Role Analysis of the scarb1 Gene in the Pigmentation of Neocaridina denticulata sinensis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Total RNA Extraction, cDNA Preparation, and Quantitative PCR (qPCR)
2.3. dsRNA Synthesis and RNAi
2.4. Calling and Filtrating SNPs
2.5. Genotyping of Candidate SNPs
3. Results
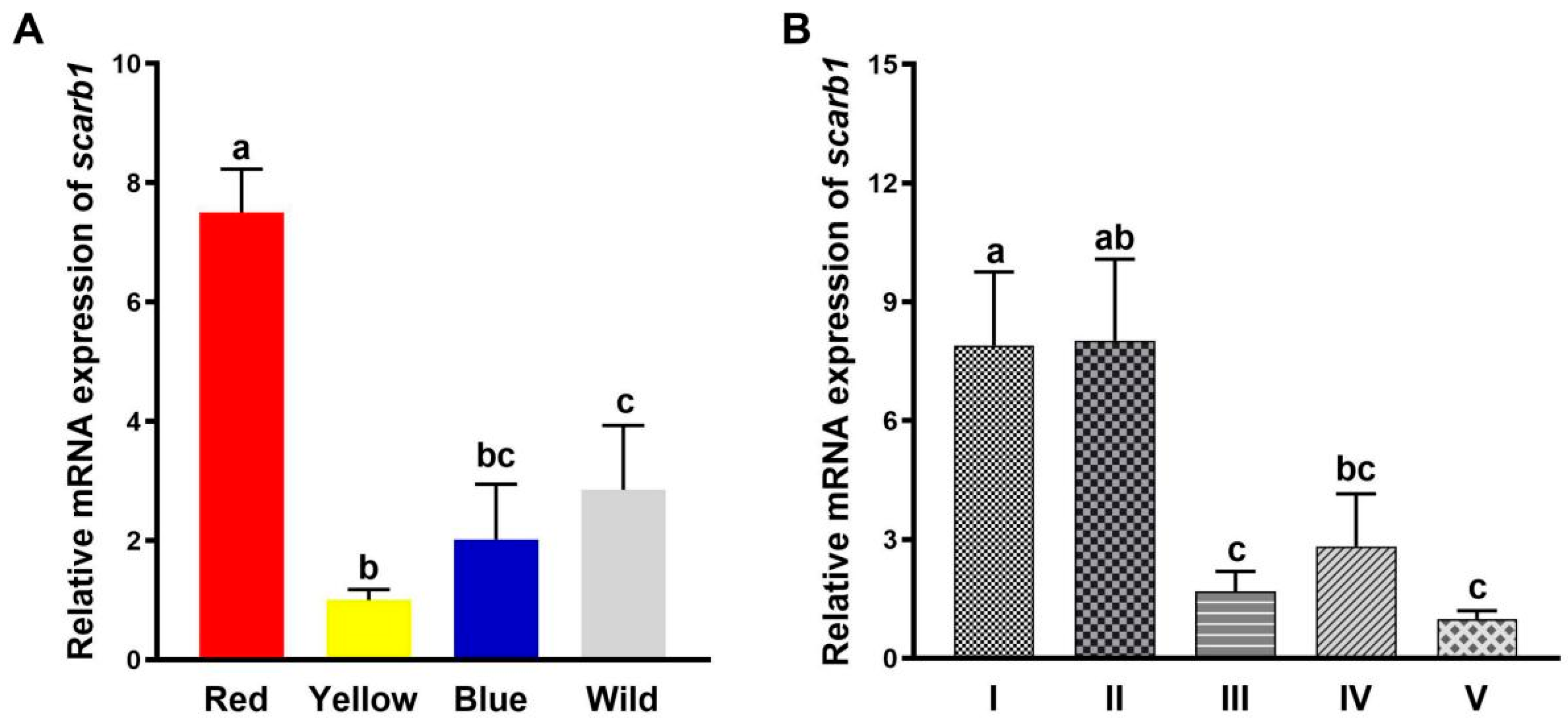
3.1. Scarb1 Expression Profiles
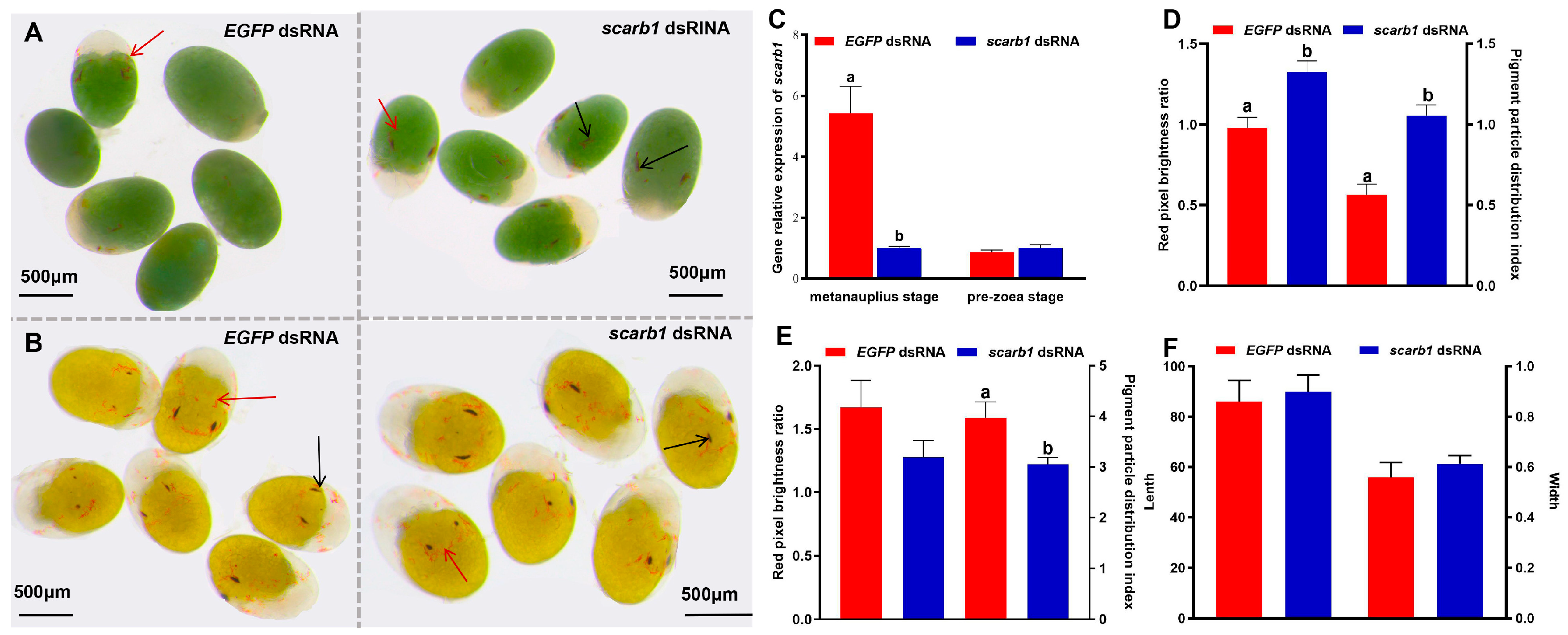
3.2. Functional Analyses of Scarb1 via RNAi
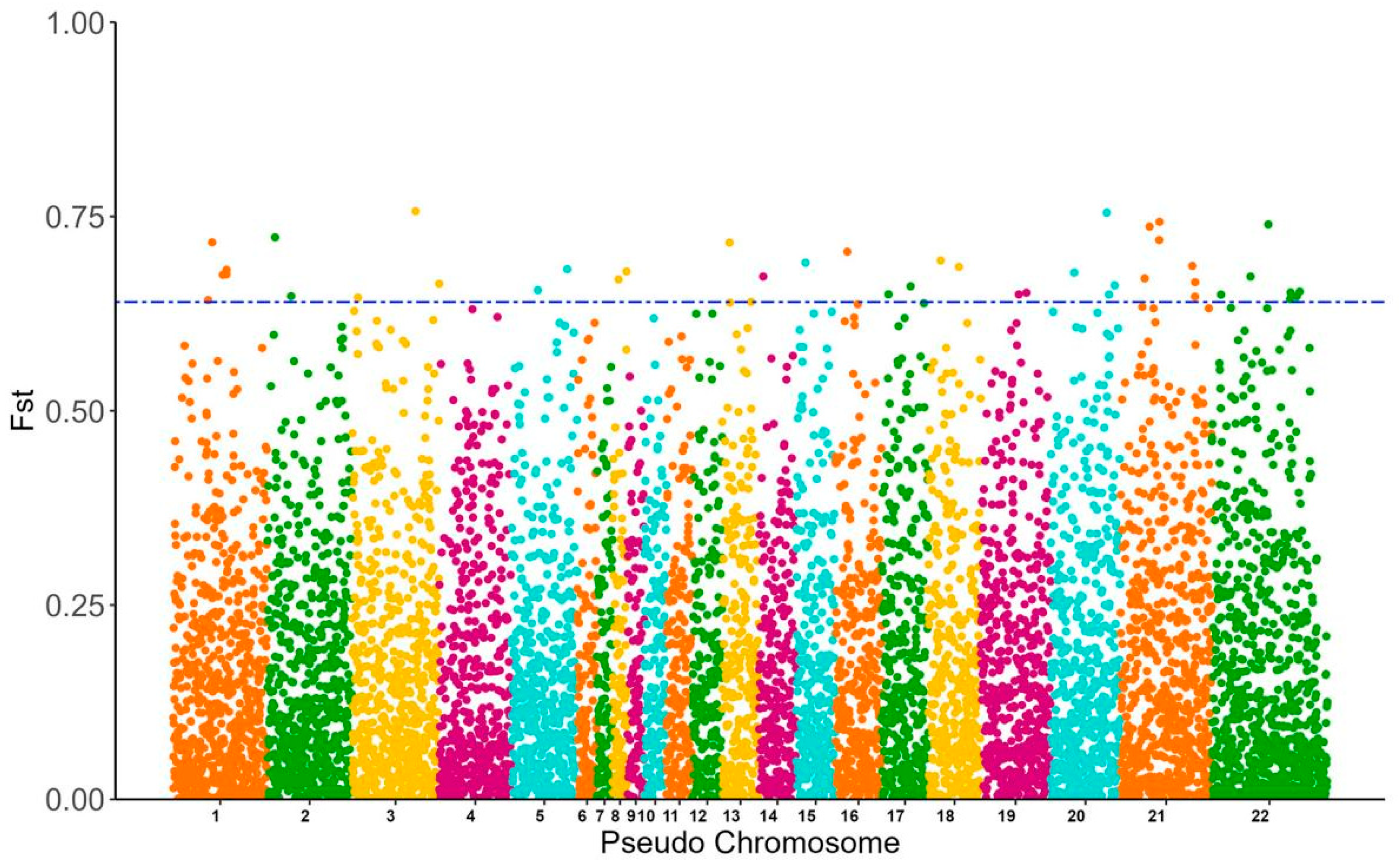
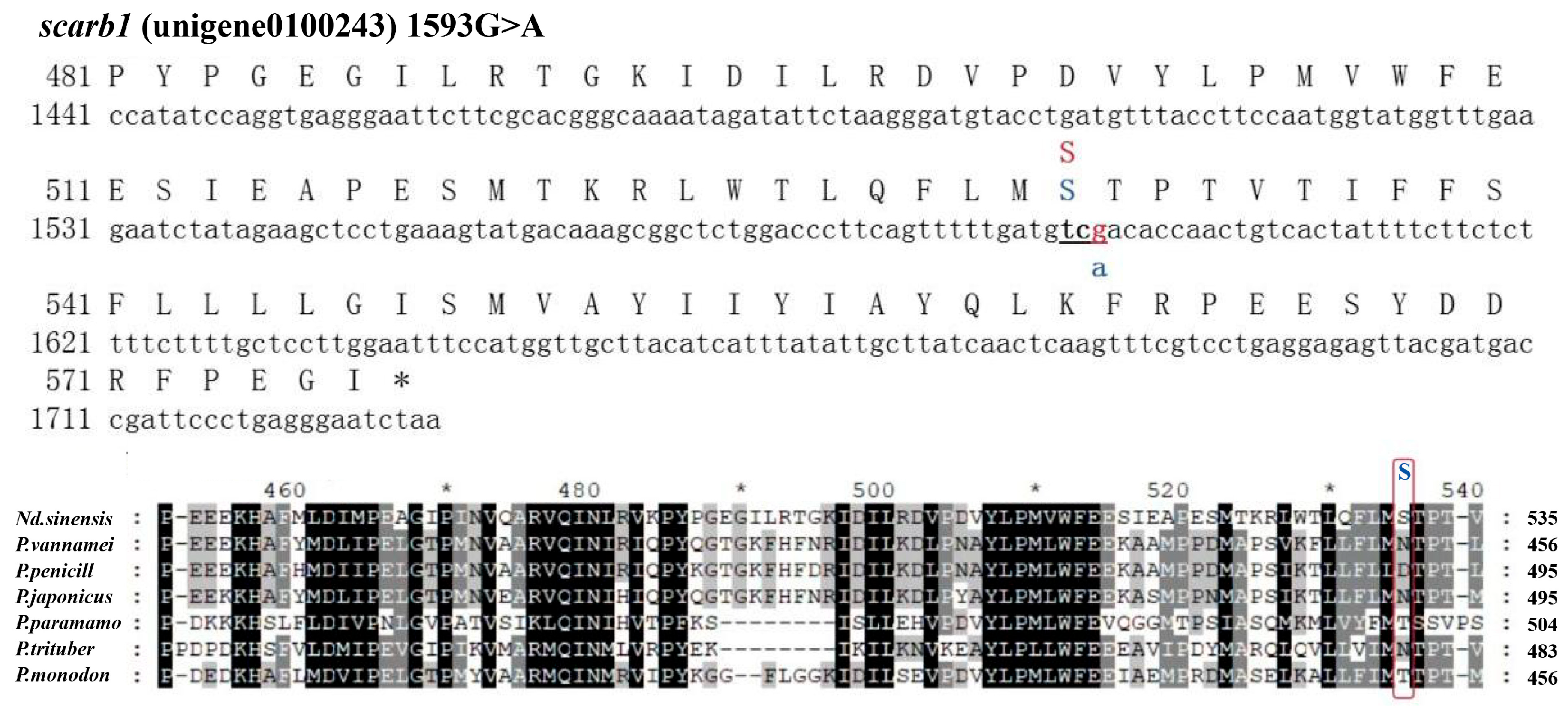
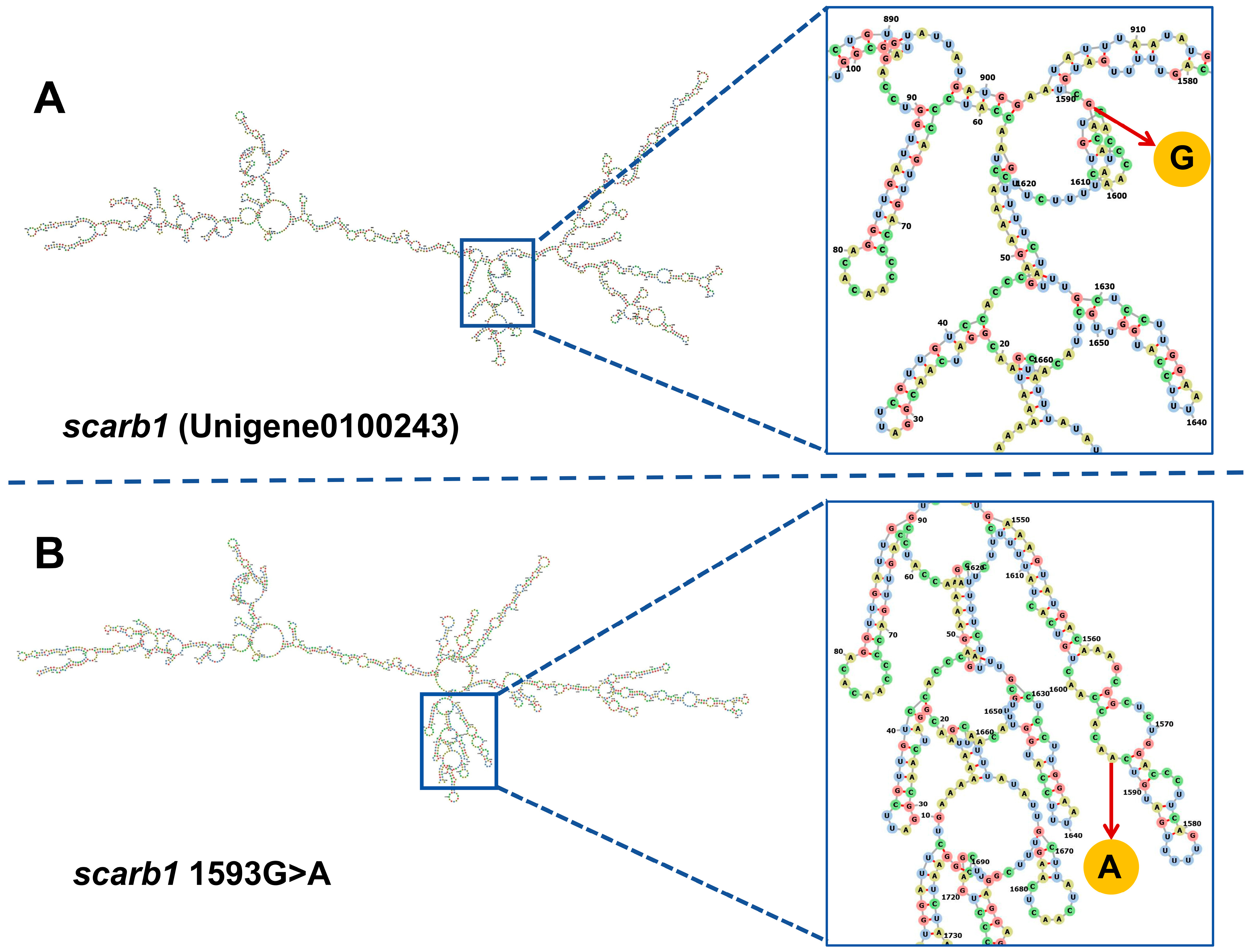
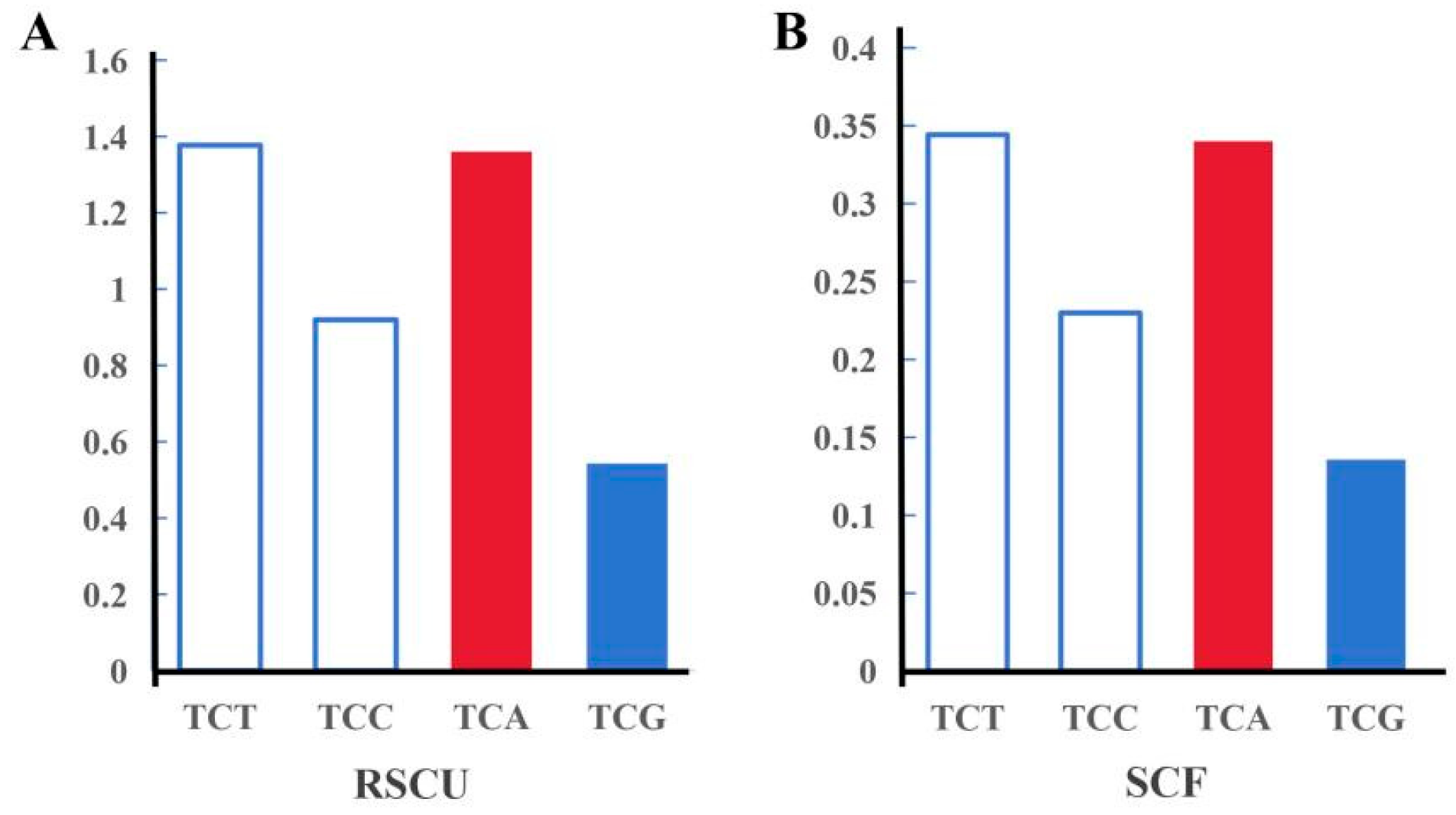
3.3. SNP Genotyping
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flores, E.E.; Chien, Y.-H. Chromatosomes in Three Phenotypes of Neocaridina denticulata Kemp, 1918: Morphological and Chromatic Differences Measured Non-Invasively. J. Crustac. Biol. 2011, 31, 590–597. [Google Scholar] [CrossRef]
- Mykles, D.L.; Hui, J.H.L. Neocaridina denticulata: A Decapod Crustacean Model for Functional Genomics. Integr. Comp. Biol. 2015, 55, 891–897. [Google Scholar] [CrossRef]
- Huang, C.-W.; Chu, P.-Y.; Wu, Y.-F.; Chan, W.-R.; Wang, Y.-H. Identification of functional SSR markers in freshwater ornamental shrimps Neocaridina denticulata using transcriptome sequencing. Mar. Biotechnol. 2020, 22, 772–785. [Google Scholar]
- Fujii, R. The regulation of motile activity in fish chromatophores. Pigment Cell Res. 2000, 13, 300–319. [Google Scholar]
- Wade, N.M.; Gabaudan, J.; Glencross, B.D. A review of carotenoid utilisation and function in crustacean aquaculture. Rev. Aquac. 2017, 9, 141–156. [Google Scholar]
- Wang, L.; Liu, Z.; Jiang, H.; Mao, X. Biotechnology advances in β-carotene production by microorganisms. Trends Food Sci. Technol. 2021, 111, 322–332. [Google Scholar]
- Jing, Y.; Guo, F.; Zhang, S.; Dong, W.; Zhou, J.; Xin, F.; Zhang, W.; Jiang, M. Recent Advances on Biological Synthesis of Lycopene by Using Industrial Yeast. Ind. Eng. Chem. Res. 2021, 60, 3485–3494. [Google Scholar]
- Fraser, P.D.; Bramley, P.M. The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 2004, 43, 228–265. [Google Scholar]
- Yonekura, L.; Nagao, A. Intestinal absorption of dietary carotenoids. Mol. Nutr. Food Res. 2007, 51, 107–115. [Google Scholar]
- Widjaja-Adhi, M.A.K.; Lobo, G.P.; Golczak, M.; Von Lintig, J. A genetic dissection of intestinal fat-soluble vitamin and carotenoid absorption. Hum. Mol. Genet. 2015, 24, 3206–3219. [Google Scholar] [CrossRef]
- Shyam, R.; Vachali, P.; Gorusupudi, A.; Nelson, K.; Bernstein, P.S. All three human scavenger receptor class B proteins can bind and transport all three macular xanthophyll carotenoids. Arch. Biochem. Biophys. 2017, 634, 21–28. [Google Scholar]
- Acton, S.; Rigotti, A.; Landschulz, K.T.; Xu, S.; Hobbs, H.H.; Krieger, M. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 1996, 271, 518–520. [Google Scholar] [CrossRef]
- Powers, H.R.; Jenjak, S.E.; Volkman, B.F.; Sahoo, D. Development and validation of a purification system for functional full-length human SR-B1 and CD36. J. Biol. Chem. 2023, 299, 105187. [Google Scholar]
- Zhang, X.; Fitzsimmons, R.L.; Cleland, L.G.; Ey, P.L.; Zannettino, A.C.; Farmer, E.-A.; Sincock, P.; Mayrhofer, G. CD36/fatty acid translocase in rats: Distribution, isolation from hepatocytes, and comparison with the scavenger receptor SR-B1. Lab. Investig. 2003, 83, 317–332. [Google Scholar]
- Silverstein, R.L.; Febbraio, M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2009, 2, re3. [Google Scholar]
- Goncalves, A.; Margier, M.; Roi, S.; Collet, X.; Niot, I.; Goupy, P.; Caris-Veyrat, C.; Reboul, E. Intestinal scavenger receptors are involved in vitamin K1 absorption. J. Biol. Chem. 2014, 289, 30743–30752. [Google Scholar]
- Reboul, E.; Klein, A.; Bietrix, F.; Gleize, B.; Malezet-Desmoulins, C.; Schneider, M.; Margotat, A.; Lagrost, L.; Collet, X.; Borel, P. Scavenger receptor class B type I (SR-BI) is involved in vitamin E transport across the enterocyte. J. Biol. Chem. 2006, 281, 4739–4745. [Google Scholar]
- Sakudoh, T.; Kuwazaki, S.; Iizuka, T.; Narukawa, J.; Yamamoto, K.; Uchino, K.; Sezutsu, H.; Banno, Y.; Tsuchida, K. CD36 homolog divergence is responsible for the selectivity of carotenoid species migration to the silk gland of the silkworm Bombyx mori [S]. J. Lipid Res. 2013, 54, 482–495. [Google Scholar] [CrossRef]
- Kiefer, C.; Sumser, E.; Wernet, M.F.; von Lintig, J. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc. Natl. Acad. Sci. USA 2002, 99, 10581–10586. [Google Scholar] [CrossRef]
- Wang, T.; Jiao, Y.; Montell, C. Dissection of the pathway required for generation of vitamin A and for Drosophila phototransduction. J. Cell Biol. 2007, 177, 305–316. [Google Scholar] [CrossRef]
- Toomey, M.B.; Lopes, R.J.; Araújo, P.M.; Johnson, J.D.; Gazda, M.A.; Afonso, S.; Mota, P.G.; Koch, R.E.; Hill, G.E.; Corbo, J.C.; et al. High-density lipoprotein receptor SCARB1 is required for carotenoid coloration in birds. Proc. Natl. Acad. Sci. USA 2017, 114, 5219–5224. [Google Scholar] [CrossRef] [PubMed]
- Yongyu, H.; Lili, Z.; Guodong, W.; Shiyu, H. De novo assembly transcriptome analysis reveals the genes associated with body color formation in the freshwater ornamental shrimps Neocaridina denticulate sinensis. Gene 2021, 806, 145929. [Google Scholar]
- Lu, X.; Zhang, L.; Wang, G.; Huang, S. Functional analysis of ABCG2 gene in pigment transport of Neocaridina denticulata sinensis. Gene 2022, 844, 146810. [Google Scholar] [PubMed]
- Li, C. Preliminary Study on Chromocytogenesis of Neocaridina denticulata sinensis. Master’s Thesis, Jimei University, Xiamen, China, 2018. [Google Scholar]
- Lu, X.; Zhang, L.; Wang, G.; Huang, S.; Zhang, Y.; Xie, Y. The occurrence process of chromatophores in three body color strains of the ornamental shrimp Neocaridina denticulata sinensis. Zoomorphology 2022, 141, 283–295. [Google Scholar]
- Hogben, L.T.; Slome, D. Containing Papers of a Biological Character. The pigmentary effector system. VI. The dual character of endocrine co-ordination in amphibian colour change. Proc. R. Soc. Lond. Ser. B Contain. Pap. A Biol. Character 1931, 108, 10–53. [Google Scholar]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J. From FastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.11–11.10.33. [Google Scholar]
- Lin, S.; Wang, G.; Zhang, L.; Huang, S. Basic biology and propagation techniques of ornamental Neocaridina denticulata sinensis. Sci. Fish Cult. 2016, 7, 80–81. [Google Scholar]
- Wang, Y.; Wang, G.; Zhang, L.; Xu, W.; Zhang, G.; Ge, H. A genome-wide association study and an accurate genotyping strategy to identify and validate the SNPs related to the larval period in bullfrog Aquarana catesbeiana. Aquac. Int. 2024, 32, 8267–8286. [Google Scholar]
- Campbell, N.R.; Harmon, S.A.; Narum, S.R. Genotyping-in-Thousands by sequencing (GT-seq): A cost effective SNP genotyping method based on custom amplicon sequencing. Mol. Ecol. Resour. 2015, 15, 855–867. [Google Scholar]
- Alquraini, A.; El Khoury, J. Scavenger receptors. Curr. Biol. 2020, 30, R790–R795. [Google Scholar] [CrossRef] [PubMed]
- Borel, P.; Lietz, G.; Goncalves, A.; de Edelenyi, F.S.; Lecompte, S.; Curtis, P.; Goumidi, L.; Caslake, M.J.; Miles, E.A.; Packard, C. CD36 and SR-BI are involved in cellular uptake of provitamin A carotenoids by Caco-2 and HEK cells, and some of their genetic variants are associated with plasma concentrations of these micronutrients in humans. J. Nutr. 2013, 143, 448–456. [Google Scholar] [PubMed]
- Fan, S.; Wang, F.; Xie, Z.; Zhao, C.; Wang, P.; Yan, L.; Wang, X.; Xu, Y.; Qiu, L. Molecular characterization and functional analysis of scavenger receptor class B from black tiger shrimp (Penaeus monodon). Electron. J. Biotechnol. 2021, 51, 40–49. [Google Scholar]
- During, A.; Doraiswamy, S.; Harrison, E.H. Xanthophylls are preferentially taken up compared with β-carotene by retinal cells via a SRBI-dependent mechanism. J. Lipid Res. 2008, 49, 1715–1724. [Google Scholar]
- Du, J.; Chen, H.; Mandal, B.K.; Wang, J.; Shi, Z.; Lu, G.; Wang, C.J.A. HDL receptor/Scavenger receptor B1-Scarb1 and Scarb1-like mediate the carotenoid-based red coloration in fish. Aquaculture 2021, 545, 737208. [Google Scholar]
- Verwilligen, R.A.; Mulder, L.; Araújo, P.M.; Carneiro, M.; Bussmann, J.; Hoekstra, M.; Van Eck, M. Zebrafish as outgroup model to study evolution of scavenger receptor class B type I functions. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2023, 1868, 159308. [Google Scholar]
- Goodman, D.S.; Huang, H.S. Biosynthesis of vitamin A with rat intestinal enzymes. Science 1965, 149, 879–880. [Google Scholar]
- Lobo, G.P.; Hessel, S.; Eichinger, A.; Noy, N.; Moise, A.R.; Wyss, A.; Palczewski, K.; Von Lintig, J. ISX is a retinoic acid-sensitive gatekeeper that controls intestinal β, β-carotene absorption and vitamin A production. FASEB J. 2010, 24, 1656. [Google Scholar]
- Mundy, N.I.; Stapley, J.; Bennison, C.; Tucker, R.; Twyman, H.; Kim, K.-W.; Burke, T.; Birkhead, T.R.; Andersson, S.; Slate, J. Red carotenoid coloration in the zebra finch is controlled by a cytochrome P450 gene cluster. Curr. Biol. 2016, 26, 1435–1440. [Google Scholar]
- Twomey, E.; Johnson, J.D.; Castroviejo-Fisher, S.; Van Bocxlaer, I. A ketocarotenoid-based colour polymorphism in the Sira poison frog Ranitomeya sirensis indicates novel gene interactions underlying aposematic signal variation. Mol. Ecol. 2020, 29, 2004–2015. [Google Scholar]
- Weaver, R.J.; Gonzalez, B.K.; Santos, S.R.; Havird, J.C. Red coloration in an anchialine shrimp: Carotenoids, genetic variation, and candidate genes. Biol. Bull. 2020, 238, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Borel, P.; Desmarchelier, C.; Nowicki, M.; Bott, R.; Morange, S.; Lesavre, N. Interindividual variability of lutein bioavailability in healthy men: Characterization, genetic variants involved, and relation with fasting plasma lutein concentration. Am. J. Clin. Nutr. 2014, 100, 168–175. [Google Scholar] [CrossRef]
- Helgadottir, A.; Sulem, P.; Thorgeirsson, G.; Gretarsdottir, S.; Thorleifsson, G.; Jensson, B.Ö.; Arnadottir, G.A.; Olafsson, I.; Eyjolfsson, G.I.; Sigurdardottir, O. Rare SCARB1 mutations associate with high-density lipoprotein cholesterol but not with coronary artery disease. Eur. Heart J. 2018, 39, 2172–2178. [Google Scholar] [PubMed]
- Vergeer, M.; Korporaal, S.J.; Franssen, R.; Meurs, I.; Out, R.; Hovingh, G.K.; Hoekstra, M.; Sierts, J.A.; Dallinga-Thie, G.M.; Motazacker, M.M. Genetic variant of the scavenger receptor BI in humans. N. Engl. J. Med. 2011, 364, 136–145. [Google Scholar] [CrossRef]
- Kimchi-Sarfaty, C.; Oh, J.M.; Kim, I.-W.; Sauna, Z.E.; Calcagno, A.M.; Ambudkar, S.V.; Gottesman, M.M. A “silent” polymorphism in the MDR 1 gene changes substrate specificity. Science 2007, 315, 525–528. [Google Scholar] [CrossRef]
- Walsh, I.M.; Bowman, M.A.; Soto Santarriaga, I.F.; Rodriguez, A.; Clark, P.L. Synonymous codon substitutions perturb cotranslational protein folding in vivo and impair cell fitness. Proc. Natl. Acad. Sci. USA 2020, 117, 3528–3534. [Google Scholar] [PubMed]
- Paul, P.; Malakar, A.K.; Chakraborty, S. Codon usage and amino acid usage influence genes expression level. Genetica 2018, 146, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Higgs, P.G.; Ran, W. Coevolution of codon usage and tRNA genes leads to alternative stable states of biased codon usage. Mol. Biol. Evol. 2008, 25, 2279–2291. [Google Scholar] [CrossRef]
- Kanaya, S.; Yamada, Y.; Kinouchi, M.; Kudo, Y.; Ikemura, T. Codon usage and tRNA genes in eukaryotes: Correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J. Mol. Evol. 2001, 53, 290–298. [Google Scholar] [CrossRef]
- Borel, P. Genetic variations involved in interindividual variability in carotenoid status. Mol. Nutr. Food Res. 2012, 56, 228–240. [Google Scholar] [CrossRef]
- Liu, F.; van Duijn, K.; Vingerling, J.R.; Hofman, A.; Uitterlinden, A.G.; Janssens, A.C.J.; Kayser, M. Eye color and the prediction of complex phenotypes from genotypes. Curr. Biol. 2009, 19, R192–R193. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, M.; Knust, E.; Hebbar, S. Drosophila melanogaster: A valuable genetic model organism to elucidate the biology of retinitis pigmentosa. In Retinal Degeneration: Methods in Molecular Biology; Humana: New York, NY, USA, 2019; pp. 221–249. [Google Scholar]
- Abdelmawla, A.; Yang, C.; Li, X.; Li, M.; Li, C.L.; Liu, Y.B.; He, X.J.; Zeng, Z.J. Feeding Asian honeybee queens with European honeybee royal jelly alters body color and expression of related coding and non-coding RNAs. Front. Physiol. 2023, 14, 1073625. [Google Scholar] [CrossRef] [PubMed]
| Primer Name | Primer Sequences (5′–3′) | Application |
|---|---|---|
| scarb1-Fq | ATGGGAGCTCCAGCCTTTAT | qPCR |
| scarb1-Rq | GCATGCTTTTCCTCTTCAGG | qPCR |
| scarb1-Fd | CGTTGTCCACCCAGAGAAAA | dsRNA generation |
| scarb1-Rd | CGTTGTCCACCCAGAGAAAA | dsRNA generation |
| scarb1-F1 | CCTTCTCTTGAGGATGCCACT | Target sequence amplification |
| scarb1-R1 | TCCTTGAACAGCCTCTCGTT | Target sequence amplification |
| scarb1-F2 | CGACAGGTTCAGAGTTCTACAGTCCGACGATCCCTTCTCTTGAGGATGCCACT | Add adapter |
| scarb1-R2 | GCTCGTCGTGACGCCATGACGTCCTTGAACAGCCTCTCGTT | Add adapter |
| ds-EGFP F | GGTGAACTTCAAGATCCGCC | dsRNA generation |
| ds-EGFP R | CTTGTACAGCTCGTCCATGC | dsRNA generation |
| GADPH-Fq | CGGTGCTGCTCAGAATATCA | qPCR |
| GADPH-Rq | TTACCAAGGCGAACGGTAAG | qPCR |
| T7F | TAATACGACTCACTATAGGG | Adapter |
| T7R | CCCTATAGTGAGTCGTATTA | Adapter |
| Population | Red | Yellow | Blue | Wild | |
|---|---|---|---|---|---|
| Genotype | AA | 345 a | 0 b | 329 a | 331 b |
| GA | 0 | 345 | 16 | 14 | |
| Allele | A | 1.000 a | 0.500 b | 0.977 a | 0.980 a |
| G | 0.000 | 0.500 | 0.023 | 0.020 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Wang, G.; Li, H.; Zhao, T. Role Analysis of the scarb1 Gene in the Pigmentation of Neocaridina denticulata sinensis. Animals 2025, 15, 901. https://doi.org/10.3390/ani15070901
Zhang L, Wang G, Li H, Zhao T. Role Analysis of the scarb1 Gene in the Pigmentation of Neocaridina denticulata sinensis. Animals. 2025; 15(7):901. https://doi.org/10.3390/ani15070901
Chicago/Turabian StyleZhang, Lili, Guodong Wang, Haifan Li, and Tanjun Zhao. 2025. "Role Analysis of the scarb1 Gene in the Pigmentation of Neocaridina denticulata sinensis" Animals 15, no. 7: 901. https://doi.org/10.3390/ani15070901
APA StyleZhang, L., Wang, G., Li, H., & Zhao, T. (2025). Role Analysis of the scarb1 Gene in the Pigmentation of Neocaridina denticulata sinensis. Animals, 15(7), 901. https://doi.org/10.3390/ani15070901




