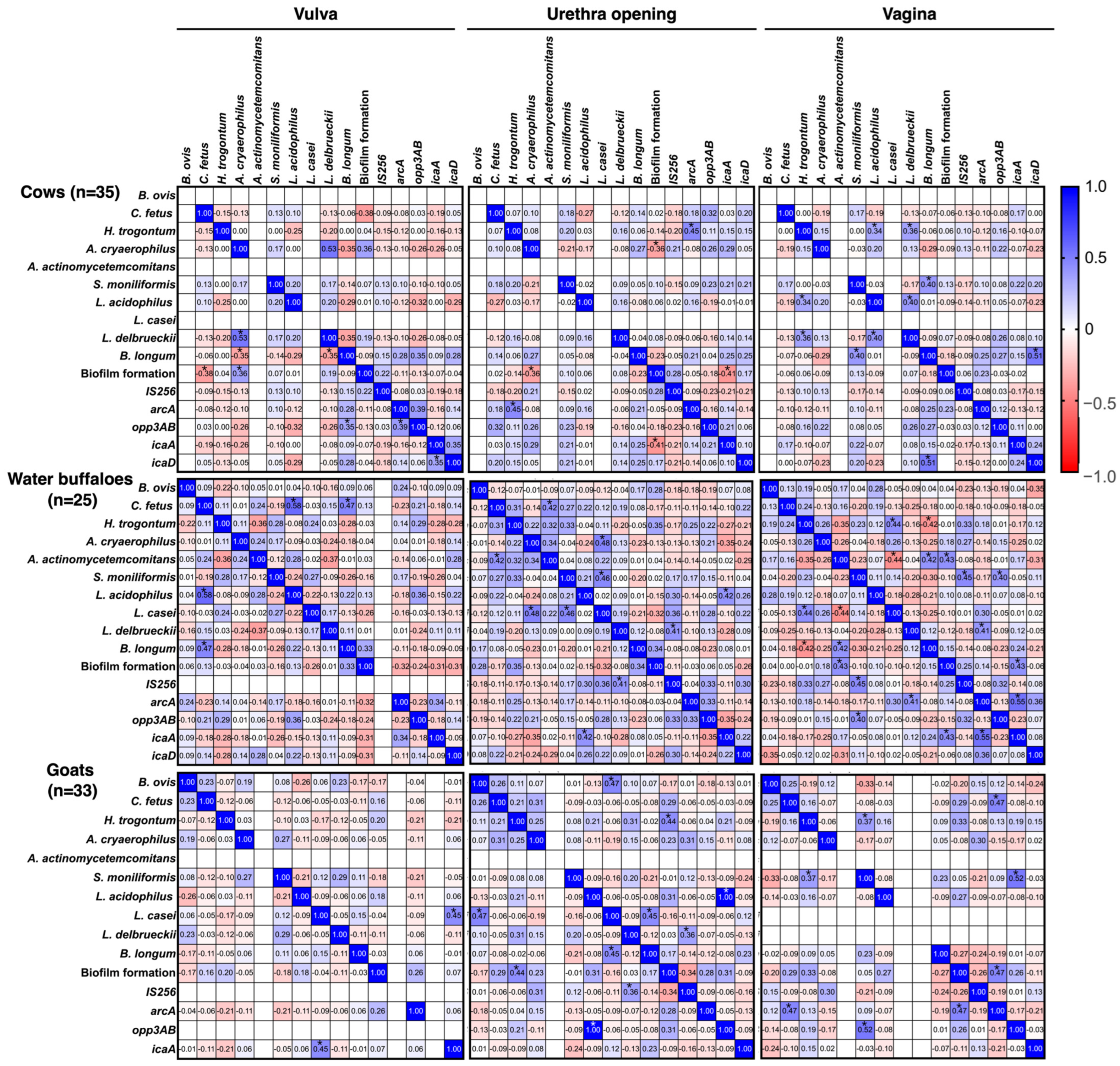
3.3. Co-Localization and Correlations of Bacteria and Biofilm-Associated Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ebani, V.V. Reproductive disorders in domestic ruminants: A one health concern. Pathogens 2022, 11, 1139. [Google Scholar] [CrossRef]
- Bekele Atoma, T.; Szonyi, B.; Haile, A.F.; Fries, R.; Baumann, M.P.O.; Randolph, D.G. Assessment of health problems of sheep and goats based on ante-mortem and post-mortem inspection at Addis Ababa Abattoir, Ethiopia. Front. Vet. Sci. 2024, 11, 1406801. [Google Scholar] [CrossRef]
- Rasmussen, P.; Barkema, H.W.; Osei, P.P.; Taylor, J.; Shaw, A.P.; Conrady, B.; Chaters, G.; Muñoz, V.; Hall, D.C.; Apenteng, O.O.; et al. Global losses due to dairy cattle diseases: A comorbidity-adjusted economic analysis. J. Dairy Sci. 2024, 107, 6945–6970. [Google Scholar] [CrossRef]
- Adnane, M.; Chapwanya, A. Microbial gatekeepers of fertility in the female reproductive microbiome of cattle. Int. J. Mol. Sci. 2024, 25, 10923. [Google Scholar] [CrossRef]
- Adnane, M.; Chapwanya, A. A review of the diversity of the genital tract microbiome and implications for fertility of cattle. Animals 2022, 12, 460. [Google Scholar] [CrossRef]
- Barrientos-Durán, A.; Fuentes-López, A.; de Salazar, A.; Plaza-Díaz, J.; García, F. Reviewing the composition of vaginal microbiota: Inclusion of nutrition and probiotic factors in the maintenance of eubiosis. Nutrients 2020, 12, 419. [Google Scholar] [CrossRef]
- Mocé, M.L.; Esteve, I.C.; Pérez-Fuentes, S.; Gómez, E.A.; Mocé, E. Microbiota in goat buck ejaculates differs between breeding and non-breeding seasons. Front. Vet. Sci. 2022, 9, 867671. [Google Scholar] [CrossRef] [PubMed]
- Wesołowska, M.; Szczuka, E. Occurrence and antimicrobial resistance among Staphylococci isolated from the skin microbiota of healthy goats and sheep. Antibiotics 2023, 12, 1594. [Google Scholar] [CrossRef] [PubMed]
- Poole, R.K.; Soffa, D.R.; McAnally, B.E.; Smith, M.S.; Hickman-Brown, K.J.; Stockland, E.L. Reproductive microbiomes in domestic livestock: Insights utilizing 16S rRNA gene amplicon community sequencing. Animals 2023, 13, 485. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Mohler, J.; Mahajan, S.D.; Schwartz, S.A.; Bruggemann, L.; Aalinkeel, R. Microbial biofilm: A review on formation, infection, antibiotic resistance, control measures, and innovative treatment. Microorganisms 2023, 11, 1614. [Google Scholar] [CrossRef]
- Wang, X.; Liu, M.; Yu, C.; Li, J.; Zhou, X. Biofilm formation: Mechanistic insights and therapeutic targets. Mol. Biomed. 2023, 4, 49. [Google Scholar] [CrossRef]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Makabenta, J.M.V.; Nabawy, A.; Li, C.H.; Schmidt-Malan, S.; Patel, R.; Rotello, V.M. Nanomaterial-based therapeutics for antibiotic-resistant bacterial infections. Nat. Rev. Microbiol. 2021, 19, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Won, M.Y.; Oyama, L.B.; Courtney, S.J.; Creevey, C.J.; Huws, S.A. Can rumen bacteria communicate to each other? Microbiome 2020, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Raheel, I.A.E.R.; Hassan, W.H.; Salem, S.S.R.; Salam, H.S.H. Biofilm forming potentiality of Escherichia coli isolated from bovine endometritis and their antibiotic resistance profiles. J. Adv. Vet. Anim. Res. 2020, 7, 442–451. [Google Scholar] [CrossRef]
- Kongphitee, K.; Sommart, K.; Phonbumrung, T.; Gunha, T.; Suzuki, T. Feed intake, digestibility and energy partitioning in beef cattle fed diets with cassava pulp instead of rice straw. Asian-Australas. J. Anim. Sci. 2018, 31, 1431–1441. [Google Scholar] [CrossRef]
- Khonkhaeng, B.; Wanapat, M.; So, S.; Lunpha, A.; Pilajun, R.; Chanjula, P.; Khejornsart, P.; Gunun, P.; Gunun, N.; Tengjaroenkul, B.; et al. Feed efficiency and growth performance in Thai beef cattle fed cricket meal as a soybean meal replacement. Vet. Med. Int. 2025, 2025, 6428834. [Google Scholar] [CrossRef]
- So-In, C.; Khankhum, S.; Khaowong, I.; Pangchai, T.; Sunthamala, N. Efficacy of Garcinia mangostana Linn. and Achyranthes aspera Linn. combined extracts in the prevention of endometritis in cattle. Trop. Anim. Sci. J. 2024, 47, 291–299. [Google Scholar] [CrossRef]
- Chopjitt, P.; Tangthong, P.; Kongkaem, J.; Wonkyai, P.; Charoenwattanamaneechai, A.; Khankhum, S.; Sunthamala, P.; Kerdsin, A.; Sunthamala, N. Molecular characterization and genotype of multi-drug resistant Staphylococcus epidermidis in nasal carriage of young population, Mahasarakham, Thailand. Biomol. Biomed. 2025, 25, 461–471. [Google Scholar] [CrossRef]
- Ruan, Q.; Geng, S.; Yu, J.; Lu, L.; Liu, Y.; Chen, J.; Liao, Q.; Guo, R. Microbial quorum sensing: Mechanisms, applications, and challenges. Biotechnol. Adv. 2025, 86, 108733. [Google Scholar] [CrossRef]
- Fu, C.; Ge, J.; Qu, M.; Ouyang, K.; Qiu, Q. Effects of 4-hydroxy-2,5-dimethyl-3(2H)-furanone supplementation on growth performance, serum antioxidant capacity, rumen fermentation characteristics, rumen bacterial quorum sensing, and microbial community in Hu sheep. Anim. Biosci. 2025, 38, 1422–1434. [Google Scholar] [CrossRef]
- Xiao, M.; Du, L.; Wei, M.; Wang, Y.; Dong, C.; Ju, J.; Zhang, R.; Peng, W.; Wang, Y.; Zheng, Y.; et al. Effects of quercetin on in vitro rumen fermentation parameters, gas production and microflora of beef cattle. Front. Microbiol. 2025, 16, 1527405. [Google Scholar] [CrossRef]
- Xavier, M.N.; Silva, T.M.; Costa, E.A.; Paixão, T.A.; Moustacas, V.S.; Carvalho, C.A., Jr.; Sant’Anna, F.M.; Robles, C.A.; Gouveia, A.M.; Lage, A.P.; et al. Development and evaluation of a species-specific PCR assay for the detection of Brucella ovis infection in rams. Vet. Microbiol. 2010, 145, 158–164. [Google Scholar] [CrossRef]
- Asakura, M.; Samosornsuk, W.; Hinenoya, A.; Misawa, N.; Nishimura, K.; Matsuhisa, A.; Yamasaki, S. Development of a cytolethal distending toxin (cdt) gene-based species-specific multiplex PCR assay for the detection and identification of Campylobacter jejuni, Campylobacter coli and Campylobacter fetus. FEMS Immunol. Med. Microbiol. 2008, 52, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Mendes, E.N.; Queiroz, D.M.; Dewhirst, F.E.; Paster, B.J.; Moura, S.B.; Fox, J.G. Helicobacter trogontum sp. nov., isolated from the rat intestine. Int. J. Syst. Bacteriol. 1996, 46, 916–921. [Google Scholar] [CrossRef]
- Houf, K.; Tutenel, A.; De Zutter, L.; Van Hoof, J.; Vandamme, P. Development of a multiplex PCR assay for the simultaneous detection and identification of Arcobacter butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii. FEMS Microbiol. Lett. 2000, 193, 89–94. [Google Scholar] [CrossRef]
- Psoter, W.J.; Ge, Y.; Russell, S.L.; Chen, Z.; Katz, R.V.; Jean-Charles, G.; Li, Y. PCR detection of Streptococcus mutans and Aggregatibacter actinomycetemcomitans in dental plaque samples from Haitian adolescents. Clin. Oral. Investig. 2011, 15, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Boot, R.; Oosterhuis, A.; Thuis, H.C. PCR for the detection of Streptobacillus moniliformis. Lab. Anim. 2002, 36, 200–208. [Google Scholar] [CrossRef]
- Sheu, S.J.; Hwang, W.Z.; Chen, H.C.; Chiang, Y.C.; Tsen, H.Y. Development and use of tuf gene-based primers for the multiplex PCR detection of Lactobacillus acidophilus, Lactobacillus casei group, Lactobacillus delbrueckii, and Bifidobacterium longum in commercial dairy products. J. Food Prot. 2009, 72, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Kozitskaya, S.; Cho, S.H.; Dietrich, K.; Marre, R.; Naber, K.; Ziebuhr, W. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: Association with biofilm formation and resistance to aminoglycosides. Infect. Immun. 2004, 72, 1210–1215. [Google Scholar] [CrossRef]
- Diemond-Hernández, B.; Solórzano-Santos, F.; Leaños-Miranda, B.; Peregrino-Bejarano, L.; Miranda-Novales, G. Production of icaADBC-encoded polysaccharide intercellular adhesin and therapeutic failure in pediatric patients with staphylococcal device-related infections. BMC Infect. Dis. 2010, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Diep, B.A.; Stone, G.G.; Basuino, L.; Graber, C.J.; Miller, A.; des Etages, S.-A.; Jones, A.; Palazzolo-Ballance, A.M.; Perdreau-Remington, F.; Sensabaugh, G.F.; et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: Convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 2008, 197, 1523–1530. [Google Scholar] [CrossRef]
- Gad, G.F.; El-Feky, M.A.; El-Rehewy, M.S.; Hassan, M.A.; Abolella, H.; El-Baky, R.M. Detection of icaA, icaD genes and biofilm production by Staphylococcus aureus and Staphylococcus epidermidis isolated from urinary tract catheterized patients. J. Infect. Dev. Ctries. 2009, 3, 342–351. [Google Scholar]
- Stepanovic, S.; Vukovic, D.; Dakic, I.; Savic, B.; Svabic-Vlahovic, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Brown, L.D.; Cai, T.T.; Das Gupta, A. Interval estimation for a binomial proportion. Stat. Sci. 2001, 16, 101–117. [Google Scholar] [CrossRef]
- Mukaka, M.M. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med. J. 2012, 24, 69–71. [Google Scholar]
- Campos-Múzquiz, L.G.; Méndez-Olvera, E.T.; Arellano-Reynoso, B.; Martínez-Gómez, D. Campylobacter fetus is internalized by bovine endometrial epithelial cells. Pol. J. Microbiol. 2019, 68, 217–224. [Google Scholar] [CrossRef]
- Caruso, M.; Latorre, L.; Santagada, G.; Fraccalvieri, R.; Difato, L.M.; Miccolupo, A.; Capozzi, L.; Bonerba, E.; Mottola, A.; Parisi, A. Arcobacter spp. in bovine milk: An emerging pathogen with potential zoonotic risk. Ital. J. Food Saf. 2019, 7, 7685. [Google Scholar] [CrossRef]
- Khurana, S.K.; Sehrawat, A.; Tiwari, R.; Prasad, M.; Gulati, B.; Shabbir, M.Z.; Chhabra, R.; Karthik, K.; Patel, S.K.; Pathak, M.; et al. Bovine brucellosis—A comprehensive review. Vet. Q. 2021, 41, 61–88. [Google Scholar] [CrossRef] [PubMed]
- Sorsa, M.; Mamo, G.; Waktole, H.; Abunna, F.; Zewude, A.; Ameni, G. Seroprevalence and associated risk factors of ovine brucellosis in South Omo Zone, Southern Ethiopia. Infect. Drug Resist. 2022, 15, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, C.A.; Maurizio, E.; Rossi, U.A. Comparative review of brucellosis in small domestic ruminants. Front. Vet. Sci. 2022, 9, 887671. [Google Scholar] [CrossRef]
- Elderbrook, M.; Schumaker, B.; Cornish, T.; Peck, D.; Sondgeroth, K. Seroprevalence and risk factors of Brucella ovis in domestic sheep in Wyoming, USA. BMC Vet. Res. 2019, 15, 246. [Google Scholar] [CrossRef]
- Gill, J.; Haydon, T.G.; Rawdon, T.G.; McFadden, A.M.; Ha, H.J.; Shen, Z.; Feng, Y.; Pang, J.; Swennes, A.G.; Paster, B.J.; et al. Helicobacter bilis and Helicobacter trogontum: Infectious causes of abortion in sheep. J. Vet. Diagn. Investig. 2016, 28, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Sabry, M.A.; Abdel-Moein, K.A.; Seleem, A. Evidence of zoonotic transmission of Helicobacter canis between sheep and human contacts. Vector Borne Zoonotic Dis. 2016, 16, 650–653. [Google Scholar] [CrossRef] [PubMed]
- Didkowska, A.; Żmuda, P.; Liberska, M.; Anusz, K. Current state of knowledge regarding bacterially-induced abortion in sheep. Ann. Agric. Environ. Med. 2022, 29, 321–325. [Google Scholar] [CrossRef]
- Mahmoud, S.F.; Fayez, M.; Swelum, A.A.; Alswat, A.S.; Alkafafy, M.; Alzahrani, O.M.; Alsunaini, S.J.; Almuslem, A.; Al Amer, A.S.; Yusuf, S. Genetic diversity, biofilm formation, and antibiotic resistance of Pseudomonas aeruginosa isolated from cow, camel, and mare with clinical endometritis. Vet. Sci. 2022, 9, 239. [Google Scholar] [CrossRef] [PubMed]
- Zangirolamo, A.F.; Souza, A.K.; Yokomizo, D.N.; Miguel, A.K.A.; Costa, M.C.D.; Alfieri, A.A.; Seneda, M.M. Updates and current callenges in reproductive microbiome: A comparative analysis between cows and women. Animals 2024, 14, 1971. [Google Scholar] [CrossRef]
- Nesse, L.L.; Osland, A.M.; Vestby, L.K. The role of biofilms in the pathogenesis of animal bacterial infections. Microorganisms 2023, 11, 608. [Google Scholar] [CrossRef]
- Javed, M.U.; Ijaz, M.; Ahmed, A.; Rasheed, H.; Jabir, A.A.; Batool, M.; Shahid, K.; Ali, A.; Talha, M. Exploring evolutionary perspectives and antibiogram analysis of biofilm-forming Staphylococcus aureus in goat mastitis. Vet. Res. Commun. 2025, 49, 209. [Google Scholar] [CrossRef]
- Shi, J.F.; Gong, Q.L.; Zhao, B.; Ma, B.Y.; Chen, Z.Y.; Yang, Y.; Sun, Y.H.; Wang, Q.; Leng, X.; Zong, Y.; et al. Seroprevalence of brucellosis in buffalo worldwide and associated risk factors: A systematic review and meta-analysis. Front. Vet. Sci. 2021, 8, 649252. [Google Scholar] [CrossRef]
- Pena-Fernández, N.; Kortabarria, N.; Hurtado, A.; Ocejo, M.; Fort, M.; Pérez-Cobo, I.; Collantes-Fernández, E.; Aduriz, G. Biochemical and molecular characterization of Campylobacter fetus isolates from bulls subjected to bovine genital campylobacteriosis diagnosis in Spain. BMC Vet. Res. 2024, 20, 131. [Google Scholar] [CrossRef]
- García, J.A.; Farace, P.; Gioffré, A.K.; Morsella, C.; Méndez, M.A.; Acuña, J.; Aller, J.F.; Signorini, M.; Paolicchi, F.A. Bovine campylobacteriosis in bulls: Insights in the conventional and molecular diagnosis. Braz. J. Microbiol. 2023, 54, 459–467. [Google Scholar] [CrossRef]
- Hänninen, M.L.; Utriainen, M.; Happonen, I.; Dewhirst, F.E. Helicobacter sp. flexispira 16S rDNA taxa 1, 4 and 5 and Finnish porcine Helicobacter isolates are members of the species Helicobacter trogontum (taxon 6). Int. J. Syst. Evol. Microbiol. 2003, 53, 425–433. [Google Scholar] [CrossRef]
- Taillieu, E.; Chiers, K.; Amorim, I.; Gärtner, F.; Maes, D.; Van Steenkiste, C.; Haesebrouck, F. Gastric Helicobacter species associated with dogs, cats and pigs: Significance for public and animal health. Vet. Res. 2022, 53, 42. [Google Scholar] [CrossRef] [PubMed]
- Masila, N.M.; Ross, K.E.; Gardner, M.G.; Whiley, H. Zoonotic and public health implications of Campylobacter species and squamates (lizards, snakes and amphisbaenians). Pathogens 2020, 9, 799. [Google Scholar] [CrossRef]
- Uddin, W.; Khan, G.; Narayan, S.; Sharma, N.; Holeyachi, B.S.; Hakeem, M.A.; Siva, A.B.; Reddy, P.A. Prevalence and diversity of Helicobacter species in captive wild carnivores, and their implications for conservation management of endangered species. BMC Vet. Res. 2025, 21, 498. [Google Scholar] [CrossRef] [PubMed]
- Yesilmen, S.; Vural, A.; Erkan, M.E.; Yildirim, I.H. Prevalence and antimicrobial susceptibility of Arcobacter species in cow milk, water buffalo milk and fresh village cheese. Int. J. Food Microbiol. 2014, 188, 11–14. [Google Scholar] [CrossRef]
- Müller, E.; Hotzel, H.; Ahlers, C.; Hänel, I.; Tomaso, H.; Abdel-Glil, M.Y. Genomic analysis and antimicrobial resistance of Aliarcobacter cryaerophilus strains from German water poultry. Front. Microbiol. 2020, 11, 1549. [Google Scholar] [CrossRef] [PubMed]
- Felipe, V.; Morgante, C.A.; Somale, P.S.; Varroni, F.; Zingaretti, M.L.; Bachetti, R.A.; Correa, S.G.; Porporatto, C. Evaluation of the biofilm forming ability and its associated genes in Staphylococcus species isolates from bovine mastitis in Argentinean dairy farms. Microb. Pathog. 2017, 104, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Getahun, A.M.; Hunderra, G.C.; Gebrezihar, T.G.; Boru, B.G.; Desta, N.T.; Ayana, T.D. Comparative study on lesions of reproductive disorders of cows and female dromedary camels slaughtered at Addis Ababa, Adama and Akaki abattoirs with bacterial isolation and characterization. BMC Vet. Res. 2021, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, E.S.; Arafa, A.A.; Dorgam, S.M.; Eid, R.H.; Atta, N.S.; El-Dabae, W.H.; Gaber Sadek, E. Molecular characterization of genes responsible for biofilm formation in Staphylococcus aureus isolated from mastitis cows. Vet. World 2022, 15, 205–212. [Google Scholar] [CrossRef]
- Kalantar-Neyestanaki, D.; Mansouri, S.; Tadjrobehkar, O.; Isaei, E. The frequency of adherence, biofilm-associated, arginine catabolic mobile element genes, and biofilm formation in clinical and healthcare worker coagulase-negative staphylococci isolates. BMC Microbiol. 2023, 23, 222. [Google Scholar] [CrossRef]
- Ghaioumy, R.; Tabatabaeifar, F.; Mozafarinia, K.; Mianroodi, A.A.; Isaei, E.; Morones-Ramírez, J.R.; Afshari, S.A.K.; Kalantar-Neyestanaki, D. Biofilm formation and molecular analysis of intercellular adhesion gene cluster (icaABCD) among Staphylococcus aureus strains isolated from children with adenoiditis. Iran. J. Microbiol. 2021, 13, 458–463. [Google Scholar] [CrossRef]
- El-Nagdy, A.H.; Abdel-Fattah, G.M.; Emarah, Z. Detection and control of biofilm formation by Staphylococcus aureus from febrile neutropenic patient. Infect. Drug Resist. 2020, 13, 3091–3101. [Google Scholar] [CrossRef]
- Armoon, M.; Babapour, E.; Mirnejad, R.; Babapour, M.; Taati Moghadam, M. Evaluation of icaA and icaD genes involved in biofilm formation in Staphylococcus aureus isolates from clinical sources using reverse transcriptase PCR. Arch. Razi. Inst. 2024, 79, 1329–1335. [Google Scholar]
- Fernández-Calderón, M.C.; Fernández-Babiano, I.; Navarro-Pérez, M.L.; Pazos-Pacheco, C.; Calvo-Cano, A. Biofilm formation and role of other pathogenic factors in the virulence of Staphylococcus epidermidis clinical isolates. Front. Cell Infect. Microbiol. 2025, 15, 1630341. [Google Scholar] [CrossRef] [PubMed]
- Paramanya, S.; Lee, J.H.; Lee, J. Antibiofilm activity of carotenoid crocetin against Staphylococcal strains. Front. Cell Infect. Microbiol. 2024, 14, 1404960. [Google Scholar] [CrossRef] [PubMed]
- Behshood, P.; Tajbakhsh, E. Systematic review and meta-analysis of the association between biofilm formation and antibiotic resistance in MRSE isolated from Iranian patients. Caspian J. Intern. Med. 2025, 16, 225–232. [Google Scholar]
- Terlizzi, M.; Speranza, B.; Sinigaglia, M.; Corbo, M.R.; Bevilacqua, A. Antibiotic resistance in Bifidobacterium animalis subsp. lactis and Bifidobacterium longum: Definition of sensitivity/resistance profiles at the species level. Microorganisms 2025, 13, 1647. [Google Scholar] [CrossRef]
- Mgomi, F.C.; Yang, Y.R.; Cheng, G.; Yang, Z.Q. Lactic acid bacteria biofilms and their antimicrobial potential against pathogenic microorganisms. Biofilm 2023, 5, 100118. [Google Scholar] [CrossRef] [PubMed]
- Che, J.; Shi, J.; Fang, C.; Zeng, X.; Wu, Z.; Du, Q.; Tu, M.; Pan, D. Elimination of pathogen biofilms via postbiotics from lactic acid bacteria: A promising method in food and biomedicine. Microorganisms 2024, 12, 704. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, S.; Baliyan, N.; Maheshwari, D.K. Appraisal of biofilm forming bacteria in developing buffalo dung-based bioformulation coupled to promote yield of Foeniculum vulgare Mill. 3 Biotech. 2022, 12, 234. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.R.; Hosen, M.; Hossain, H.; Islam, R.; Uddin, B.; Rahman, M.; Hossain, M.; Rahman, M. Biofilm production and virulence traits among extensively drug-resistant and methicillin-resistant Staphylococcus aureus from buffalo subclinical mastitis in Bangladesh. Sci. Rep. 2025, 15, 34425. [Google Scholar] [CrossRef] [PubMed]
| Category | Organisms and Primers | Annealing Temperature (°C) | PCR Product Size (Base Pair) | References |
|---|---|---|---|---|
| Pathogenic bacteria strains | Brucella ovis | 55 | 228 | [23] |
| FW: GCCTACGCTGAAACTTGCTTTTG RV: ATCCCCCCATCACCATAACCGAAG | ||||
| Campylobacter fetus | 55 | 485 | [24] | |
| FW: AACGACAAATGTAAGCACTC RV: TATTTATGCAAGTCGTGCGA | ||||
| Helicobacter trogontum | 50 | 887 | [25] | |
| FW: CATAGGTAACATGCCCCA RV: CTGTTTCAAGCTCCCC | ||||
| Arcobacter cryaerophilus | 50 | 255 | [26] | |
| FW: TGCTGGAGCGGATAGAAGTA RV: AACAACCTACGTCCTTCGAC | ||||
| Microbiota bacteria strains | Aggregatibacter actinomycetemcomitans | 58 | 194 | [27] |
| FW: ATTGGGGTTTAGCCCTGGT RV: GGCACAAACCCATCTCTGA | ||||
| Streptobacillus moniliformis | 57 | 296 | [28] | |
| FW: GCTTAACACATGCAAATCTAT RV: AGTAAGGGCCGTATCTCA | ||||
| Lactobacillus acidophilus | 57 | 397 | [29] | |
| FW: GGAAGCTCAAGACCAAATCATG RV: CTTCTTCAAAACATAAACTTGTG | ||||
| Lactobacillus casei | 60 | 202 | [29] | |
| FW: ATCATGGAATTGATGGATACCA RV: TAGACTTGATAACATCTGGCTT | ||||
| Lactobacillus delbrueckii | 65 | 230 | [29] | |
| FW: TACTGTTAAGGTTGGCGACAGC RV: TGTAGACTTGGCCCTTGAAAGT | ||||
| Bifidobacterium longum | 57 | 161 | [29] | |
| FW: GTATCCGTCCGACCCAGCAG RV: GGTGACGGAGCCCGGCTTG | ||||
| Biofilm associated genes | IS256 | 55 | 1102 | [30,31] |
| FW: TGAAAAGCGAAGAGATTCAAAGC RV: ATGTAGGTCCATAAGAACGGC | ||||
| arcA | 51 | 1942 | [32] | |
| FW: CTAACACTGAACCCCAATG RV: GAGCCAGAAGTACGCGAG | ||||
| Opp3AB | 52 | 1183 | [32] | |
| FW: GCAAATCTGTAAATGGTCTGTTC RV: GAAGATTGGCAGCACAAAGTG | ||||
| icaA | 54 | 188 | [33] | |
| FW: TCTCTTGCAGGAGCAATCAA RV: TCAGGCACTAACATCCAGCA | ||||
| icaD | 50 | 198 | [33] | |
| FW: ATGGTCAAGCCCAGACAGAG RV: CGTGTTTTCAACATTTAATGCAA |
| Collection Sites | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cattle | Vulva | Urethra Opening | Vagina | |||||||||
| Cows (n = 35) | Water Buffaloes (n = 25) | Goats (n = 33) | Fisher’s Exact Test (p ≤ 0.05) | Cows (n = 35) | Water Buffaloes (n = 25) | Goats (n = 33) | Fisher’s Exact Test (p ≤ 0.05) | Cows (n = 35) | Water Buffaloes (n = 25) | Goats (n = 33) | Fisher’s Exact Test (p ≤ 0.05) | |
| Pathogens (%) [95% CI] | ||||||||||||
| B. ovis | 0.00 (0–9.93) | 36.00 (20.16–55.53) | 36.36 (21.35–54.22) | 0.000000 | 0.00 (0–9.89) | 44.00 (26.67–62.93) | 30.30 (17.38–47.34) | 0.000000 | 0.00 (0–9.89) | 28.00 (14.28–47.58) | 33.33 (19.08–51.50) | 0.000000 |
| C. fetus | 8.57 (2.96–22.34) | 28.00 (14.11–47.18) | 3.03 (0.54–15.46) | 0.000000 | 20.00 (10.04–35.89) | 24.00 (11.50–43.43) | 3.03 (0.54–15.32) | 0.000085 | 14.29 (6.27–29.91) | 20.00 (8.86–39.13) | 3.03 (0.54–15.32) | 0.001065 |
| H. trogontum | 20.00 (10–35.6) | 48.00 (30–66.3) | 27.27 (14.56–45.21) | 0.000059 | 22.86 (12.07–39.02) | 40.00 (23.40–59.26) | 39.39 (24.68–56.32) | 0.014846 | 20.00 (10.04–35.89) | 56.00 (36.77–73.31) | 54.55 (37.24–70.96) | 0.000000 |
| A. cryaerophilus | 14.29 (6.27–29.91) | 28.00 (14.11–47.18) | 9.09 (3.16–23.74) | 0.001201 | 8.57 (2.96–22.38) | 28.00 (14.28–47.58) | 24.24 (12.83–41.02) | 0.001402 | 17.14 (8.33–31.72) | 32.00 (17.20–51.59) | 12.12 (5.24–25.51) | 0.001395 |
| Microbiota (%) [95% CI] | ||||||||||||
| A. actinomycetemcomitans | 0.00 (0–9.93) | 52.00 (33.5–70) | 0.00 (0–10.45) | 0.000000 | 0.00 (0–9.89) | 32.00 (17.2–51.6) | 0.00 (0–10.43) | 0.000000 | 0.00 (0–9.89) | 44.00 (26.67–62.93) | 0.00 (0–10.43) | 0.000000 |
| S. moniliformis | 85.71 (70.74–93.82) | 44.00 (25.6–63.9) | 27.27 (14.56–45.21) | 0.000000 | 88.57 (74.05–95.46) | 60.00 (40.74–76.6) | 18.18 (8.61–34.39) | 0.000000 | 85.71 (70.74–93.82) | 40.00 (23.40–59.26) | 15.15 (6.65–30.92) | 0.000000 |
| L. acidophilus | 20.00 (10–35.6) | 20.00 (9.45–37.73) | 9.09 (3.16–23.74) | 0.055023 | 22.86 (12.07–39.02) | 32.00 (17.2–51.6) | 3.03 (0.54–15.32) | 0.000001 | 17.14 (8.33–31.72) | 24.00 (11.50–43.43) | 3.03 (0.54–15.32) | 0.000111 |
| L. casei | 0.00 (0–9.93) | 16.00 (6.37–34.74) | 6.06 (1.69–19.57) | 0.000069 | 0.00 (0–9.89) | 24.00 (11.50–43.43) | 9.09 (3.14–23.57) | 0.000000 | 0.00 (0–9.89) | 32.00 (17.20–51.59) | 0.00 (0–10.43) | 0.000000 |
| L. delbrueckii | 14.29 (6.27–29.91) | 32.00 (16.65–51.98) | 3.03 (0.54–15.46) | 0.000000 | 5.71 (1.58–18.61) | 20.00 (8.86–39.13) | 6.06 (1.68–19.61) | 0.000898 | 8.57 (2.96–22.38) | 20.00 (8.86–39.13) | 0.00 (0–10.43) | 0.000008 |
| B. longum | 42.86 (27.33–59.55) | 8.00 (2.22–25.00) | 24.24 (12.36–41.92) | 0.000000 | 57.14 (40.86–72.02) | 12.00 (4.17–29.96) | 15.15 (6.65–30.92) | 0.000000 | 48.57 (32.26–65.20) | 12.00 (4.17–29.96) | 18.18 (8.61–34.39) | 0.000000 |
| Collection Sites | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cattle | Vulva | Urethra Opening | Vagina | |||||||||
| Cows (n = 35) | Water Buffaloes (n = 25) | Goats (n = 33) | Fisher’s Exact Test (p ≤ 0.05) | Cows (n = 35) | Water Buffaloes (n = 25) | Goats (n = 33) | Fisher’s Exact Test (p ≤ 0.05) | Cows (n = 35) | Water Buffaloes (n = 25) | Goats (n = 33) | Fisher’s Exact Test (p ≤ 0.05) | |
| Biofilm associated genes (%) [95% CI] | ||||||||||||
| IS256 | 8.57 (2.96–22.34) | 0.00 (0–13.3) | 0.00 (0–10.2) | 0.481901 | 11.43 (4.53–25.6) | 4.00 (0.7–19.5) | 9.09 (3.16–23.7) | 0.145876 | 8.57 (2.96–22.34) | 12.00 (4.25–29.9) | 18.18 (9.0–33.7) | 0.121238 |
| arcA | 5.71 (1.58–18.6) | 12.00 (4.25–29.9) | 9.09 (3.16–23.7) | 0.293758 | 5.71 (1.58–18.6) | 4.00 (0.7–19.5) | 3.03 (0.54–15.5) | 0.965605 | 5.71 (1.58–18.6) | 4.00 (0.7–19.5) | 9.09 (3.16–23.7) | 0.319819 |
| opp3AB | 28.57 (16.6–44.8) | 28.00 (14.1–47.2) | 0.00 (0–10.2) | 0.000000 | 28.57 (16.6–44.8) | 28.00 (14.1–47.2) | 3.03 (0.54–15.5) | 0.000002 | 28.57 (16.6–44.8) | 28.00 (14.1–47.2) | 15.15 (7.2–29.4) | 0.042214 |
| icaA | 28.57 (16.6–44.8) | 8.00 (2.2–25) | 24.24 (13–40.6) | 0.000693 | 25.71 (14.6–41.5) | 24.00 (11.3–42.5) | 18.18 (9.0–33.7) | 0.410656 | 22.86 (12–38.5) | 12.00 (4.25–29.9) | 21.21 (11–37.4) | 0.104874 |
| icaD | 25.71 (14.6–41.5) | 8.00 (2.2–25) | 0.00 (0–10.2) | 0.000000 | 25.71 (14.6–41.5) | 32.00 (17–51) | 0.00 (0–10.2) | 0.000000 | 20.00 (10–35.6) | 24.00 (11.3–42.5) | 0.00 (0–10.2) | 0.000002 |
| Biofilm-forming capacity (%) [95% CI] | ||||||||||||
| Non-adherant | 91.43 (77–97) | 64.00 (44–80) | 0.00 (0–10.2) | 0.000000 | 77.14 (61–88) | 52.00 (33–70) | 0.00 (0–10.2) | 0.000000 | 80.00 (64–90) | 64.00 (44–80) | 0.00 (0–10.2) | 0.000000 |
| Weak | 8.57 (2.9–22.3) | 36.00 (20–55) | 100.00 (89–100) | 0.000000 | 22.86 (12–38.5) | 48.00 (30–66) | 100.00 (89–100) | 0.000000 | 20.00 (0–10.2) | 36.00 (10–36) | 96.97 (20–55) | 0.000000 |
| Moderate | 0.00 (0–9.9) | 0.00 (0–13.3) | 0.00 (0–10.2) | ns | 0.00 (0–9.9) | 0.00 (0–13.3) | 0.00 (0–10.2) | ns | 0.00 (85–99.8) | 0.00 (0–9.9) | 3.03 (0–15.5) | ns |
| Strong | 0.00 (0–9.9) | 0.00 (0–13.3) | 0.00 (0–10.2) | ns | 0.00 (0–9.9) | 0.00 (0–13.3) | 0.00 (0–10.2) | ns | 0.00 (0–9.9) | 0.00 (0–13.3) | 0.00 (0–10.2) | ns |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
So-In, C.; Piamalung, N.; Kongkaew, A.; Sriarun, P.; Kammungkun, B.; Phongchaiwasin, S.; Somwaeng, B.; Haputon, W.; Wadmuang, T.; Khankhum, S.; et al. Microbial Distribution and Biofilm-Forming Capacity in the Reproductive Tract of Farm Ruminants. Animals 2026, 16, 133. https://doi.org/10.3390/ani16010133
So-In C, Piamalung N, Kongkaew A, Sriarun P, Kammungkun B, Phongchaiwasin S, Somwaeng B, Haputon W, Wadmuang T, Khankhum S, et al. Microbial Distribution and Biofilm-Forming Capacity in the Reproductive Tract of Farm Ruminants. Animals. 2026; 16(1):133. https://doi.org/10.3390/ani16010133
Chicago/Turabian StyleSo-In, Charinya, Natchaporn Piamalung, Aomsab Kongkaew, Phiyakorn Sriarun, Benyapa Kammungkun, Sawarod Phongchaiwasin, Bongkodkanok Somwaeng, Wichayada Haputon, Thanchanok Wadmuang, Surasak Khankhum, and et al. 2026. "Microbial Distribution and Biofilm-Forming Capacity in the Reproductive Tract of Farm Ruminants" Animals 16, no. 1: 133. https://doi.org/10.3390/ani16010133
APA StyleSo-In, C., Piamalung, N., Kongkaew, A., Sriarun, P., Kammungkun, B., Phongchaiwasin, S., Somwaeng, B., Haputon, W., Wadmuang, T., Khankhum, S., & Sunthamala, N. (2026). Microbial Distribution and Biofilm-Forming Capacity in the Reproductive Tract of Farm Ruminants. Animals, 16(1), 133. https://doi.org/10.3390/ani16010133





