Genomic Characterization of Crossbred-Driven Adaptation in the Endangered Yangba Cattle of China
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection and Whole-Genome Resequencing
2.3. Data Quality Control and Alignment
2.4. Single Nucleotide Polymorphism (SNP) Detection and Annotation
2.5. Detection of Genetic Diversity and Runs of Homozygosity (ROHs)
2.6. Population Structure and Phylogenetic Analysis
2.7. Detection of Gene Flow
2.8. Analysis of Selection Signals
3. Results
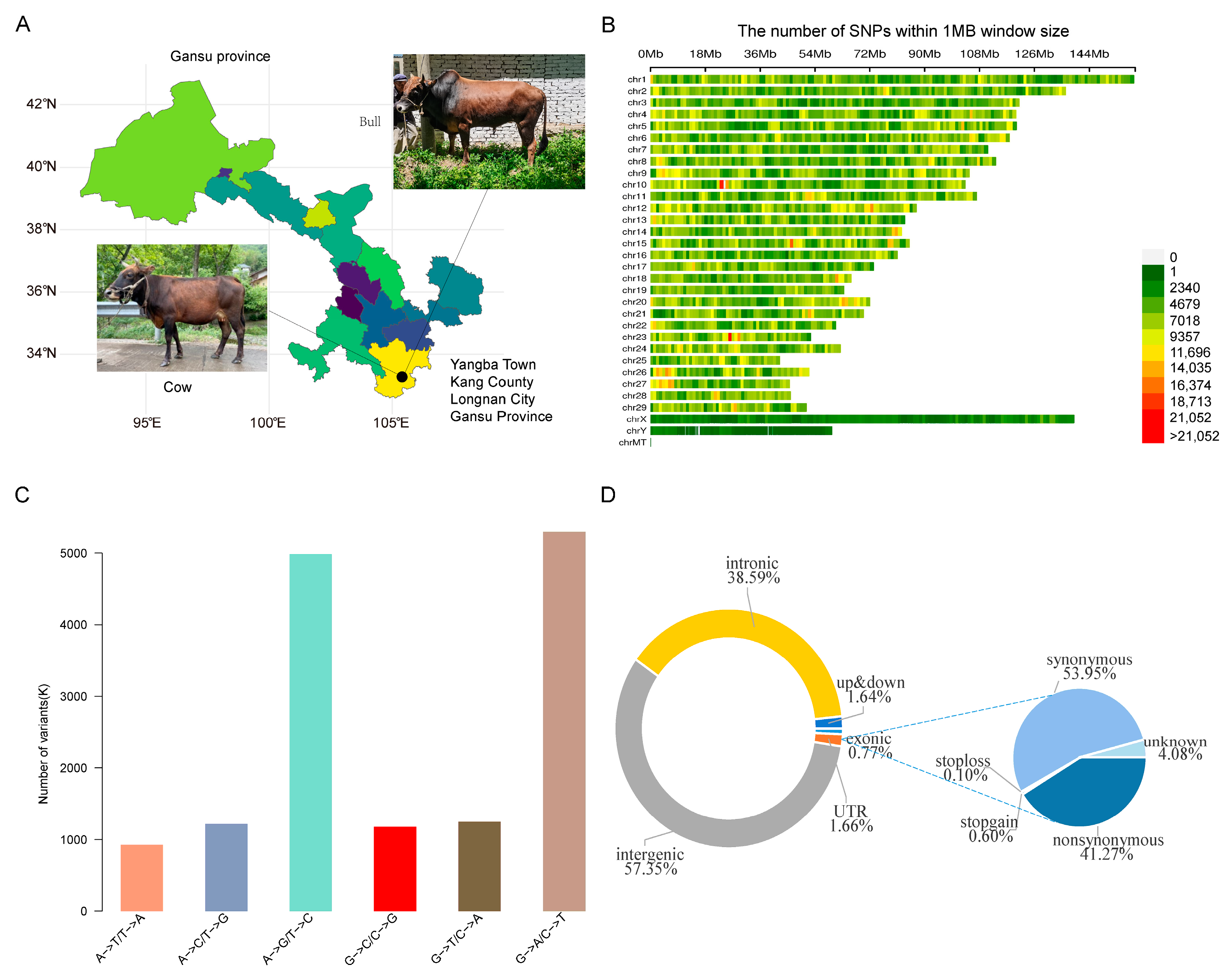
3.1. SNP Identification and Analysis of Genetic Diversity
3.2. Analysis of Population Structure
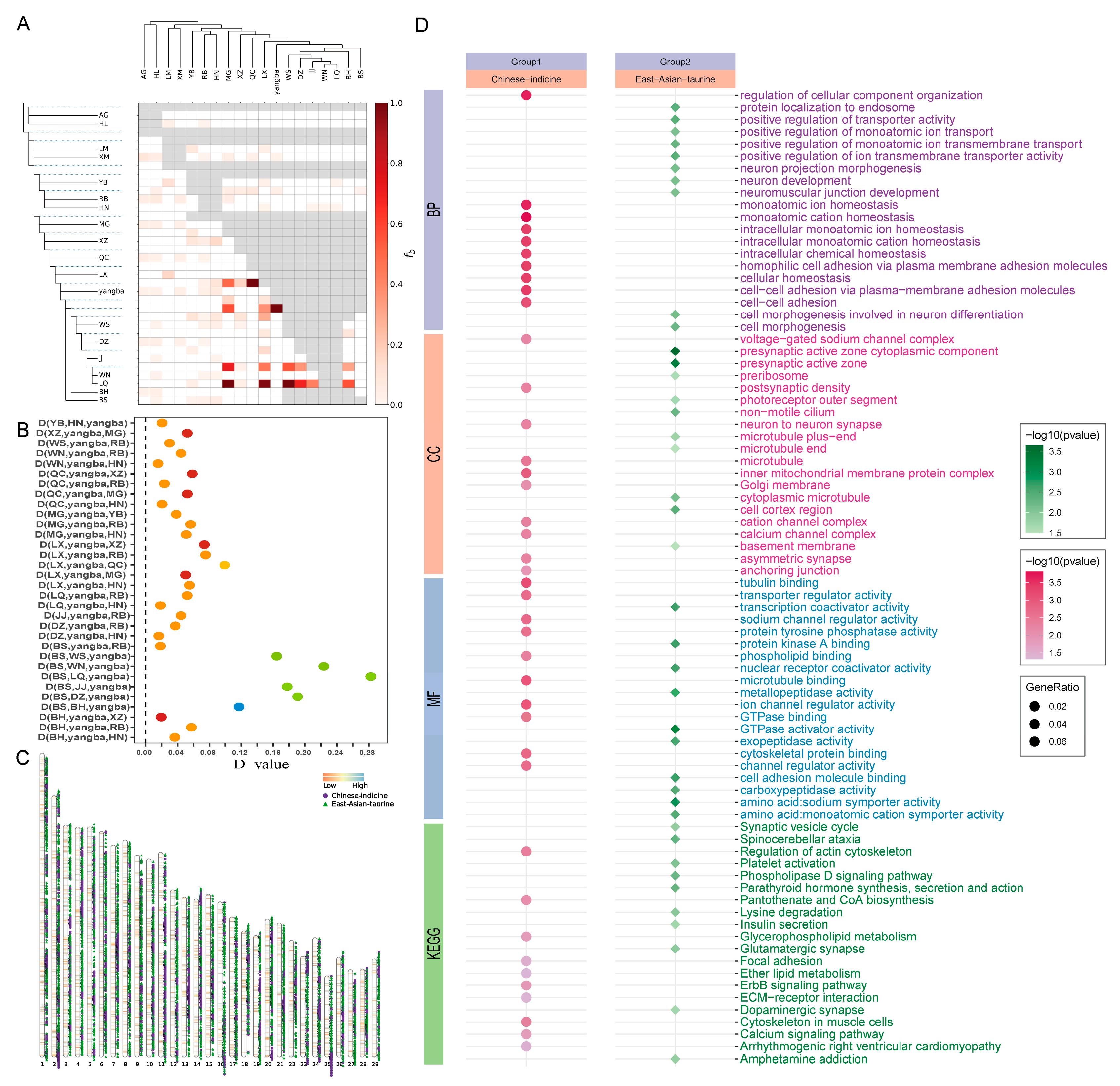
3.3. Analysis of Gene Introgression
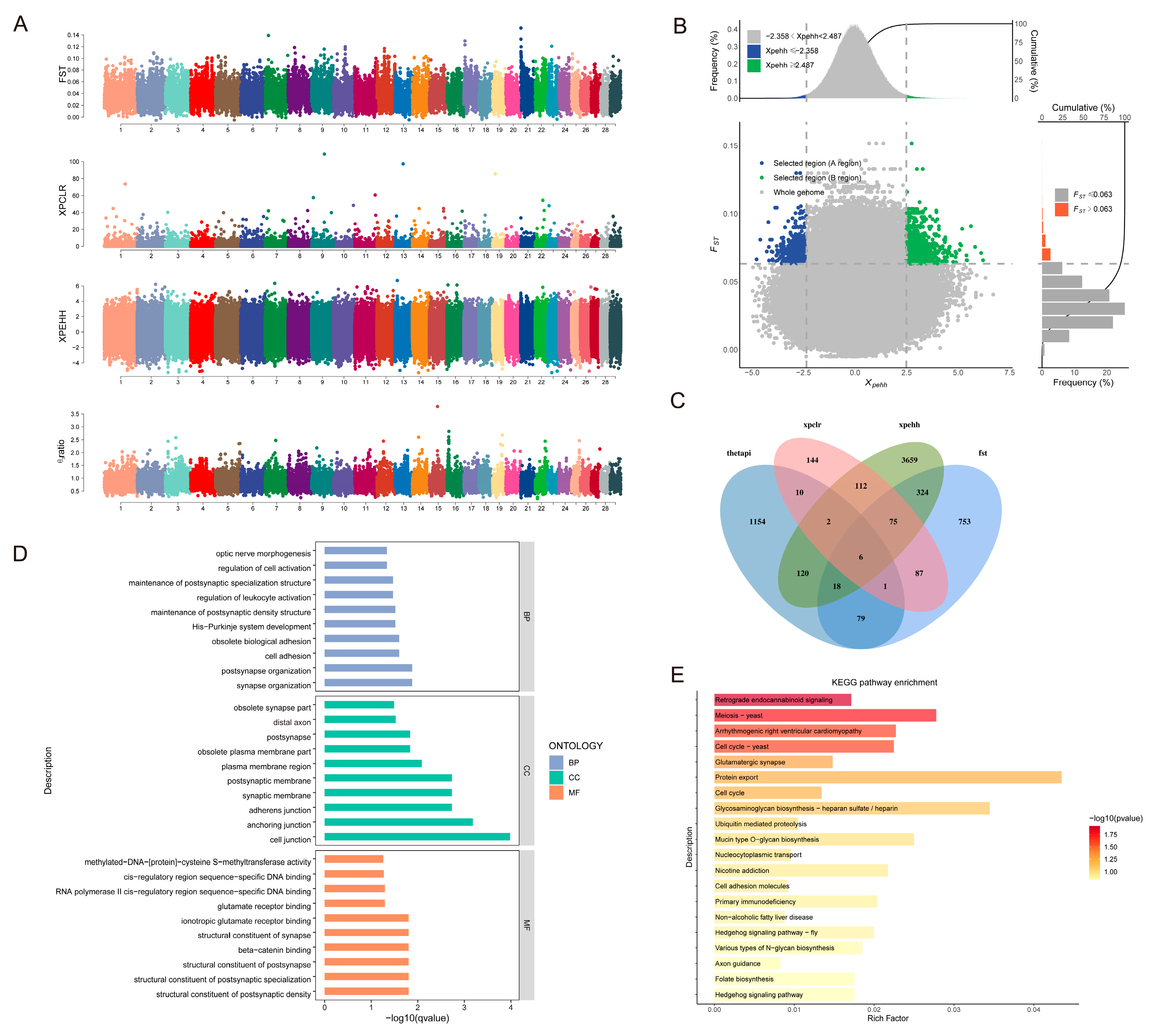
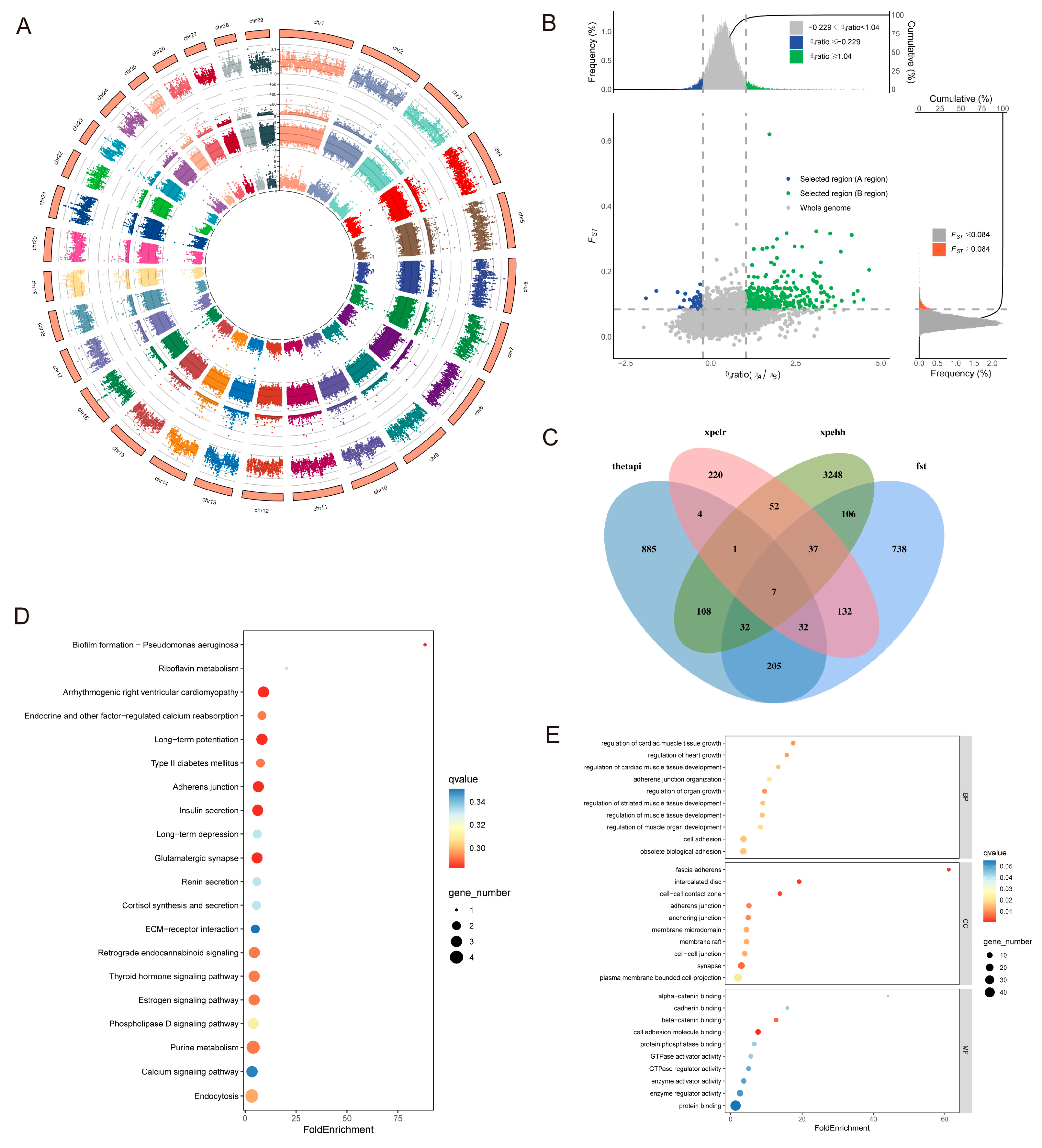
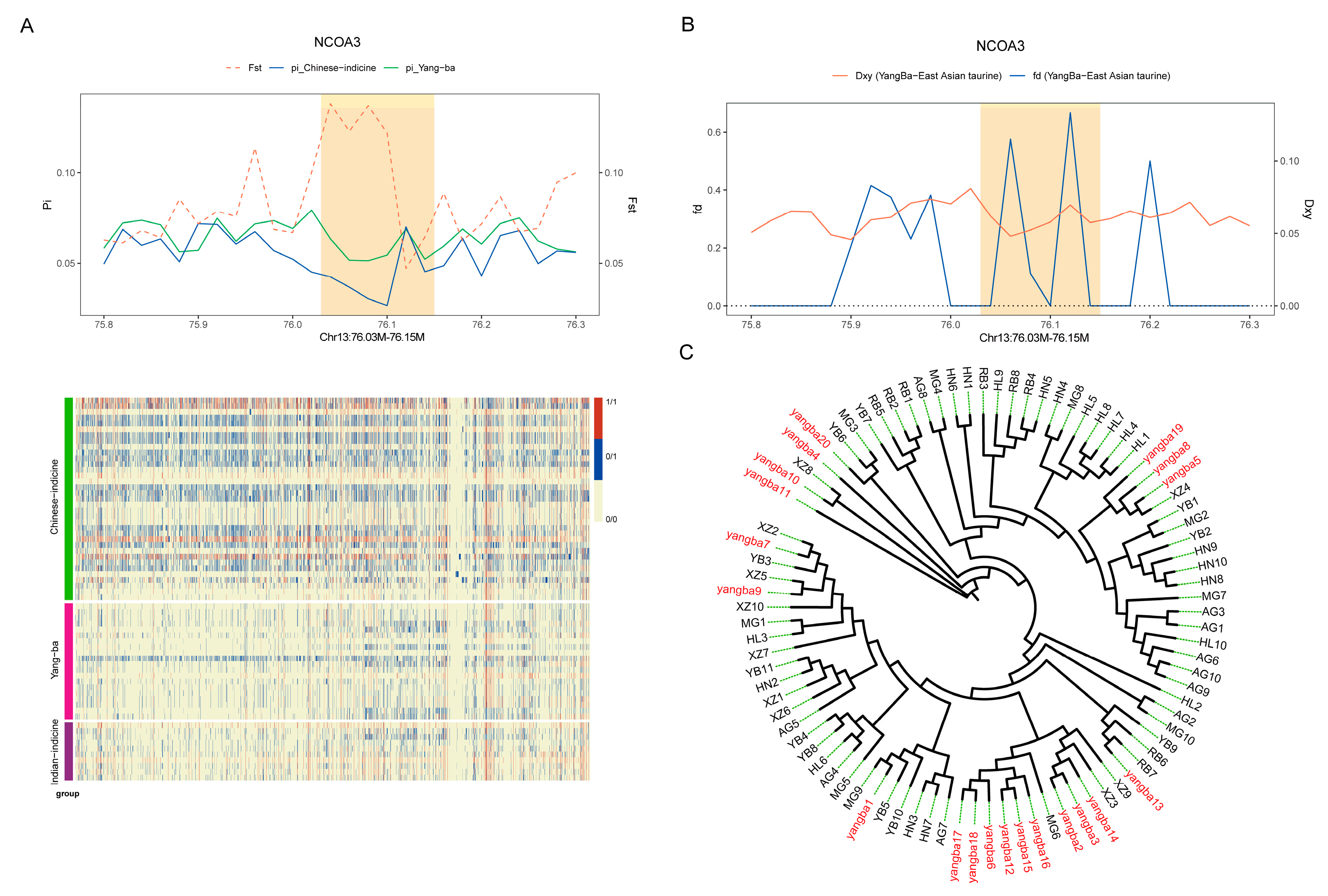
3.4. Analysis of Selection Signals
3.4.1. Adaptive Analysis of Indicine Lineage
3.4.2. Adaptive Analysis of Taurine Lineage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SNP | Single nucleotide polymorphism |
| MAF | Minor allele frequency |
| WGS | Whole-genome sequencing |
| θπ | Nucleotide diversity |
| XP-EHH | Cross-population extended haplotype homozygosity |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| NJ | Neighbor-joining |
| PCA | Principal component analysis |
| ROH | Runs of homozygosity |
| LD | Linkage disequilibrium |
| Fst | Fixation index |
| fd | Gene flow values |
| Dxy | Absolute divergence |
| AG | Angus cattle |
| HL | Hereford cattle |
| ZX | Jersey cattle |
| LM | Limousin cattle |
| XM | Simmental cattle |
| MG | Mongolian cattle |
| XZ | Tibetan cattle |
| YB | Yanbian cattle |
| HN | Korean cattle |
| RB | Japanese cattle |
| QC | Qinchuan cattle |
| LX | Luxi cattle |
| Yang Ba | Yangba cattle |
| NY | Nanyang cattle |
| BS | Bashan cattle |
| DZ | Dianzhong cattle |
| BH | Indian cattle |
| WS | Wenshan cattle |
| JJ | Jinjiang cattle |
| WN | Wannan cattle |
| LQ | Leiqiong cattle |
References
- Xia, X.T.; Achilli, A.; Lenstra, J.A.; Tong, B.; Ma, Y.; Huang, Y.Z.; Han, J.L.; Sun, Z.Y.; Chen, H.; Lei, C.Z.; et al. Mitochondrial genomes from modern and ancient Turano-Mongolian cattle reveal an ancient diversity of taurine maternal lineages in East Asia. Heredity 2021, 126, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Gautier, M.; Ding, X.; Zhang, H.; Wang, Y.; Wang, X.; Faruque, M.O.; Li, J.; Ye, S.; Gou, X.; et al. Species composition and environmental adaptation of indigenous Chinese cattle. Sci. Rep. 2017, 7, 16196. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, L.; Yang, L.; Zhao, G.; Li, J.; Liu, D.; Li, Y. Discovery of Genomic Characteristics and Selection Signatures in Southern Chinese Local Cattle. Front. Genet. 2020, 11, 533052. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Zhao, L.; Wang, J.; Jiang, Q.; Ju, Z.; Wang, X.; Yang, C.; Gao, Y.; Wei, X.; Zhang, Y.; et al. Signatures of selection in indigenous Chinese cattle genomes reveal adaptive genes and genetic variations to cold climate. J. Anim. Sci. 2023, 101, skad006. [Google Scholar] [CrossRef]
- Xu, L.; Yang, L.; Zhu, B.; Zhang, W.; Wang, Z.; Chen, Y.; Zhang, L.; Gao, X.; Gao, H.; Liu, G.E.; et al. Genome-wide scan reveals genetic divergence and diverse adaptive selection in Chinese local cattle. BMC Genomics 2019, 20, 494. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.; Yao, T.; Chen, Z.; Huangfu, R.; Cheng, H.; Ma, W.; Qi, X.; Li, F.; Chen, N.; Lei, C. Genomic characterization of dryland adaptation in endangered Anxi cattle in China. Anim. Genet. 2024, 55, 352–361. [Google Scholar] [CrossRef]
- Wang, T.; Ma, X.; Ma, C.; Wu, X.; ZhaXi, T.; Yin, L.; Li, W.; Li, Y.; Liang, C.; Yan, P. Whole genome resequencing-based analysis of plateau adaptation in Meiren yak (Bos grunniens). Anim. Biotechnol. 2024, 35, 2298406. [Google Scholar] [CrossRef]
- Jiang, L.; Kon, T.; Chen, C.; Ichikawa, R.; Zheng, Q.; Pei, L.; Takemura, I.; Nsobi, L.H.; Tabata, H.; Pan, H.; et al. Whole-genome sequencing of endangered Zhoushan cattle suggests its origin and the association of MC1R with black coat colour. Sci. Rep. 2021, 11, 17359. [Google Scholar] [CrossRef]
- Wolf, M.J.; Neumann, G.B.; Kokuc, P.; Yin, T.; Brockmann, G.A.; Konig, S.; May, K. Genetic evaluations for endangered dual-purpose German Black Pied cattle using 50K SNPs, a breed-specific 200K chip, and whole-genome sequencing. J. Dairy Sci. 2023, 106, 3345–3358. [Google Scholar] [CrossRef]
- Harish, A.; Lopes Pinto, F.A.; Eriksson, S.; Johansson, A.M. Genetic diversity and recent ancestry based on whole-genome sequencing of endangered Swedish cattle breeds. BMC Genomics 2024, 25, 89. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, H.; Lai, W.; Hu, M.; Zhang, Y.; Bai, C.; Liu, J.; Ren, H.; Li, F.; Yan, S. Genome-wide re-sequencing reveals population structure and genetic diversity of Bohai Black cattle. Anim. Genet. 2022, 53, 133–136. [Google Scholar] [CrossRef]
- Xia, X.; Zhang, S.; Zhang, H.; Zhang, Z.; Chen, N.; Li, Z.; Sun, H.; Liu, X.; Lyu, S.; Wang, X.; et al. Assessing genomic diversity and signatures of selection in Jiaxian Red cattle using whole-genome sequencing data. BMC Genomics 2021, 22, 43. [Google Scholar] [CrossRef]
- Yu, H.; Zhang, K.; Cheng, G.; Mei, C.; Wang, H.; Zan, L. Genome-wide analysis reveals genomic diversity and signatures of selection in Qinchuan beef cattle. BMC Genomics 2024, 25, 558. [Google Scholar] [CrossRef]
- Cheng, H.; Lyu, Y.; Liu, Z.; Li, C.; Qu, K.; Li, S.; Ahmed, Z.; Ma, W.; Qi, X.; Chen, N.; et al. A Whole-Genome Scan Revealed Genomic Features and Selection Footprints of Mengshan Cattle. Genes 2024, 15, 1113. [Google Scholar] [CrossRef]
- Peripolli, E.; Reimer, C.; Ha, N.T.; Geibel, J.; Machado, M.A.; Panetto, J.; do Egito, A.A.; Baldi, F.; Simianer, H.; da Silva, M. Genome-wide detection of signatures of selection in indicine and Brazilian locally adapted taurine cattle breeds using whole-genome re-sequencing data. BMC Genomics 2020, 21, 624. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Tarasov, A.; Vilella, A.J.; Cuppen, E.; Nijman, I.J.; Prins, P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics 2015, 31, 2032–2034. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Narasimhan, V.; Danecek, P.; Scally, A.; Xue, Y.; Tyler-Smith, C.; Durbin, R. BCFtools/RoH: A hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics 2016, 32, 1749–1751. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics 2011, 12, 246. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Malinsky, M.; Matschiner, M.; Svardal, H. Dsuite—Fast D-statistics and related admixture evidence from VCF files. Mol. Ecol. Resour. 2021, 21, 584–595. [Google Scholar] [CrossRef]
- Petr, M.; Vernot, B.; Kelso, J. admixr-R package for reproducible analyses using ADMIXTOOLS. Bioinformatics 2019, 35, 3194–3195. [Google Scholar] [CrossRef]
- Szpiech, Z.A.; Hernandez, R.D. selscan: An efficient multithreaded program to perform EHH-based scans for positive selection. Mol. Biol. Evol. 2014, 31, 2824–2827. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Chen, N.; Cai, Y.; Chen, Q.; Li, R.; Wang, K.; Huang, Y.; Hu, S.; Huang, S.; Zhang, H.; Zheng, Z.; et al. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat. Commun. 2018, 9, 2337. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Qu, K.; Liu, Y.; Ma, X.; Chen, N.; Zhang, J.; Huang, B.; Lei, C. Assessing genomic diversity and selective pressures in Bashan cattle by whole-genome sequencing data. Anim. Biotechnol. 2023, 34, 835–846. [Google Scholar] [CrossRef]
- Lyu, Y.; Ren, Y.; Qu, K.; Quji, S.; Zhuzha, B.; Lei, C.; Chen, N. Local ancestry and selection in admixed Sanjiang cattle. Stress Biol. 2023, 3, 30. [Google Scholar] [CrossRef]
- Zhao, L.; Li, F.; Yuan, L.; Zhang, X.; Zhang, D.; Li, X.; Zhang, Y.; Zhao, Y.; Song, Q.; Wang, J.; et al. Expression of ovine CTNNA3 and CAP2 genes and their association with growth traits. Gene 2022, 807, 145949. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, Q.; Zhang, X.; Li, C.; Zhang, D.; Li, G.; Zhang, Y.; Zhao, Y.; Shi, Z.; Wang, W.; et al. Whole-Genome Resequencing to Study Brucellosis Susceptibility in Sheep. Front. Genet. 2021, 12, 653927. [Google Scholar] [CrossRef]
- Wang, Y.; Gan, X.; Cheng, X.; Jia, Y.; Wang, G.; Tang, X.; Du, H.; Li, X.; Liu, X.; Xing, X.; et al. ABCC2 induces metabolic vulnerability and cellular ferroptosis via enhanced glutathione efflux in gastric cancer. Clin. Transl. Med. 2024, 14, e1754. [Google Scholar] [CrossRef]
- Xie, K.; Ning, C.; Yang, A.; Zhang, Q.; Wang, D.; Fan, X. Resequencing Analyses Revealed Genetic Diversity and Selection Signatures during Rabbit Breeding and Improvement. Genes 2024, 15, 433. [Google Scholar] [CrossRef]
- Aboul-Naga, A.M.; Alsamman, A.M.; El Allali, A.; Elshafie, M.H.; Abdelal, E.S.; Abdelkhalek, T.M.; Abdelsabour, T.H.; Mohamed, L.G.; Hamwieh, A. Genome-wide analysis identified candidate variants and genes associated with heat stress adaptation in Egyptian sheep breeds. Front. Genet. 2022, 13, 898522. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, G.; Yang, Y.; Yang, J.; Qin, B.; Gu, L. miR-217-5p regulates myogenesis in skeletal muscle stem cells by targeting FGFR2. Mol. Med. Rep. 2020, 22, 850–858. [Google Scholar] [CrossRef]
- Lee, S.W.; Won, J.Y.; Yang, J.; Lee, J.; Kim, S.Y.; Lee, E.J.; Kim, H.S. AKAP6 inhibition impairs myoblast differentiation and muscle regeneration: Positive loop between AKAP6 and myogenin. Sci. Rep. 2015, 5, 16523. [Google Scholar] [CrossRef] [PubMed]
- Ash, G.I.; Kostek, M.A.; Lee, H.; Angelopoulos, T.J.; Clarkson, P.M.; Gordon, P.M.; Moyna, N.M.; Visich, P.S.; Zoeller, R.F.; Price, T.B.; et al. Glucocorticoid Receptor (NR3C1) Variants Associate with the Muscle Strength and Size Response to Resistance Training. PLoS ONE 2016, 11, e0148112. [Google Scholar] [CrossRef][Green Version]
- Sayson, S.L.; Fan, J.N.; Ku, C.L.; Lo, J.F.; Chou, S.H. DNAJA3 regulates B cell development and immune function. Biomed. J. 2024, 47, 100628. [Google Scholar] [CrossRef] [PubMed]
- Serranito, B.; Cavalazzi, M.; Vidal, P.; Taurisson-Mouret, D.; Ciani, E.; Bal, M.; Rouvellac, E.; Servin, B.; Moreno-Romieux, C.; Tosser-Klopp, G.; et al. Local adaptations of Mediterranean sheep and goats through an integrative approach. Sci. Rep. 2021, 11, 21363. [Google Scholar] [CrossRef]
- Lu, X.; Abdalla, I.M.; Nazar, M.; Fan, Y.; Zhang, Z.; Wu, X.; Xu, T.; Yang, Z. Genome-Wide Association Study on Reproduction-Related Body-Shape Traits of Chinese Holstein Cows. Animals 2021, 11, 1927. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lu, J.; Wang, Y.; Liu, Z.; Li, D.; Deng, K.; Zhang, G.; Zhao, B.; You, P.; Fan, Y.; et al. Genome-Wide Scans for Selection Signatures in Haimen Goats Reveal Candidate Genes Associated with Growth Traits. Biology 2025, 14, 40. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Z.; Fang, Y.; Cao, C.; Zhang, Z.; Pan, Y.; Wang, Q. Runs of Homozygosity Revealed Reproductive Traits of Hu Sheep. Genes 2022, 13, 1848. [Google Scholar] [CrossRef]
- Li, Y.; Liang, J.; Dang, H.; Zhang, R.; Chen, P.; Shao, Y. NCOA3 is a critical oncogene in thyroid cancer via the modulation of major signaling pathways. Endocrine 2022, 75, 149–158. [Google Scholar] [CrossRef]
- de Semir, D.; Bezrookove, V.; Nosrati, M.; Dar, A.A.; Miller, J.R., 3rd; Leong, S.P.; Kim, K.B.; Liao, W.; Soroceanu, L.; McAllister, S.; et al. Nuclear Receptor Coactivator NCOA3 Regulates UV Radiation-Induced DNA Damage and Melanoma Susceptibility. Cancer Res. 2021, 81, 2956–2969. [Google Scholar] [CrossRef]
- Wen, Z.; Zhu, H.; Zhang, A.; Lin, J.; Zhang, G.; Liu, D.; Xiao, Y.; Ye, C.; Sun, D.; Wu, B.; et al. Cdc14a has a role in spermatogenesis, sperm maturation and male fertility. Exp. Cell Res. 2020, 395, 112178. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sun, L.; Sun, J.; Jiang, H. Genome-wide association research on the reproductive traits of Qianhua Mutton Merino sheep. Anim. Biosci. 2024, 37, 1535–1547. [Google Scholar] [CrossRef] [PubMed]
- Khalkhali-Evrigh, R.; Hedayat, N.; Ming, L.; Jirimutu. Identification of selection signatures in Iranian dromedary and Bactrian camels using whole genome sequencing data. Sci. Rep. 2022, 12, 9653. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, B.; Kang, Y.; Ding, Z.; Guo, S.; Cao, M.; Hu, L.; Zhang, B.; Wang, X.; Pei, J.; Ge, Q.; et al. Genomic Characterization of Crossbred-Driven Adaptation in the Endangered Yangba Cattle of China. Animals 2025, 15, 1065. https://doi.org/10.3390/ani15071065
Cai B, Kang Y, Ding Z, Guo S, Cao M, Hu L, Zhang B, Wang X, Pei J, Ge Q, et al. Genomic Characterization of Crossbred-Driven Adaptation in the Endangered Yangba Cattle of China. Animals. 2025; 15(7):1065. https://doi.org/10.3390/ani15071065
Chicago/Turabian StyleCai, Bao, Yandong Kang, Ziqiang Ding, Shaoke Guo, Mengli Cao, Liyan Hu, Ben Zhang, Xingdong Wang, Jie Pei, Qianyun Ge, and et al. 2025. "Genomic Characterization of Crossbred-Driven Adaptation in the Endangered Yangba Cattle of China" Animals 15, no. 7: 1065. https://doi.org/10.3390/ani15071065
APA StyleCai, B., Kang, Y., Ding, Z., Guo, S., Cao, M., Hu, L., Zhang, B., Wang, X., Pei, J., Ge, Q., Xiong, L., Wu, X., & Guo, X. (2025). Genomic Characterization of Crossbred-Driven Adaptation in the Endangered Yangba Cattle of China. Animals, 15(7), 1065. https://doi.org/10.3390/ani15071065





