Geographical Variation of Diet Composition of Cervus nippon kopschi in Jiangxi, China Based on DNA Metabarcoding
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
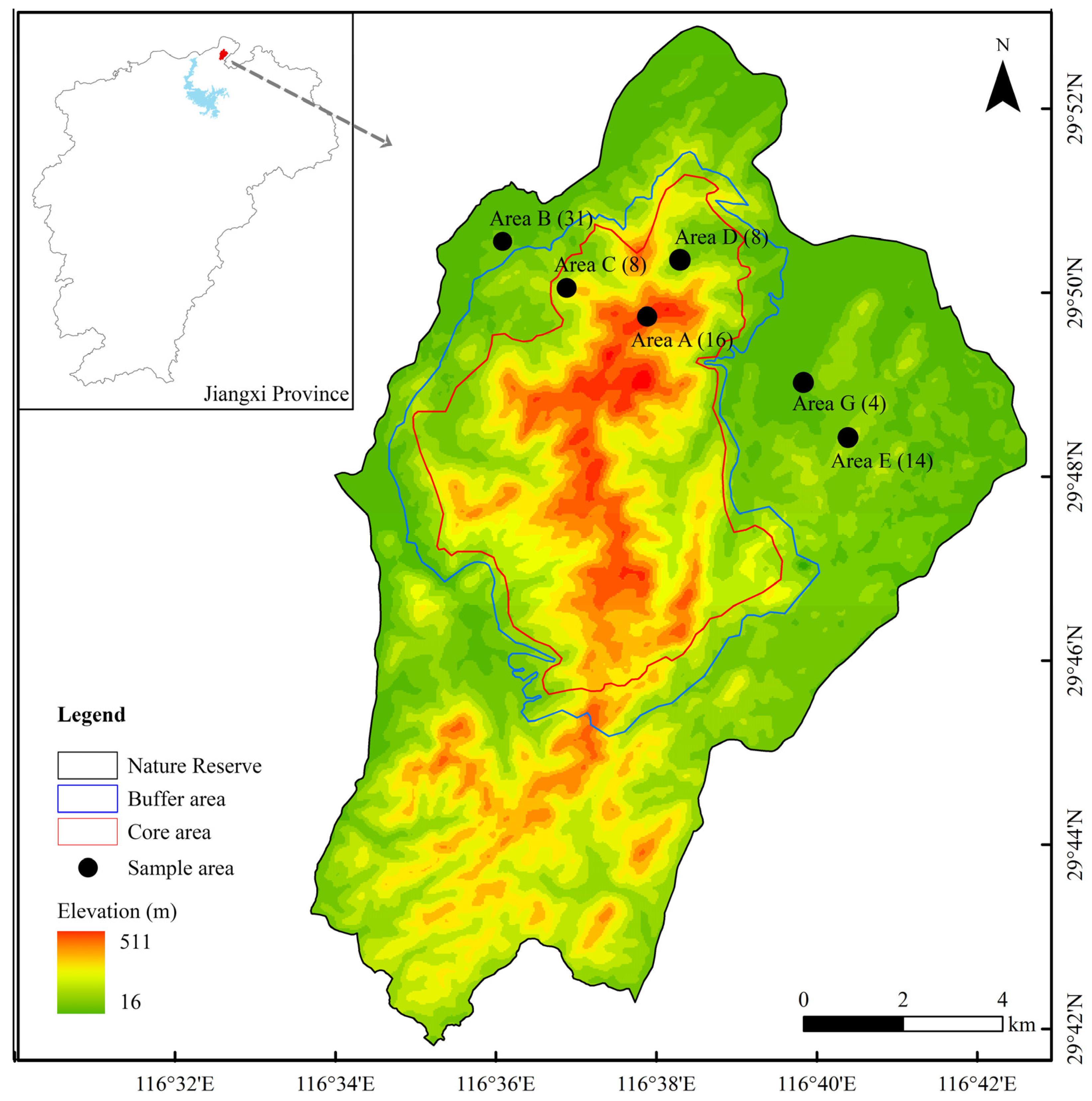
2.1. Study Area
2.2. Sample Collection
2.3. DNA Processing and High-Throughput Sequencing
2.4. Bioinformatics and Statistics Analysis
3. Results
3.1. Analysis of Sequencing Results
3.2. Relative Abundance of the Diet Composition in C. n. kopschi and C. n. hortulorum
3.3. Alpha Diversity of the Diet Composition Between C. n. kopschi and C. n. hortulorum
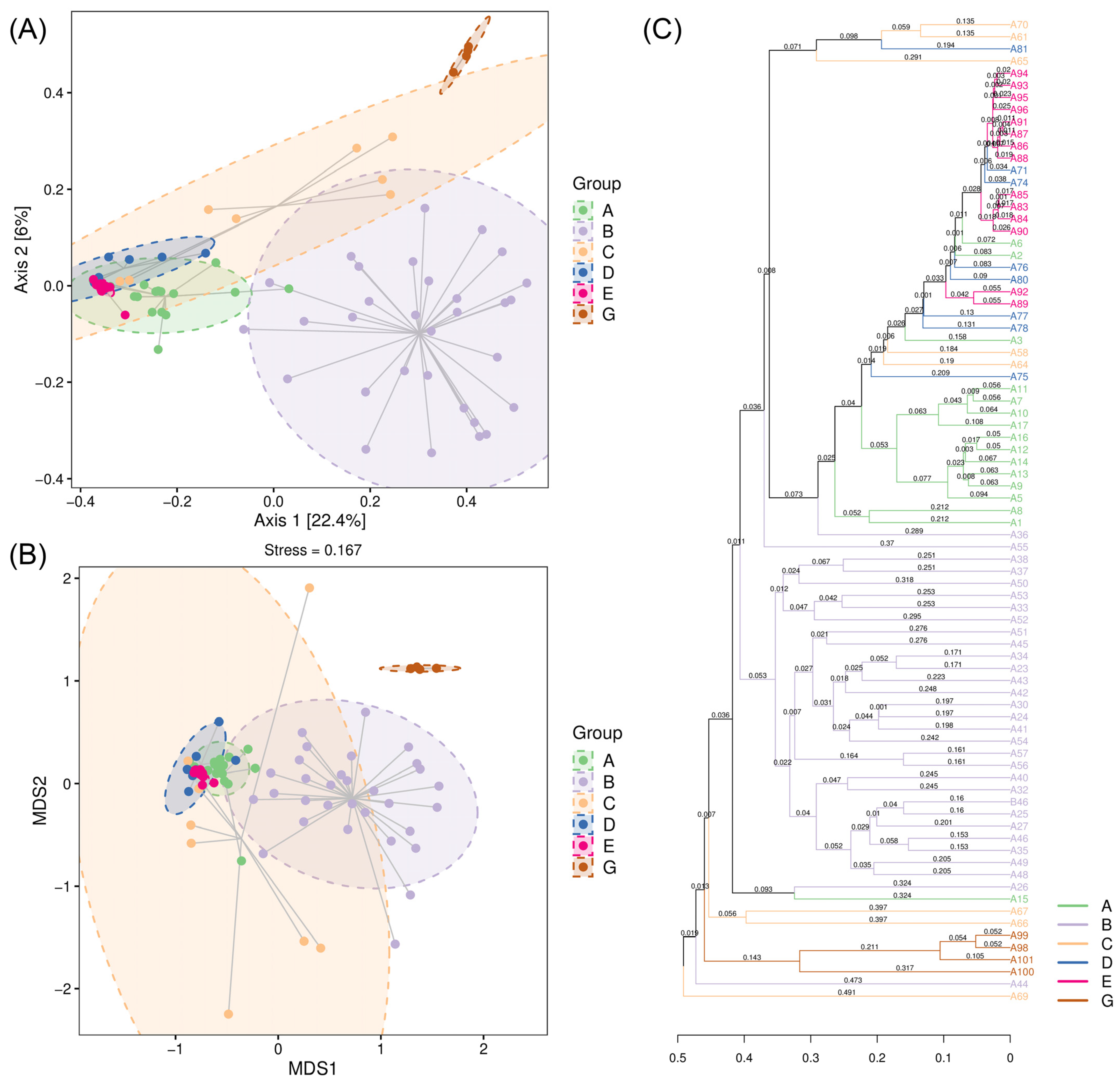
3.4. Beta Diversity of the Diet Composition Between C. n. kopschi and C. n. hortulorum
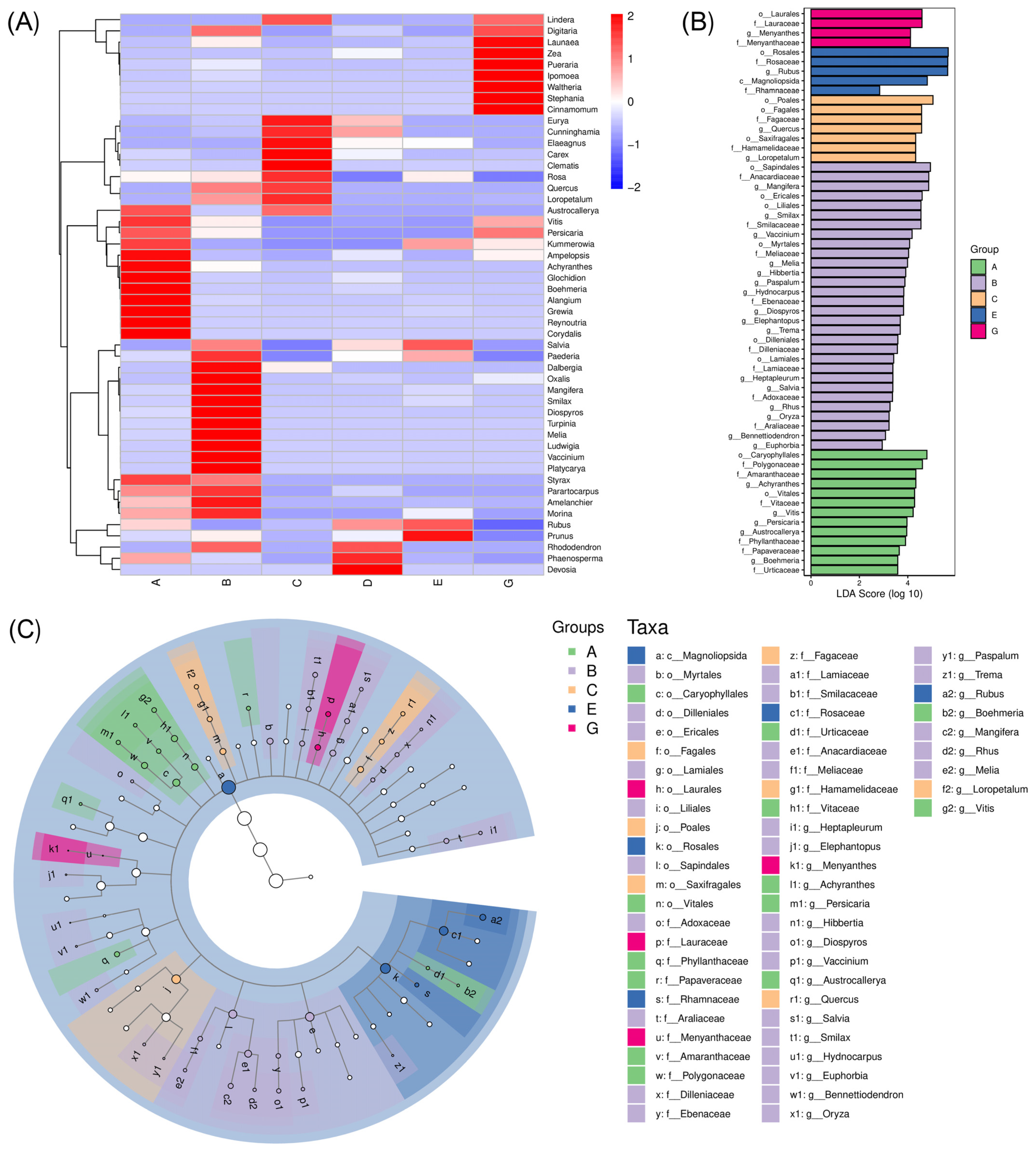
3.5. Differences in the Diet Composition Between C. n. kopschi and C. n. hortulorum
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Whitehead, G.K. Deer of the World; Constable and Company: London, UK, 1972. [Google Scholar]
- Liu, H.; Ju, Y.; Tamate, H.; Wang, T.; Xing, X. Phylogeography of sika deer (Cervus nippon) inferred from mitochondrial cytochrome-b gene and microsatellite DNA. Gene 2021, 772, 145375. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.S. Winter Diet Composition and Non-Invasive Health Assessment of Feral Sika Deer. Ph.D. Thesis, Northeast Forestry University, Harbin, China, 2012. (In Chinese). [Google Scholar]
- Dong, Y.; Li, Y.; Wang, T.; Liu, H.; Zhang, R.; Ju, Y.; Su, W.; Tamate, H.; Xing, X. Complete mitochondrial genome and phylogenetic analysis of eight sika deer subspecies in northeast Asia. J. Genet. 2022, 101, 35. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.G.; Xu, X.R.; Liu, W.H.; Li, C.L.; Li, C.W.; Lu, X.L.; Xiao, J.P.; Li, Y.K.; Tang, S.H.; Ping, X.G.; et al. Population status of south China sika deer in Taohongling National Nature Reserve. Chin. J. Wildl. 2012, 33, 305–308+332. (In Chinese) [Google Scholar] [CrossRef]
- Liu, W.H.; Yu, B. The Population Dynamics and Conservation Strategies of Sika Deer at Taohongling Natural Reserve. Jiangxi Sci. 2010, 28, 458–460. [Google Scholar] [CrossRef]
- Sheppard, S.K.; Harwood, J.D. Advances in molecular ecology: Tracking trophic links through predator–prey food-webs. Funct. Ecol. 2005, 19, 751–762. [Google Scholar] [CrossRef]
- Duffy, J.E.; Cardinale, B.J.; France, K.E.; Mcintyre, P.B.; Thébault, E.; Loreau, M. The functional role of biodiversity in ecosystems: Incorporating trophic complexity. Ecol. Lett. 2007, 10, 522–538. [Google Scholar] [CrossRef]
- Severud, W.J.; Windels, S.K.; Belant, J.L.; Bruggink, J.G. The role of forage availability on diet choice and body condition in American beavers (Castor canadensis). Mamm. Biol. 2013, 78, 87–93. [Google Scholar] [CrossRef]
- Kartzinel, T.R.; Chen, P.A.; Coverdale, T.C.; Erickson, D.L.; Kress, W.J.; Kuzmina, M.L.; Rubenstein, D.I.; Wang, W.; Pringle, R.M. DNA metabarcoding illuminates dietary niche partitioning by African large herbivores. Proc. Natl. Acad. Sci. USA 2015, 112, 8019–8024. [Google Scholar] [CrossRef]
- Klare, U.; Kamler, J.F.; Macdonald, D.W. A comparison and critique of different scat-analysis methods for determining carnivore diet. Mammal Rev. 2011, 41, 294–312. [Google Scholar] [CrossRef]
- Gosselin, E.N.; Lonsinger, R.C.; Waits, L.P. Comparing Morphological and Molecular Diet Analyses and Fecal DNA Sampling Protocols for a Terrestrial Carnivore. Wildl. Soc. B 2017, 41, 362–369. [Google Scholar] [CrossRef]
- Spaulding, R.; Ballard, K.W.B. Observer Bias and Analysis of Gray Wolf Diets from Scats. Wildl. Soc. B 2000, 28, 947–950. Available online: https://www.jstor.org/stable/3783852 (accessed on 20 January 2025).
- Pompanon, F.; Deagle, B.E.; Symondson, W.O.C.; Brown, D.S.; Jarman, S.N.; Taberlet, P. Who is eating what: Diet assessment using next generation sequencing. Mol. Ecol. 2012, 21, 1931–1950. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Brochmann, C.; Willerslev, E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 2012, 21, 2045–2050. [Google Scholar] [CrossRef] [PubMed]
- Monterroso, P.; Godinho, R.; Oliveira, T.; Ferreras, P.; Kelly, M.J.; Morin, D.J.; Waits, L.P.; Alves, P.C.; Mills, L.S. Feeding ecological knowledge: The underutilised power of faecal DNA approaches for carnivore diet analysis. Mammal Rev. 2019, 49, 97–112. [Google Scholar] [CrossRef]
- Xiong, M.; Wang, D.; Bu, H.; Shao, X.; Zhang, D.; Li, S.; Wang, R.; Yao, M. Molecular dietary analysis of two sympatric felids in the Mountains of Southwest China biodiversity hotspot and conservation implications. Sci. Rep. 2017, 7, 41909. [Google Scholar] [CrossRef]
- Buzan, E.; Potočnik, H.; Pokorny, B.; Potušek, S.; Iacolina, L.; Gerič, U.; Urzi, F.; Kos, I. Molecular analysis of scats revealed diet and prey choice of grey wolves and Eurasian lynx in the contact zone between the Dinaric Mountains and the Alps. Front. Zool. 2024, 21, 9. [Google Scholar] [CrossRef]
- Marcolin, F.; Iordan, F.; Pizzul, E.; Pallavicini, A.; Torboli, V.; Manfrin, C.; Quaglietta, L. Otter diet and prey selection in a recently recolonized area assessed using microscope analysis and DNA barcoding. Hystrix 2020, 32, 64–72. [Google Scholar] [CrossRef]
- Kaiser, S.A.; Forg, L.E.; Stillman, A.N.; Deitsch, J.F.; Sillett, T.S.; Clucas, G.V. Black-throated blue warblers (Setophaga caerulescens) exhibit diet flexibility and track seasonal changes in insect availability. Ecol. Evol. 2024, 14, e70340. [Google Scholar] [CrossRef]
- Ma, R.; Ma, S.; Liu, H.; Hu, L.; Li, Y.; He, K.; Zhu, Y. Seasonal changes in invertebrate diet of breeding black-necked cranes (Grus nigricollis). Ecol. Evol. 2024, 14, e70234. [Google Scholar] [CrossRef]
- Nichols, R.V.; Åkesson, M.; Kjellander, P. Diet Assessment Based on Rumen Contents: A Comparison between DNA Metabarcoding and Macroscopy. PLoS ONE 2016, 11, e0157977. [Google Scholar] [CrossRef]
- Kowalczyk, R.; Taberlet, P.; Coissac, E.; Valentini, A.; Miquel, C.; Kamiński, T.; Wójcik, J.M. Influence of management practices on large herbivore diet—Case of European bison in Białowieża Primeval Forest (Poland). For. Ecol. Manag. 2011, 261, 821–828. [Google Scholar] [CrossRef]
- Mata, J.C.; Davison, C.W.; Frøslev, T.G.; Buitenwerf, R.; Svenning, J.C. Resource partitioning in a novel herbivore assemblage in South America. J. Anim. Ecol. 2024, 93, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Putman, R.J. Ungulates in temperate forest ecosystems: Perspectives and recommendations for future research. For. Ecol. Manag. 1996, 88, 205–214. [Google Scholar] [CrossRef]
- Reimoser, F.; Gossow, H. Impact of ungulates on forest vegetation and its dependence on the silvicultural system. For. Ecol. Manag. 1996, 88, 107–119. [Google Scholar] [CrossRef]
- Valentini, A.; Miquel, C.; Nawaz, M.A.; Bellemain, E.; Taberlet, P. New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: The trnL approach. Mol. Ecol. Resour. 2010, 9, 51–60. [Google Scholar] [CrossRef]
- Valentini, A.; Pompanon, F.; Taberlet, P. DNA barcoding for ecologists. Trends Ecol. Evol. 2009, 24, 110–117. [Google Scholar] [CrossRef]
- Pierre, T.; Eric, C.; François, P.; Ludovic, G.; Christian, M.; Alice, V.; Thierry, V.; Gérard, C.; Christian, B.; Eske, W. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007, 35, e14. [Google Scholar] [CrossRef]
- Hofreiter, M.; Poinar, H.N.; Spaulding, W.G.; Bauer, K.; Martin, P.S.; Possnert, G.; Pääbo, S. A molecular analysis of ground sloth diet through the last glaciation. Mol. Ecol. 2000, 9, 1975–1984. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBO J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Ryberg, M.; Kristiansson, E.; Abarenkov, K.; Larsson, K.H.; Kõljalg, U. Taxonomic reliability of DNA sequences in public sequence databases: A fungal perspective. PLoS ONE 2006, 1, e59. [Google Scholar] [CrossRef]
- Kemp, P.F.; Aller, J.Y. Bacterial diversity in aquatic and other environments: What 16S rDNA libraries can tell us. FEMS Microbiol. Ecol. 2004, 47, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 623–656. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Pielou, E.C.J. The Measurement of Diversity in Different Types of Biological Collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Chao, A.; Shen, T.J. Nonparametric prediction in species sampling. J. Agric. Biol. Environ. Stat. 2004, 9, 253–269. [Google Scholar] [CrossRef]
- Good, I.J. The population frequencies of species and the estimation of population parameters. Biometrika 1953, 40, 237–264. [Google Scholar] [CrossRef]
- Esty, W.W. The efficiency of Good’s nonparametric coverage estimator. Ann. Stat. 1986, 14, 1257–1260. [Google Scholar] [CrossRef]
- Li, R.; Wang, D.; Cao, Z.; Liu, Y.; Wu, W.; Liu, W.; Zhan, J.; Xu, Y. DNA metabarcoding reveals diet diversity and niche partitioning by two sympatric herbivores in summer. PeerJ 2024, 12, e18665. [Google Scholar] [CrossRef]
- Cao, Z.; Wang, D.; Hu, X.; He, J.; Liu, Y.; Liu, W.; Zhan, J.; Bao, Z.; Guo, C.; Xu, Y. Comparison and association of winter diets and gut microbiota using trnL and 16S rRNA gene sequencing for three herbivores in Taohongling, China. Glob. Ecol. Conserv. 2024, 53, e03041. [Google Scholar] [CrossRef]
- Wallach, A.D.; Shanas, U.; Inbar, M. Feeding activity and dietary composition of roe deer at the southern edge of their range. Eur. J. Wildl. Res. 2010, 56, 1–9. [Google Scholar] [CrossRef]
- Dong, L.J.; You, W.Y.; Zhou, Q.; Weng, D.M.; Zhang, S.Y.; Wang, W.G. Study on Diet of Cervus nippon kopschi in Qingliangfeng Natural Reserve. J. Zhejiang Sci. Technol. 2009, 29, 41–46. (In Chinese) [Google Scholar]
- Jiang, Z.G. Biodiversity Research in Jiangxi Taohongling Sika Deer National Nature Reserve; Tsinghua University Press: Beijing, China, 2009. [Google Scholar]
- Wu, F.; Zhu, D.; Wen, P.; Tang, Z.; Bao, L.; Guan, Y.; Ge, J.; Wang, H. Domestic Cattle in a National Park Restricting the Sika Deer Due to Diet Overlap. Animals 2023, 13, 561. [Google Scholar] [CrossRef]
- Huang, N.Y. Based on the Molecular Fecal Method and Fecal Microscopy Method for a Comparative Study of the Feeding Habits of Sika Deer and Roe Deer. Master’s Thesis, Northeast Forestry University, Harbin, China, 2023. (In Chinese). [Google Scholar] [CrossRef]
- Guan, Y.; Yang, H.; Han, S.; Feng, L.; Wang, T.; Ge, J. Comparison of the gut microbiota composition between wild and captive sika deer (Cervus nippon hortulorum) from feces by high-throughput sequencing. AMB Express 2017, 7, 212. [Google Scholar] [CrossRef]
- Duparc, A.; Garel, M.; Marchand, P.; Dubray, D.; Loison, A. Through the taste buds of a large herbivore: Foodscape modeling contributes to an understanding of forage selection processes. Oikos 2020, 129, 170–183. [Google Scholar] [CrossRef]
- Parker, K.J. Advances in the nutritional ecology of cervids at different scales. Ecoscience 2003, 10, 395–411. [Google Scholar] [CrossRef]
- Wam, H.K.; Felton, A.M.; Stolter, C.; Nybakken, L.; Hjeljord, O. Moose selecting for specific nutritional composition of birch places limits on food acceptability. Ecol. Evol. 2018, 8, 1117–1130. [Google Scholar] [CrossRef]
- Felton, A.M.; Felton, A.; Raubenheimer, D.; Simpson, S.J.; Krizsan, S.J.; Hedwall, P.O.; Stolter, C. The Nutritional Balancing Act of a Large Herbivore: An Experiment with Captive Moose (Alces alces L). PLoS ONE 2016, 11, e0150870. [Google Scholar] [CrossRef]
- Simpson, S.J.; Sibly, R.M.; Lee, K.P.; Behmer, S.T.; Raubenheimer, D. Optimal foraging when regulating intake of multiple nutrients. Anim. Behav. 2004, 68, 1299–1311. [Google Scholar] [CrossRef]
- Gaillard, J.M.; Danilkin, A.A. Behavioral Ecology of Siberian and European Roe Deer. J. Wildl. Manag. 1996, 62, 424. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, M.; Xia, F.; Li, S. Human activities reshape the spatial overlap between North Chinese leopard and its wild ungulate prey. Front. Zool. 2024, 21, 24. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T.R.; DeVivo, M.T.; Wirsing, A.J.; Bassing, S.B.; Kertson, B.N.; Walker, S.L.; Prugh, L.R. Cougars, wolves, and humans drive a dynamic landscape of fear for elk. Ecology 2024, 105, e4255. [Google Scholar] [CrossRef] [PubMed]
- Wrege, P.H.; Bambi, F.B.; Malonga, P.J.F.; Samba, O.J.; Brncic, T. Early detection of human impacts using acoustic monitoring: An example with forest elephants. PLoS ONE 2024, 19, e0306932. [Google Scholar] [CrossRef]
- Gaynor, K.M.; McInturff, A.; Brashares, J.S. Contrasting patterns of risk from human and non-human predators shape temporal activity of prey. J. Anim. Ecol. 2022, 91, 46–60. [Google Scholar] [CrossRef]
- Lacroux, C.; Robira, B.; Kane-Maguire, N.; Guma, N.; Krief, S. Between forest and croplands: Nocturnal behavior in wild chimpanzees of Sebitoli, Kibale National Park, Uganda. PLoS ONE 2022, 17, e0268132. [Google Scholar] [CrossRef]
- Langvatn, R.; Hanley, T.A. Feeding-patch choice by red deer in relation to foraging efficiency. Oecologia 1993, 95, 164–170. [Google Scholar] [CrossRef]
- Yuto, H.; Shun, K.H.; Yoshihisa, S.; Tetsukazu, Y. Geographical and seasonal variation of plant taxa detected in faces of Cervus nippon yakushimae based on plant DNA analysis in Yakushima Island. Ecol. Res. 2022, 37, 582–597. [Google Scholar] [CrossRef]
- Zhang, X.; Zou, Y.; Zou, X.; Xu, Z.; Nan, X.; Han, C. DNA metabarcoding uncovers the diet of subterranean rodents in China. PLoS ONE 2022, 17, e0258078. [Google Scholar] [CrossRef]
- Osman, N.A.; Abdul-Latiff, M.A.B.; Mohd-Ridwan, A.R.; Yaakop, S.; Nor, S.M.; Md-Zain, B.M. Diet Composition of the Wild Stump-Tailed Macaque (Macaca arctoides) in Perlis State Park, Peninsular Malaysia, Using a Chloroplast tRNL DNA Metabarcoding Approach: A Preliminary Study. Animals 2020, 10, 2215. [Google Scholar] [CrossRef]
- Esmaeili, S.; King, S.R.B.; Schoenecker, K.A. Browsers or Grazers? New Insights into Feral Burro Diet Using a Non-Invasive Sampling and Plant DNA Metabarcoding Approach. Animals 2023, 13, 2683. [Google Scholar] [CrossRef] [PubMed]
- Lazic, T.; Pierri, C.; Corriero, G.; Balech, B.; Cardone, F.; Deflorio, M.; Fosso, B.; Gissi, C.; Marzano, M.; Nonnis Marzano, F.; et al. Evaluating the Efficiency of DNA Metabarcoding to Analyze the Diet of Hippocampus guttulatus (Teleostea: Syngnathidae). Life 2021, 11, 998. [Google Scholar] [CrossRef]
- Domènech, M.; Wangensteen, O.S.; Enguídanos, A.; Malumbres-Olarte, J.; Arnedo, M.A. For all audiences: Incorporating immature stages into standardised spider inventories has a major impact on the assessment of biodiversity patterns. Mol. Ecol. Resour. 2022, 22, 2319–2332. [Google Scholar] [CrossRef] [PubMed]
- Elbrecht, V.; Leese, F. Can DNA-Based Ecosystem Assessments Quantify Species Abundance? Testing Primer Bias and Biomass--Sequence Relationships with an Innovative Metabarcoding Protocol. PLoS ONE 2015, 10, e0130324. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, X.; Xie, Y.; Song, C.; Zhang, Y.; Yu, H.; Burton, G.A. Zooplankton Community Profiling in a Eutrophic Freshwater Ecosystem-Lake Tai Basin by DNA Metabarcoding. Sci. Rep. 2017, 7, 1773. [Google Scholar] [CrossRef]
- Banerji, A.; Bagley, M.; Elk, M.; Pilgrim, E.; Marinson, J.; Santo Domingo, J. Spatial and temporal dynamics of a freshwater eukaryotic plankton community revealed via 18S rRNA gene metabarcoding. Hydrobiologia 2018, 818, 71–86. [Google Scholar] [CrossRef]

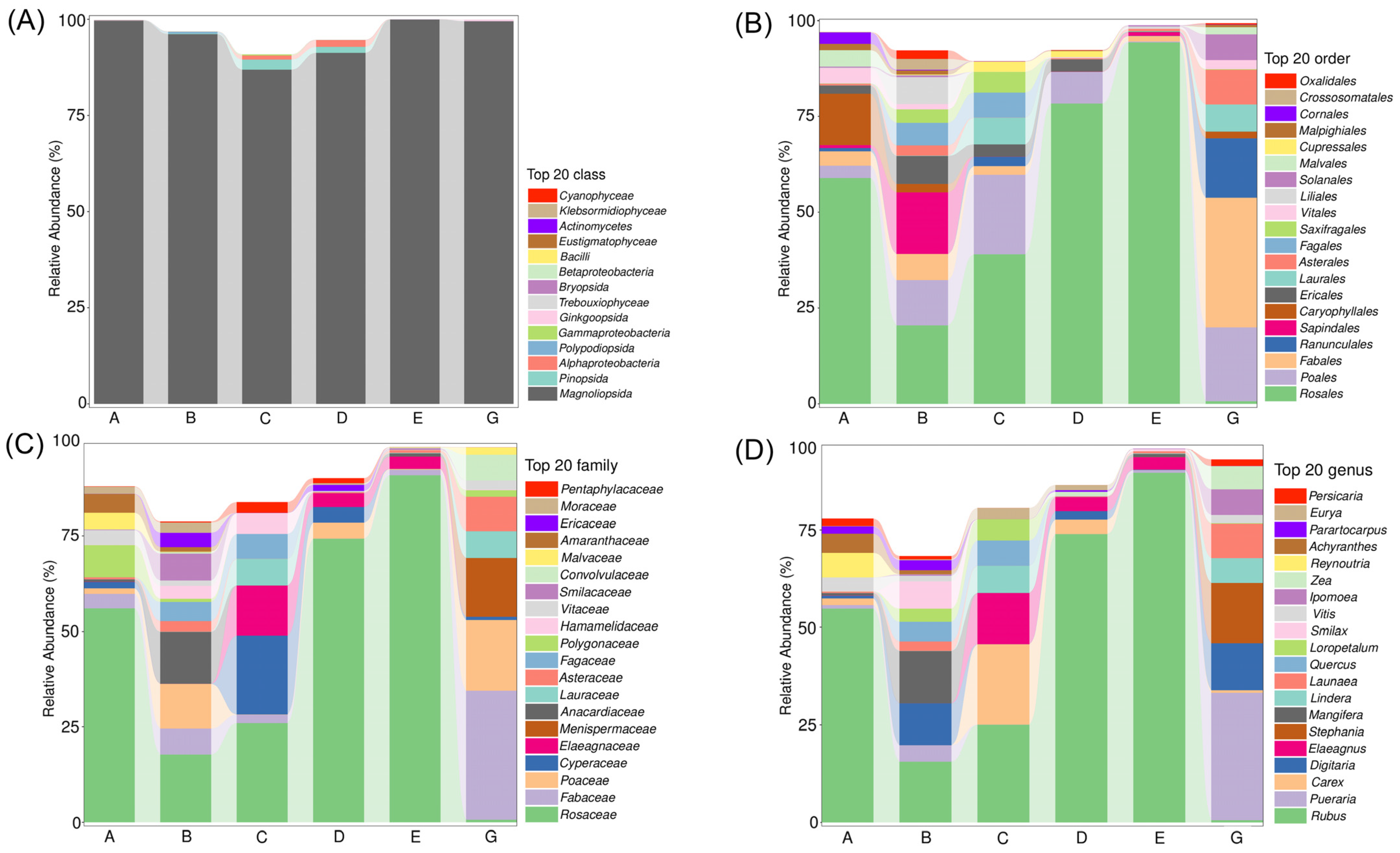
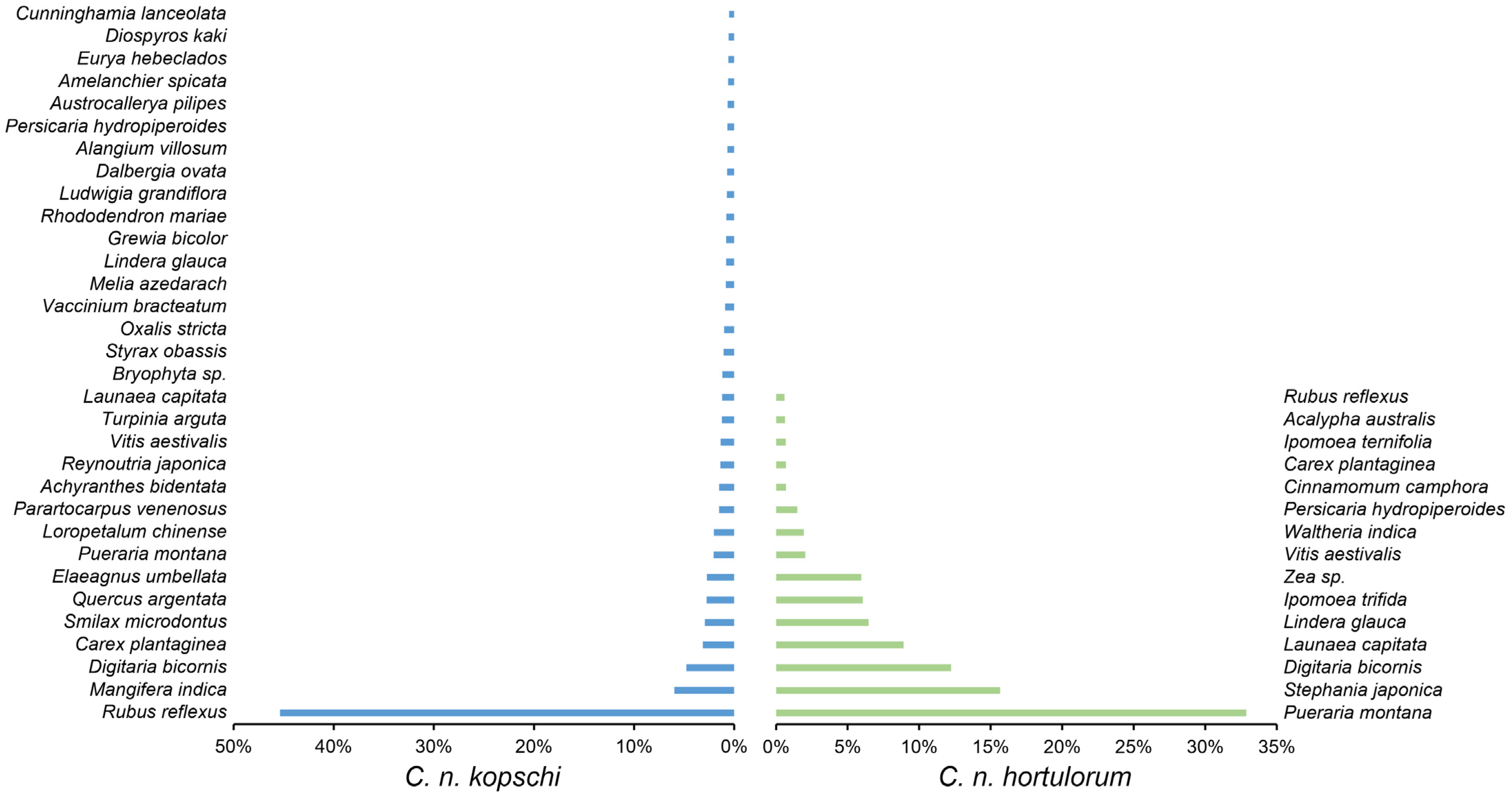

| Rank | C. n. kopschi | C. n. hortulorum | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Order | RA (%) | Family | RA (%) | Genus | RA (%) | Order | RA (%) | Family | RA (%) | Genus | RA (%) | |
| 1 | Rosales | 51.26 | Rosaceae | 46.73 | Rubus | 45.43 | Fabales | 33.89 | Fabaceae | 33.89 | Pueraria | 32.87 |
| 2 | Poales | 8.72 | Anacardiaceae | 6.02 | Mangifera | 5.98 | Poales | 19.29 | Poaceae | 18.43 | Stephania | 15.65 |
| 3 | Sapindales | 7.02 | Poaceae | 5.54 | Digitaria | 4.77 | Ranunculales | 15.65 | Menispermaceae | 15.65 | Digitaria | 12.23 |
| 4 | Ericales | 4.06 | Fabaceae | 3.92 | Carex | 3.13 | Asterales | 8.94 | Asteraceae | 8.94 | Launaea | 8.91 |
| 5 | Fabales | 3.92 | Cyperaceae | 3.19 | Smilax | 3.00 | Laurales | 7.15 | Lauraceae | 7.15 | Ipomoea | 6.73 |
| 6 | Caryophyllales | 3.76 | Smilacaceae | 3.00 | Quercus | 2.74 | Solanales | 6.82 | Convolvulaceae | 6.82 | Lindera | 6.45 |
| 7 | Fagales | 3.17 | Fagaceae | 2.76 | Elaeagnus | 2.74 | Vitales | 2.26 | Vitaceae | 2.26 | Zea | 5.96 |
| 8 | Liliales | 3.00 | Elaeagnaceae | 2.74 | Pueraria | 2.05 | Malvales | 1.94 | Malvaceae | 1.94 | Vitis | 2.03 |
| 9 | Saxifragales | 2.06 | Polygonaceae | 2.16 | Loropetalum | 2.04 | Caryophyllales | 1.75 | Polygonaceae | 1.75 | Waltheria | 1.94 |
| 10 | Vitales | 1.48 | Hamamelidaceae | 2.04 | Parartocarpus | 1.50 | Rosales | 0.68 | Cyperaceae | 0.87 | Persicaria | 1.73 |
| Comparison | Chao1 | Observed Species | Simpson | Shannon | Pielou’s Evenness | Good’s Coverage |
|---|---|---|---|---|---|---|
| A-B | 0.0669 | 0.0876 | 0.0077 | 0.0915 | 0.1216 | 0.0469 |
| A-C | 0.6217 | 0.7745 | 1.0000 | 0.8564 | 0.9988 | 0.6117 |
| A-D | 0.2809 | 0.2492 | 0.8443 | 0.1887 | 0.1887 | 0.3210 |
| A-E | 1.0000 | 1.0000 | 0.0133 | 0.0047 | 0.0025 | 1.0000 |
| A-G | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| B-C | 0.0004 | 0.0008 | 0.0133 | 0.0052 | 0.0100 | 0.0003 |
| B-D | 0.0001 | 0.0001 | 0.0002 | 0.0001 | 0.0002 | 0.0001 |
| B-E | 0.0007 | 0.0002 | 0.0000 | 0.0000 | 0.0000 | 0.0008 |
| B-G | 0.9935 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.6088 |
| C-D | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| C-E | 1.0000 | 1.0000 | 0.2540 | 0.6491 | 0.4106 | 1.0000 |
| C-G | 1.0000 | 1.0000 | 0.8443 | 0.5789 | 0.4100 | 1.0000 |
| D-E | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| D-G | 0.9935 | 0.7745 | 0.2479 | 0.1607 | 0.0972 | 1.0000 |
| E-G | 1.0000 | 1.0000 | 0.0128 | 0.0192 | 0.0063 | 1.0000 |
| Comparison | Permanova | Anosim | ||
|---|---|---|---|---|
| Pseudo-F | p-Value | R2 | p-Value | |
| A-B | 17.979 | 0.001 | 0.556 | 0.001 |
| A-C | 7.321 | 0.001 | 0.631 | 0.002 |
| A-D | 5.291 | 0.002 | 0.361 | 0.002 |
| A-E | 18.098 | 0.001 | 0.538 | 0.001 |
| A-G | 25.430 | 0.001 | 1.000 | 0.003 |
| B-C | 6.182 | 0.001 | 0.721 | 0.001 |
| B-D | 11.770 | 0.001 | 0.574 | 0.001 |
| B-E | 25.222 | 0.001 | 0.615 | 0.001 |
| B-G | 6.558 | 0.001 | 0.721 | 0.001 |
| C-D | 3.879 | 0.002 | 0.281 | 0.008 |
| C-E | 11.895 | 0.001 | 0.725 | 0.001 |
| C-G | 5.541 | 0.004 | 0.654 | 0.003 |
| D-E | 2.950 | 0.006 | 0.483 | 0.002 |
| D-G | 23.373 | 0.006 | 1.000 | 0.001 |
| E-G | 106.586 | 0.001 | 1.000 | 0.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Lv, F.; Hu, X.; Tian, J.; Yang, R.; Yao, J.; Huang, Z.; Zhai, J. Geographical Variation of Diet Composition of Cervus nippon kopschi in Jiangxi, China Based on DNA Metabarcoding. Animals 2025, 15, 940. https://doi.org/10.3390/ani15070940
Sun X, Lv F, Hu X, Tian J, Yang R, Yao J, Huang Z, Zhai J. Geographical Variation of Diet Composition of Cervus nippon kopschi in Jiangxi, China Based on DNA Metabarcoding. Animals. 2025; 15(7):940. https://doi.org/10.3390/ani15070940
Chicago/Turabian StyleSun, Xiao, Feiyan Lv, Xueqin Hu, Jun Tian, Ruijie Yang, Jie Yao, Zhiqiang Huang, and Jiancheng Zhai. 2025. "Geographical Variation of Diet Composition of Cervus nippon kopschi in Jiangxi, China Based on DNA Metabarcoding" Animals 15, no. 7: 940. https://doi.org/10.3390/ani15070940
APA StyleSun, X., Lv, F., Hu, X., Tian, J., Yang, R., Yao, J., Huang, Z., & Zhai, J. (2025). Geographical Variation of Diet Composition of Cervus nippon kopschi in Jiangxi, China Based on DNA Metabarcoding. Animals, 15(7), 940. https://doi.org/10.3390/ani15070940






