Genome-Wide Identification and Evolutionary Analysis of Ionotropic Receptors Gene Family: Insights into Olfaction Ability Evolution and Antennal Expression Patterns in Oratosquilla oratoria
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. Genomic Data Acquisition
2.3. Sequence Search and Chromosomal Mapping Analysis of the IR Gene Family in O. oratoria
2.4. Physicochemical Properties and Subcellular Localization Analysis of the IR Gene Family
2.5. Multiple Sequence Alignment and Phylogenetic Tree Construction Analysis of the IR Gene Family
2.6. Characterization of the IR Gene Family: Motif, Gene Structure, and Domain Prediction Analysis
2.7. Protein–Protein Interaction (PPI) Network Analysis of the IR Gene Family
2.8. Fluorescence In Situ Hybridization of the IR Gene Family
2.9. RT-qPCR Validation of the Expression of Five OratIRs in O. oratoria
3. Results
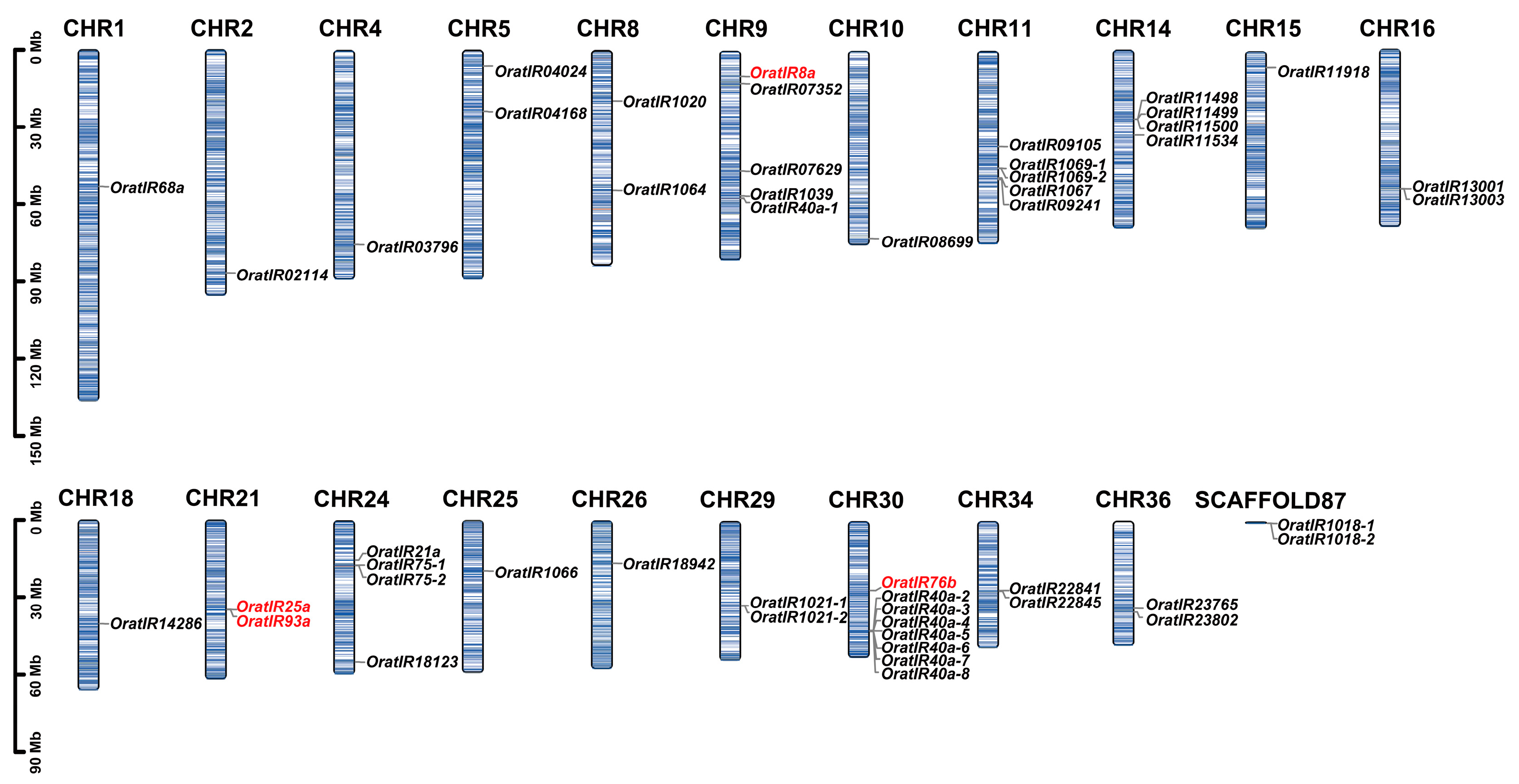
3.1. Identification, Classification, and Chromosomal Distribution of the IR Gene Family in O. oratoria
3.2. Physicochemical Properties and Subcellular Localization Analysis of the IR Gene Family
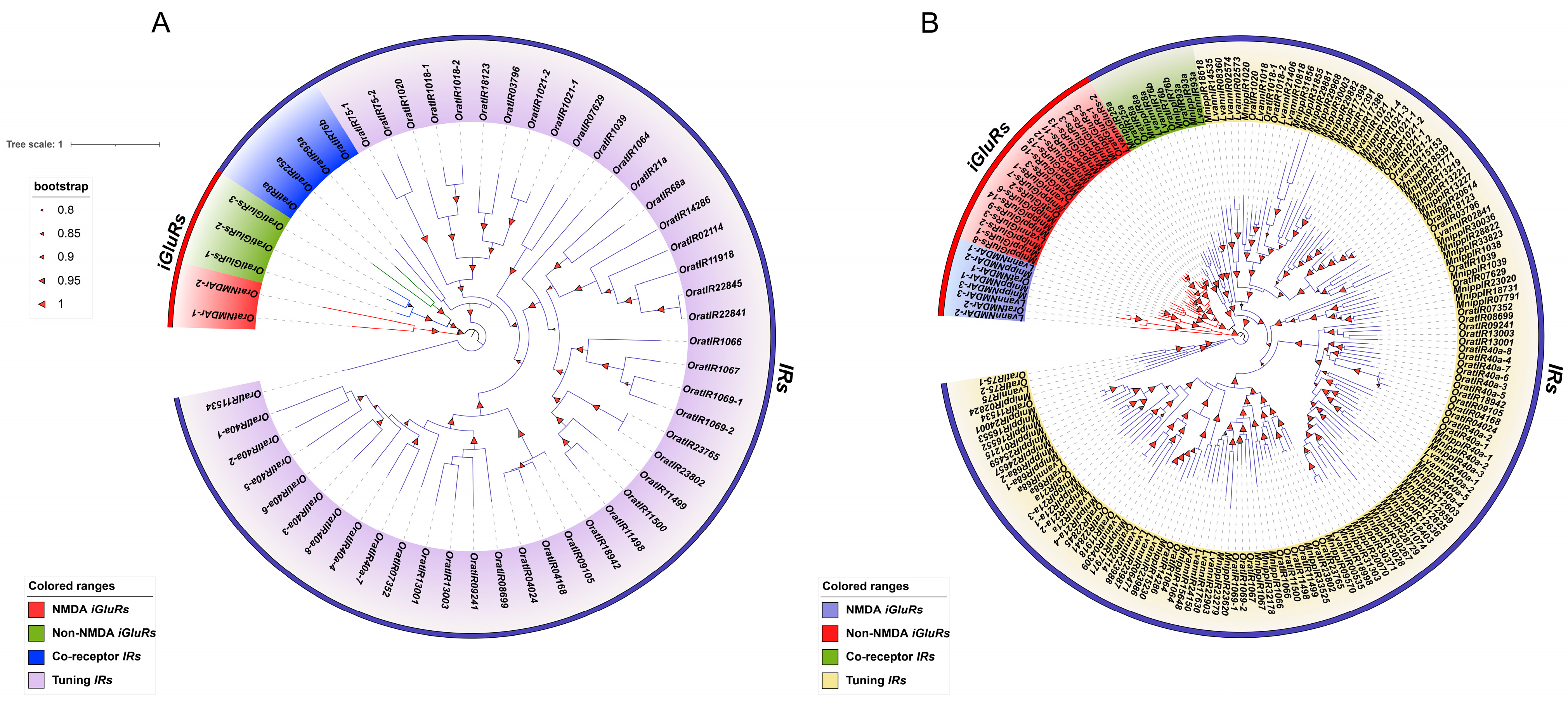
3.3. Interspecific and Intraspecific Phylogenetic Analysis of the IR Gene Family
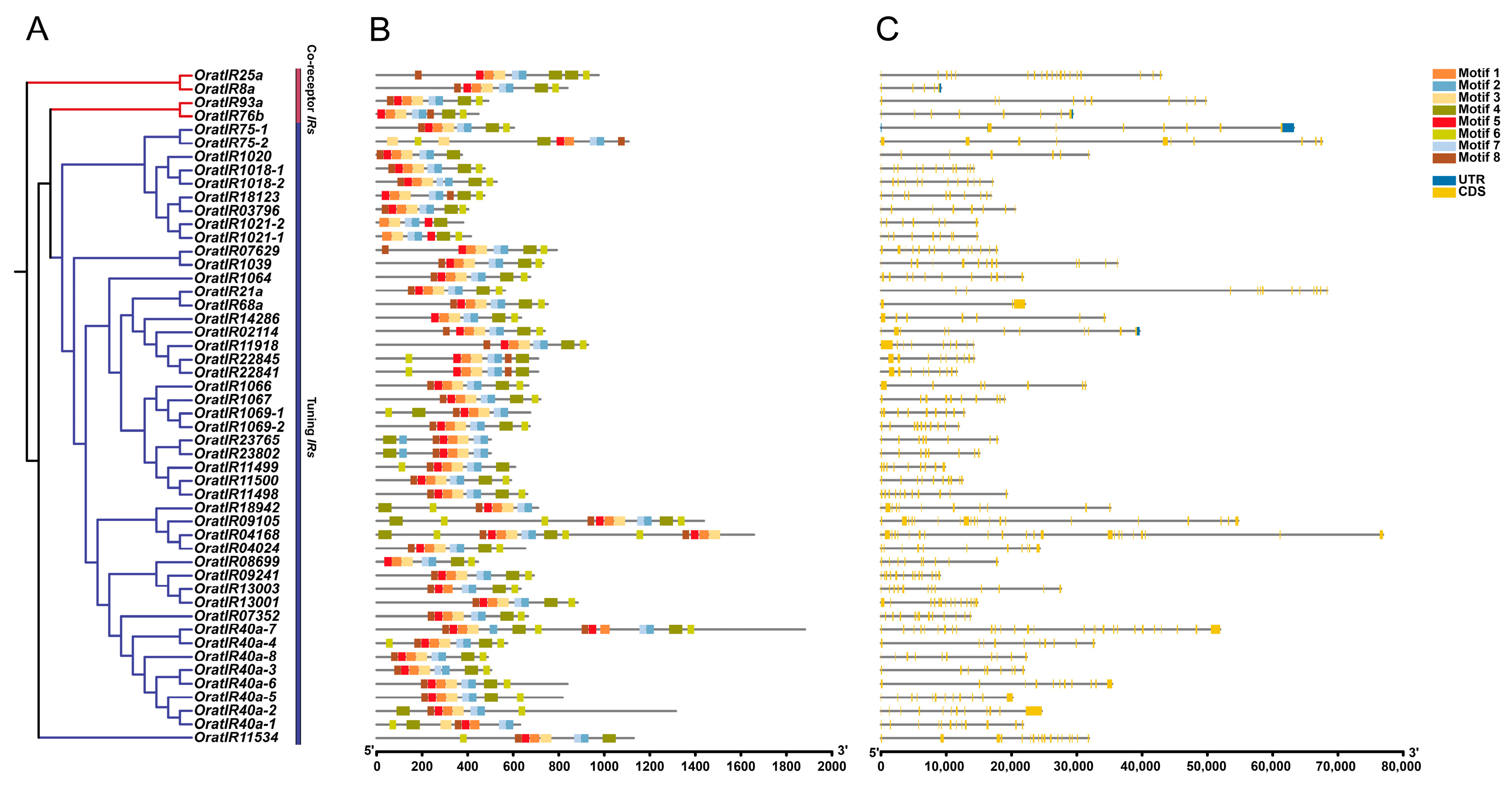
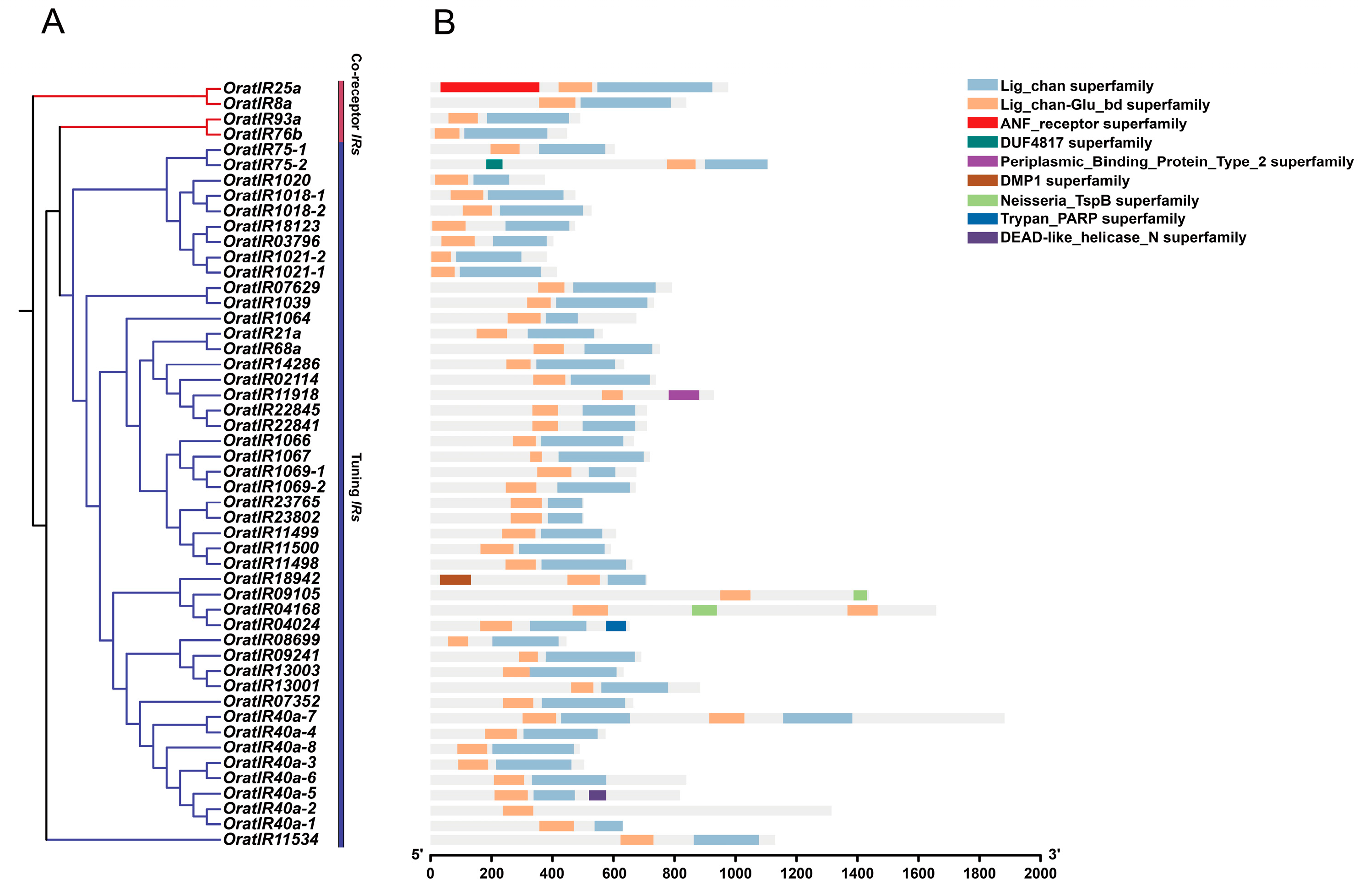
3.4. Motif, Gene Structure, and Domain Prediction Analysis of the IR Gene Family
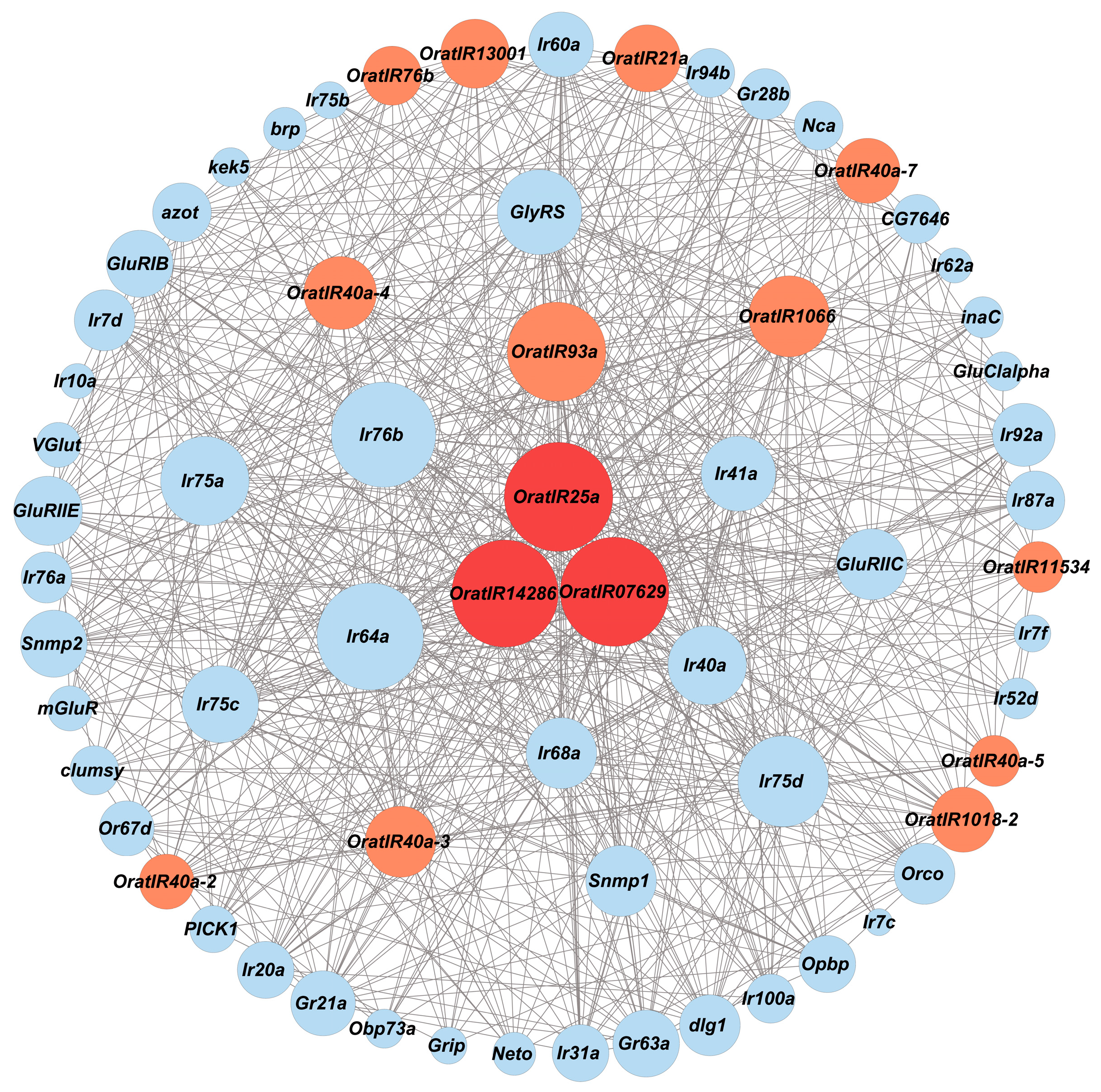
3.5. Protein Interaction Network of Genes Related to IRs
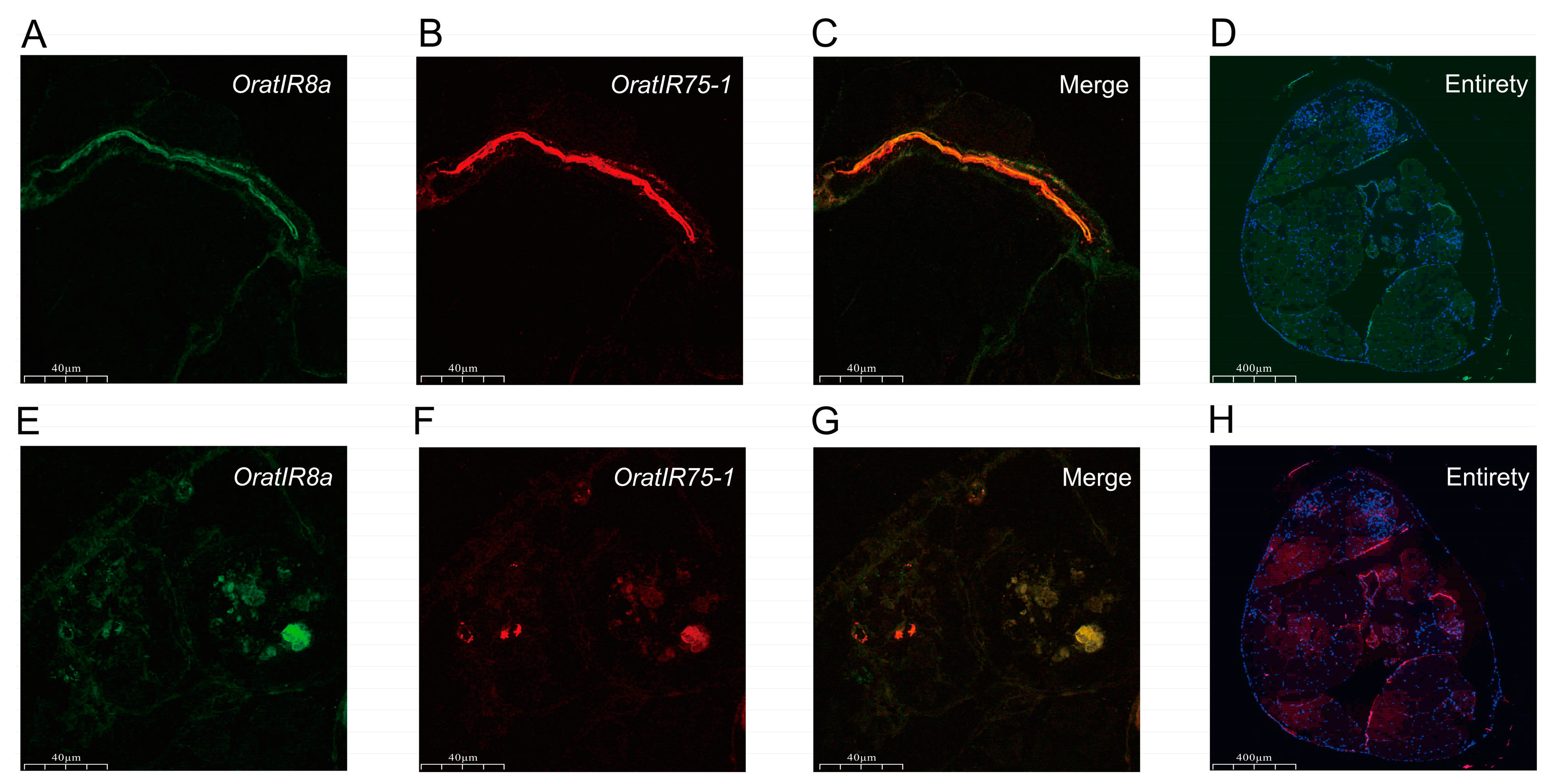
3.6. FISH Detection of OratIR8a and OratIR75-1
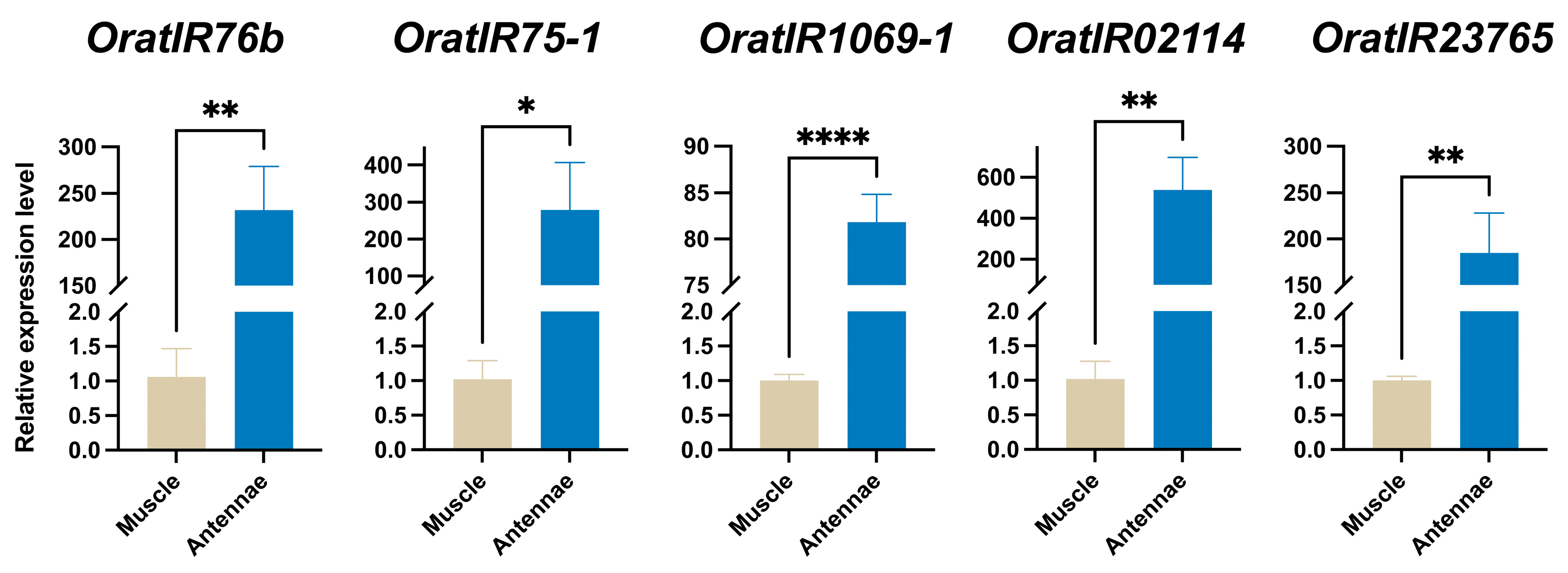
3.7. Expression Levels of Five OratIRs in O. oratoria Muscle and Tentacles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chivers, D.P.; Brown, G.E.; Smith, R.J.F. The Evolution of Chemical Alarm Signals: Attracting Predators Benefits Alarm Signal Senders. Am. Nat. 1996, 148, 649–659. [Google Scholar] [CrossRef]
- Derby, C.D. Learning from Spiny Lobsters about Chemosensory Coding of Mixtures. Physiol. Behav. 2000, 69, 203–209. [Google Scholar] [CrossRef]
- Harzsch, S.; Krieger, J. Crustacean Olfactory Systems: A Comparative Review and a Crustacean Perspective on Olfaction in Insects. Prog. Neurobiol. 2018, 161, 23–60. [Google Scholar] [CrossRef]
- Derby, C.D. Structure and Function of Cuticular Sensilla of the Lobster Homarus americanus. J. Crustac. Biol. 1982, 2, 1–21. [Google Scholar] [CrossRef]
- Wyatt, T.D. Fifty Years of Pheromones. Nature 2009, 457, 262–263. [Google Scholar] [CrossRef]
- Su, C.Y.; Menuz, K.; Carlson, J.R. Olfactory Perception: Receptors, Cells, and Circuits. Cell 2009, 139, 45–59. [Google Scholar] [CrossRef]
- Atema, J. Chemical Signals in the Marine Environment: Dispersal, Detection, and Temporal Signal Analysis. Proc. Natl. Acad. Sci. USA 1995, 92, 62–66. [Google Scholar] [CrossRef]
- Joseph, R.M.; Carlson, J.R. Drosophila Chemoreceptors: A Molecular Interface between the Chemical World and the Brain. Trends Genet. 2015, 31, 683–695. [Google Scholar] [CrossRef]
- Kamio, M.; Derby, C.D. Finding Food: How Marine Invertebrates Use Chemical Cues to Track and Select Food. Nat. Prod. Rep. 2017, 34, 514–528. [Google Scholar] [CrossRef]
- Bhandawat, V.; Reisert, J.; Yau, K.W. Elementary Response of Olfactory Receptor Neurons to Odorants. Science 2005, 308, 1931–1934. [Google Scholar] [CrossRef]
- Fulton, K.A.; Zimmerman, D.; Samuel, A.; Vogt, K.; Datta, S.R. Common Principles for Odour Coding across Vertebrates and Invertebrates. Nat. Rev. Neurosci. 2024, 25, 453–472. [Google Scholar] [CrossRef]
- Touhara, K.; Vosshall, L.B. Sensing Odorants and Pheromones with Chemosensory Receptors. Annu. Rev. Physiol. 2009, 71, 307–332. [Google Scholar] [CrossRef]
- Abuin, L.; Bargeton, B.; Ulbrich, M.H.; Isacoff, E.Y.; Kellenberger, S.; Benton, R. Functional Architecture of Olfactory Ionotropic Glutamate Receptors. Neuron 2011, 69, 44–60. [Google Scholar] [CrossRef]
- Benton, R.; Vannice, K.S.; Gomez-Diaz, C.; Vosshall, L.B. Variant Ionotropic Glutamate Receptors as Chemosensory Receptors in Drosophila. Cell 2009, 136, 149–162. [Google Scholar] [CrossRef]
- Croset, V.; Rytz, R.; Cummins, S.F.; Budd, A.; Brawand, D.; Kaessmann, H.; Gibson, T.J.; Benton, R. Ancient Protostome Origin of Chemosensory Ionotropic Glutamate Receptors and the Evolution of Insect Taste and Olfaction. PLoS Genet. 2010, 6, e1001064. [Google Scholar] [CrossRef]
- Rytz, R.; Croset, V.; Benton, R. Ionotropic Receptors (IRs): Chemosensory Ionotropic Glutamate Receptors in Drosophila and Beyond. Insect Biochem. Mol. Biol. 2013, 43, 888–897. [Google Scholar] [CrossRef]
- Chen, C.; Buhl, E.; Xu, M.; Croset, V.; Rees, J.S.; Lilley, K.S.; Benton, R.; Hodge, J.J.L.; Stanewsky, R. Drosophila Ionotropic Receptor 25a Mediates Circadian Clock Resetting by Temperature. Nature 2015, 527, 516–520. [Google Scholar] [CrossRef]
- Mayer, M.L. Glutamate Receptors at Atomic Resolution. Nature 2006, 440, 456–462. [Google Scholar] [CrossRef]
- Fleischer, J.; Pregitzer, P.; Breer, H.; Krieger, J. Access to the Odor World: Olfactory Receptors and Their Role for Signal Transduction in Insects. Cell Mol. Life Sci. 2017, 75, 485–508. [Google Scholar] [CrossRef]
- Abuin, L.; Prieto-Godino, L.L.; Pan, H.; Gutierrez, C.; Huang, L.; Jin, R.; Benton, R. In Vivo Assembly and Trafficking of Olfactory Ionotropic Receptors. BMC Biol. 2019, 17, 34. [Google Scholar] [CrossRef]
- van Giesen, L.; Garrity, P.A. More than Meets the IR: The Expanding Roles of Variant Ionotropic Glutamate Receptors in Sensing Odor, Taste, Temperature and Moisture. F1000Research 2017, 6, 1753. [Google Scholar] [CrossRef] [PubMed]
- Robertson, H.M. Molecular Evolution of the Major Arthropod Chemoreceptor Gene Families. Annu. Rev. Entomol. 2019, 64, 227–242. [Google Scholar] [CrossRef]
- Sánchez-Alcañiz, J.A.; Silbering, A.F.; Croset, V.; Zappia, G.; Sivasubramaniam, A.K.; Abuin, L.; Sahai, S.Y.; Münch, D.; Steck, K.; Auer, T.O.; et al. An Expression Atlas of Variant Ionotropic Glutamate Receptors Identifies a Molecular Basis of Carbonation Sensing. Nat. Commun. 2018, 9, 4252. [Google Scholar] [CrossRef] [PubMed]
- Silbering, A.F.; Rytz, R.; Grosjean, Y.; Abuin, L.; Ramdya, P.; Jefferis, G.S.; Benton, R. Complementary Function and Integrated Wiring of the Evolutionarily Distinct Drosophila Olfactory Subsystems. J. Neurosci. 2011, 31, 13357–13375. [Google Scholar] [CrossRef]
- Wicher, D.; Miazzi, F. Functional Properties of Insect Olfactory Receptors: Ionotropic Receptors and Odorant Receptors. Cell Tissue Res. 2021, 383, 7–19. [Google Scholar] [CrossRef]
- Ni, L.; Klein, M.; Svec, K.V.; Budelli, G.; Chang, E.C.; Ferrer, A.J.; Benton, R.; Samuel, A.D.; Garrity, P.A. The Ionotropic Receptors IR21a and IR25a Mediate Cool Sensing in Drosophila. eLife 2016, 5, e13254. [Google Scholar] [CrossRef]
- Enjin, A.; Zaharieva, E.E.; Frank, D.D.; Mansourian, S.; Suh, G.S.; Gallio, M.; Stensmyr, M.C. Humidity Sensing in Drosophila. Curr. Biol. 2016, 26, 1352–1358. [Google Scholar] [CrossRef]
- Knecht, Z.A.; Silbering, A.F.; Ni, L.; Klein, M.; Budelli, G.; Bell, R.; Abuin, L.; Ferrer, A.J.; Samuel, A.D.; Benton, R.; et al. Distinct Combinations of Variant Ionotropic Glutamate Receptors Mediate Thermosensation and Hygrosensation in Drosophila. eLife 2016, 5, e17879. [Google Scholar] [CrossRef]
- Knecht, Z.A.; Silbering, A.F.; Cruz, J.; Yang, L.; Croset, V.; Benton, R.; Garrity, P.A. Ionotropic Receptor-Dependent Moist and Dry Cells Control Hygrosensation in Drosophila. eLife 2017, 6, e26654. [Google Scholar] [CrossRef]
- Groh, K.C.; Vogel, H.; Stensmyr, M.C.; Grosse-Wilde, E.; Hansson, B.S. The Hermit Crab’s Nose—Antennal Transcriptomics. Front. Neurosci. 2014, 7, 266. [Google Scholar] [CrossRef]
- Kozma, M.T.; Schmidt, M.; Ngo-Vu, H.; Sparks, S.D.; Senatore, A.; Derby, C.D. Chemoreceptor Proteins in the Caribbean Spiny Lobster, Panulirus argus: Expression of Ionotropic Receptors, Gustatory Receptors, and TRP Channels in Two Chemosensory Organs and Brain. PLoS ONE 2018, 13, e0203935. [Google Scholar] [CrossRef]
- Kozma, M.T.; Ngo-Vu, H.; Rump, M.T.; Bobkov, Y.V.; Ache, B.W.; Derby, C.D. Single Cell Transcriptomes Reveal Expression Patterns of Chemoreceptor Genes in Olfactory Sensory Neurons of the Caribbean Spiny Lobster, Panulirus argus. BMC Genom. 2020, 21, 649. [Google Scholar] [CrossRef]
- Kelly, T.R.; Fitzgibbon, Q.P.; Smith, G.G.; Banks, T.M.; Ventura, T. Tropical Rock Lobster (Panulirus ornatus) Uses Chemoreception via the Antennular Lateral Flagellum to Identify Conspecific Ecdysis. Sci. Rep. 2023, 13, 12409. [Google Scholar] [CrossRef]
- Shen, C.; Tang, D.; Zhang, Y.; Wu, L.; Luo, Y.; Tang, B.; Wang, Z. Identification of Putative Ingestion-Related Olfactory Receptor Genes in the Chinese Mitten Crab (Eriocheir Japonica Sinensis). Genes. Genom. 2021, 43, 479–490. [Google Scholar] [CrossRef]
- Kozma, M.T.; Ngo-Vu, H.; Wong, Y.Y.; Shukla, N.S.; Pawar, S.D.; Senatore, A.; Schmidt, M.; Derby, C.D. Comparison of Transcriptomes from Two Chemosensory Organs in Four Decapod Crustaceans Reveals Hundreds of Candidate Chemoreceptor Proteins. PLoS ONE 2020, 15, e0230266. [Google Scholar] [CrossRef]
- Maynou, F.; Abelló, P.; Sartor, P. A Review of the Fisheries Biology of the Mantis Shrimp, Squilla Mantis (L., 1758) (Stomatopoda, Squillidae) in the Mediterranean. Crustaceana 2004, 77, 1081–1099. [Google Scholar] [CrossRef]
- Kodama, K.; Oyama, M.; Lee, J.; Akaba, Y.; Tajima, Y.; Shimizu, T.; Shiraishi, H.; Horiguchi, T. Interannual Variation in Quantitative Relationships among Egg Production and Densities of Larvae and Juveniles of the Japanese Mantis Shrimp Oratosquilla oratoria in Tokyo Bay, Japan. Fish. Sci. 2009, 75, 875–886. [Google Scholar] [CrossRef]
- Lou, F.; Gao, T.; Han, Z. Effect of Salinity Fluctuation on the Transcriptome of the Japanese Mantis Shrimp Oratosquilla oratoria. Int. J. Biol. Macromol. 2019, 140, 1202–1213. [Google Scholar] [CrossRef]
- Sun, X.; Wang, G.; Yang, J.; Yu, W.; Xu, J.; Tang, B.; Ding, G.; Zhang, D. Whole Genome Evaluation Analysis and Preliminary Assembly of Oratosquilla oratoria (Stomatopoda: Squillidae). Mol. Biol. Rep. 2023, 50, 4165–4173. [Google Scholar] [CrossRef]
- Luo, F.; Zhou, J.; Celia, S.; Wang, L.; Tang, Y.; Han, Z.; Kang, B. How Oratosquilla oratoria Compound Eye Response to the Polarization of Light: In the Perspective of Vision Genes and Related Proteins. Int. J. Biol. Macromol. 2024, 259, 129053. [Google Scholar] [CrossRef]
- Zhang, D.; Ding, G.; Ge, B.; Zhang, H.; Tang, B. Development and Characterization of Microsatellite Loci of Oratosquilla oratoria (Crustacea: Squillidae). Conserv. Genet. Resour. 2012, 4, 147–150. [Google Scholar] [CrossRef]
- Zhang, D.; Ding, G.; Ge, B.; Zhang, H.; Tang, B.; Yang, G. Comparative Phylogeography of Two Marine Species of Crustacean: Recent Divergence and Expansion Due to Environmental Changes. Gene 2014, 550, 141–147. [Google Scholar] [CrossRef]
- Lou, F.; Gao, T.; Cai, S.; Han, Z. De Novo Assembly and Annotation of the Whole Transcriptome of Oratosquilla oratoria. Mar. Genom. 2018, 38, 17–20. [Google Scholar] [CrossRef]
- Jia, B.; Chen, J.; Yang, G.; Bi, J.; Guo, J.; Shang, K.; Wang, S.; Wu, Z.; Zhang, K. Improvement of Solubility, Gelation and Emulsifying Properties of Myofibrillar Protein from Mantis Shrimp (Oratosquilla oratoria) by Phosphorylation Modification under Low Ionic Strength of KCl. Food Chem. 2023, 403, 134497. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Sha, Z.-l. Cryptic Diversity in the Japanese Mantis Shrimp Oratosquilla oratoria (Crustacea: Squillidae): Allopatric Diversification, Secondary Contact and Hybridization. Sci. Rep. 2017, 7, 1972. [Google Scholar] [CrossRef]
- Zhang, D.; Sun, X.; Chen, L.; Lin, L.; Yin, C.; Yang, W.; Liu, J.; Liu, Q.; Zhang, H.; Jiang, S.; et al. The Chromosome-Level Genome Provides Insights into the Adaptive Evolution of the Visual System in Oratosquilla oratoria. BMC Biol. 2025, 23, 38. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, J.; Sun, Y.; Li, S.; Gao, Y.; Yu, Y.; Liu, C.; Wang, Q.; Lv, X.; Zhang, X.; et al. Penaeid Shrimp Genome Provides Insights into Benthic Adaptation and Frequent Molting. Nat. Commun. 2019, 10, 356. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Bian, C.; Jiang, S.; Han, K.; Xiong, Y.; Zhang, W.; Shi, C.; Qiao, H.; Gao, Z.; Li, R.; et al. A Chromosome-Level Genome Assembly of the Oriental River Prawn, Macrobrachium nipponense. Gigascience 2021, 10, giaa160. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as Designed by Its Users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent Updates to the Phylogenetic Tree Display and Annotation Tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional Classification of Proteins via Subfamily Domain Architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ai, M.; Blais, S.; Park, J.; Min, S.; Neubert, T.A.; Suh, G.S.B. Ionotropic Glutamate Receptors IR64a and IR8a Form a Functional Odorant Receptor Complex In Vivo in Drosophila. J. Neurosci. 2013, 33, 10741–10749. [Google Scholar] [CrossRef]
- Tang, R.; Jiang, N.-J.; Ning, C.; Li, G.-C.; Huang, L.-Q.; Wang, C.-Z. The Olfactory Reception of Acetic Acid and Ionotropic Receptors in the Oriental Armyworm, Mythimna separata Walker. Insect Biochem. Mol. Biol. 2020, 118, 103312. [Google Scholar] [CrossRef]
- Hou, X.Q.; Zhang, D.D.; Powell, D.; Wang, H.L.; Andersson, M.N.; Löfstedt, C. Ionotropic Receptors in the Turnip Moth Agrotis segetum Respond to Repellent Medium-Chain Fatty Acids. BMC Biol. 2022, 20, 34. [Google Scholar] [CrossRef]
- Ache, B.W.; Young, J.M. Olfaction: Diverse Species, Conserved Principles. Neuron 2005, 48, 417–430. [Google Scholar] [CrossRef] [PubMed]
- Hansson, B.S.; Stensmyr, M.C. Evolution of Insect Olfaction. Neuron 2011, 72, 698–711. [Google Scholar] [CrossRef]
- Koehl, M.A.; Koseff, J.R.; Crimaldi, J.P.; McCay, M.G.; Cooper, T.; Wiley, M.B.; Moore, P.A. Lobster Sniffing: Antennule Design and Hydrodynamic Filtering of Information in an Odor Plume. Science 2001, 294, 1948–1951. [Google Scholar] [CrossRef]
- Corey, E.A.; Bobkov, Y.; Ukhanov, K.; Ache, B.W. Ionotropic Crustacean Olfactory Receptors. PLoS ONE 2013, 8, e60551. [Google Scholar] [CrossRef] [PubMed]
- Groh-Lunow, K.C.; Getahun, M.N.; Grosse-Wilde, E.; Hansson, B.S. Expression of Ionotropic Receptors in Terrestrial Hermit Crab’s Olfactory Sensory Neurons. Front. Cell Neurosci. 2015, 8, 448. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, W.; Zhang, F.; Ma, C.; Liu, Z.; Yang, M.H.; Chen, W.; Li, Q.; Cui, M.; Jiang, K.; et al. A Chromosome-Level Genome of the Mud Crab (Scylla paramamosain Estampador) Provides Insights into the Evolution of Chemical and Light Perception in This Crustacean. Mol. Ecol. Resour. 2021, 21, 1299–1317. [Google Scholar] [CrossRef]
- Liu, Z.; Kou, G.; Nan, Y.; Wang, K.; Zhou, Y. Identification and Expression Profiling of Ionotropic receptors Gene in Gynaephora qinghaiensis (Lepidoptera: Lymantriidae). Acta Agrestia Sin. 2024, 32, 1382–1400. [Google Scholar] [CrossRef]
- Derby, C.D.; Sorensen, P.W. Neural Processing, Perception, and Behavioral Responses to Natural Chemical Stimuli by Fish and Crustaceans. J. Chem. Ecol. 2008, 34, 898–914. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.Y.; Xu, W.; Dong, S.L.; Zhu, J.Y.; Xu, Y.X.; Anderson, A. Genome-Wide Analysis of Ionotropic Receptor Gene Repertoire in Lepidoptera with an Emphasis on Its Functions of Helicoverpa armigera. Insect Biochem. Mol. Biol. 2018, 99, 37–53. [Google Scholar] [CrossRef]
- Wang, T.T.; Si, F.L.; He, Z.B.; Chen, B. Genome-Wide Identification, Characterization and Classification of Ionotropic Glutamate Receptor Genes (iGluRs) in the Malaria Vector Anopheles sinensis (Diptera: Culicidae). Parasites Vectors 2018, 11, 34. [Google Scholar] [CrossRef]
- Biswas, T.; Vogel, H.; Biedermann, P.H.W.; Lehenberger, M.; Yuvaraj, J.K.; Andersson, M.N. Few Chemoreceptor Genes in the Ambrosia Beetle Trypodendron lineatum May Reflect Its Specialized Ecology. BMC Genom. 2024, 25, 764. [Google Scholar] [CrossRef] [PubMed]
- Moleirinho, A.; Carneiro, J.; Matthiesen, R.; Silva, R.M.; Amorim, A.; Azevedo, L. Gains, Losses and Changes of Function after Gene Duplication: Study of the Metallothionein Family. PLoS ONE 2011, 6, e18487. [Google Scholar] [CrossRef]
- Crow, K.D.; Wagner, G.P. What Is the Role of Genome Duplication in the Evolution of Complexity and Diversity? Mol. Biol. Evol. 2006, 23, 887–892. [Google Scholar] [CrossRef]
- van Schooten, B.; Jiggins, C.D.; Briscoe, A.D.; Papa, R. Genome-Wide Analysis of Ionotropic Receptors Provides Insight into Their Evolution in Heliconius butterflies. BMC Genom. 2016, 17, 254. [Google Scholar] [CrossRef] [PubMed]
- Sobolevsky, A.I.; Rosconi, M.P.; Gouaux, E. X-Ray Structure of AMPA-Subtype Glutamate Receptor: Symmetry and Mechanism. Nature 2009, 462, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Singh, S.K.; Gu, S.; Furukawa, H.; Sobolevsky, A.I.; Zhou, J.; Jin, Y.; Gouaux, E. Crystal Structure and Association Behaviour of the GluR2 Amino-Terminal Domain. EMBO J. 2009, 28, 1812–1823. [Google Scholar] [CrossRef]
- Mayer, M.L.; Armstrong, N. Structure and Function of Glutamate Receptor Ion Channels. Annu. Rev. Physiol. 2004, 66, 161–181. [Google Scholar] [CrossRef]
- Liu, J.W. Bioinformatics Analysis of Structure and Function of OATP1B1 Protein. Mol. Plant Breed. 2023, 21, 2230–2235. [Google Scholar] [CrossRef]
- Rimal, S.; Lee, Y. The Multidimensional Ionotropic Receptors of Drosophila melanogaster. Insect Mol. Biol. 2018, 27, 1–7. [Google Scholar] [CrossRef]
- Ai, M.; Min, S.; Grosjean, Y.; Leblanc, C.; Bell, R.; Benton, R.; Suh, G.S.B. Acid Sensing by the Drosophila Olfactory System. Nature 2010, 468, 691–695. [Google Scholar] [CrossRef]
- Koh, T.; He, Z.; Gorur-Shandilya, S.; Menuz, K.; Larter, N.K.; Stewart, S.; Carlson, J.R. The Drosophila IR20a Clade of Ionotropic Receptors Are Candidate Taste and Pheromone Receptors. Neuron 2014, 83, 850–865. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.V.; Ni, J.; Montell, C. The Molecular Basis for Attractive Salt Taste Coding in Drosophila. Science 2013, 340, 1334–1338. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Zhang, M.; Üçpunar, H.K.; Svensson, T.; Quillery, E.; Gompel, N.; Ignell, R.; Grunwald Kadow, I.C. Ionotropic Chemosensory Receptors Mediate the Taste and Smell of Polyamines. PLoS Biol. 2016, 14, e1002454. [Google Scholar] [CrossRef]
- Ganguly, A.; Pang, L.; Duong, V.K.; Lee, A.; Schoniger, H.; Varady, E.; Dahanukar, A. A Molecular and Cellular Context-Dependent Role for Ir76b in Detection of Amino Acid Taste. Cell Rep. 2017, 18, 737–750. [Google Scholar] [CrossRef]
- Chen, Y.; Amrein, H. Ionotropic Receptors Mediate Drosophila Oviposition Preference through Sour Gustatory Receptor Neurons. Curr. Biol. 2017, 27, 2741–2750.e4. [Google Scholar] [CrossRef] [PubMed]
| Probe Name | Primer Sequence (5′~3′) | Usage |
|---|---|---|
| OratIR8a | CCGTCACTGATTGGTGAAGACAGG | OratIR8a in situ hybridization probe utilized FAM (488) fluorochrome for visualization, with the excitation light-emitting green fluorescence. |
| CTTGATCCAGCTCTGCTTCATTGC GAGCTGTTTGGGTCATGTCCCTGC CCAGTGACCACAAGAGCAGCAAC GGTTGTTCAGCAAAGGGCTCACC | ||
| OratIR75-1 | GGCATCACTCAAAGACTTCGGAGC | OratIR75-1 in situ hybridization probe utilized Cy3 fluorochrome for visualization, with the excitation light-emitting red fluorescence. |
| CGGAGGTGGAGCCCAGTAAGGTTC | ||
| GGTGTCGGGAAGTCGACGTACTG | ||
| GAGGTTACTCGTGTAGAAGGCCAG | ||
| CGTAGGTCCAGGAGAACATCACTG |
| Primer Name | Forward Primer (5′~3′) | Reverse Primer (5′~3′) |
|---|---|---|
| OratIR76b | GTATGGTGGGAATGGTCAA | TCGTAGTAGGCGTGGGTG |
| OratIR75-1 | TCAACTACGGAGCAGGAA | GAGAAGAAGGAAGAGGATGG |
| OratIR1069-1 | GTGGTGGTGATGATGATGA | CGTCCTTCTCCTCCTCTT |
| OratIR02114 | GATGCTGCTGATGCTACTA | TGTGAAGGAGGAGGTTGT |
| OratIR23765 | AGACTGACCACGACAACGG | GGAGTATAAGACCCAAGCACC |
| β-actin | ATCGTTCGTGACATTAAGGA | CAAGGAATGAAGGCTGGAA |
| Gene Name | Amino Acid Length (aa) | Molecular Weights (kDa) | Isoelectric Point (pI) | Instability Index | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| OratIR8a | 839 | 93.47 | 4.9 | 39.84 | 82.3 | −0.182 | Plasma membrane |
| OratIR25a | 976 | 109.73 | 4.88 | 39.43 | 83.05 | −0.178 | Plasma membrane |
| OratIR93a | 491 | 56.15 | 5.91 | 38.98 | 81.14 | −0.117 | Endoplasmic reticulum |
| OratIR76b | 448 | 52.3 | 9.08 | 37.11 | 86.14 | −0.16 | Golgi |
| OratIR21a | 565 | 62.27 | 6.57 | 42.43 | 105.88 | 0.188 | Plasma membrane |
| OratIR40a-1 | 631 | 70.84 | 5.91 | 50.29 | 92.69 | −0.208 | Plasma membrane |
| OratIR40a-2 | 1315 | 151.95 | 4.98 | 76.62 | 80.87 | −0.119 | Endoplasmic reticulum |
| OratIR40a-3 | 504 | 56.18 | 5.47 | 38.54 | 88.81 | 0.061 | Plasma membrane |
| OratIR40a-4 | 574 | 64.06 | 5.57 | 36.1 | 104.23 | 0.207 | Plasma membrane |
| OratIR40a-5 | 818 | 93.00 | 7.56 | 41.92 | 99.66 | −0.087 | Plasma membrane |
| OratIR40a-6 | 839 | 92.98 | 4.00 | 27.98 | 83.03 | −0.553 | Mitochondrial |
| OratIR40a-7 | 1882 | 219.30 | 4.22 | 87.04 | 70.65 | −0.902 | Extracellular |
| OratIR40a-8 | 489 | 54.30 | 8.61 | 38.49 | 96.67 | 0.065 | Golgi |
| OratIR68a | 752 | 85.18 | 8.37 | 44.96 | 91.9 | −0.042 | Plasma membrane |
| OratIR75-1 | 604 | 68.58 | 9.4 | 52.13 | 85.91 | −0.273 | Golgi |
| OratIR75-2 | 1107 | 127.23 | 9.32 | 56.41 | 73.21 | −0.584 | Plasma membrane |
| OratIR1018-1 | 475 | 53.49 | 7.63 | 43.45 | 97.77 | 0.176 | Plasma membrane |
| OratIR1018-2 | 528 | 58.59 | 5.35 | 47.3 | 93.41 | 0.082 | Plasma membrane |
| OratIR1020 | 375 | 42.32 | 6.38 | 31.37 | 95.76 | 0.113 | Plasma membrane |
| OratIR1021-1 | 415 | 46.31 | 8.18 | 42.58 | 91.33 | 0.028 | Plasma membrane |
| OratIR1021-2 | 381 | 42.88 | 5.65 | 45.99 | 89.55 | 0.021 | Plasma membrane |
| OratIR1039 | 733 | 83.05 | 6.79 | 41.43 | 94.13 | −0.048 | Plasma membrane |
| OratIR1064 | 675 | 75.20 | 6.41 | 37.38 | 91.41 | −0.035 | Endoplasmic reticulum |
| OratIR1066 | 667 | 76.17 | 8.32 | 40.3 | 95.13 | 0.01 | Plasma membrane |
| OratIR1067 | 720 | 82.01 | 8.01 | 44.37 | 91.51 | −0.137 | Endoplasmic reticulum |
| OratIR1069-1 | 675 | 76.12 | 4.21 | 30.81 | 75.24 | −0.577 | Golgi |
| OratIR1069-2 | 673 | 75.48 | 5.92 | 35.6 | 98.96 | −0.02 | Plasma membrane |
| OratIR02114 | 739 | 83.72 | 7.27 | 48.36 | 99.01 | 0.017 | Plasma membrane |
| OratIR03796 | 403 | 44.04 | 8.63 | 33.91 | 106.92 | 0.229 | Plasma membrane |
| OratIR04024 | 653 | 73.52 | 7.67 | 38.57 | 95.93 | −0.076 | Plasma membrane |
| OratIR04168 | 1658 | 184.00 | 6.68 | 48.18 | 87.22 | −0.222 | Plasma membrane |
| OratIR07352 | 665 | 73.96 | 5.97 | 39.36 | 96.03 | 0.070 | Vacuolar |
| OratIR07629 | 792 | 88.69 | 6.36 | 42.34 | 87.37 | −0.093 | Plasma membrane |
| OratIR08699 | 446 | 49.18 | 6.39 | 36.01 | 90.47 | −0.017 | Plasma membrane |
| OratIR09105 | 1438 | 159.58 | 7.42 | 49.87 | 87.20 | −0.248 | Plasma membrane |
| OratIR09241 | 691 | 77.18 | 8.97 | 35.37 | 100.85 | 0.045 | Mitochondrial |
| OratIR11498 | 662 | 75.03 | 5.79 | 43.36 | 92.51 | −0.026 | Plasma membrane |
| OratIR11499 | 609 | 68.88 | 8.11 | 39 | 85.81 | −0.057 | Plasma membrane |
| OratIR11500 | 591 | 66.83 | 5.71 | 34.15 | 87.11 | −0.052 | Endoplasmic reticulum |
| OratIR11534 | 1130 | 127.38 | 4.77 | 72.91 | 68.18 | −0.756 | Plasma membrane |
| OratIR11918 | 929 | 105.23 | 6.62 | 43.4 | 89.89 | −0.174 | Golgi |
| OratIR13001 | 884 | 97.37 | 5.62 | 46.85 | 91.07 | −0.147 | Plasma membrane |
| OratIR13003 | 633 | 71.53 | 6.14 | 39.38 | 102.56 | 0.089 | Plasma membrane |
| OratIR14286 | 635 | 73.69 | 5.42 | 43.88 | 89.94 | 0.026 | Endoplasmic reticulum |
| OratIR18123 | 474 | 53.44 | 6.54 | 45.6 | 90.46 | −0.185 | Plasma membrane |
| OratIR18942 | 710 | 79.4 | 6.67 | 50.78 | 90.01 | −0.183 | Plasma membrane |
| OratIR22841 | 710 | 80.31 | 5.31 | 47.53 | 91.46 | −0.083 | Plasma membrane |
| OratIR22845 | 710 | 80.27 | 5.39 | 47.6 | 91.61 | −0.081 | Plasma membrane |
| OratIR23765 | 502 | 56.35 | 6.24 | 35.34 | 90.28 | −0.034 | Plasma membrane |
| OratIR23802 | 502 | 56.3 | 6.08 | 35.85 | 90.08 | −0.027 | Plasma membrane |
| Gene Name | Gene ID | Alpha Helix (Hh) | Extended Strand (Ee) | Random Coil (Cc) |
|---|---|---|---|---|
| OratIR8a | Orat_gene07298 | 318/37.90% | 163/19.43% | 358/42.67% |
| OratIR25a | Orat_gene16135 | 349/35.76% | 198/20.29% | 429/43.95% |
| OratIR93a | Orat_gene16136 | 106/21.59% | 152/30.96% | 233/47.45% |
| OratIR76b | Orat_gene21236 | 130/29.02% | 98/21.88% | 220/49.11% |
| OratIR21a | Orat_gene17732 | 185/32.74% | 99/17.52% | 281/49.73% |
| OratIR40a-1 | Orat_gene07733 | 242/38.35% | 106/16.80% | 283/44.85% |
| OratIR40a-2 | Orat_gene21360 | 161/12.24% | 145/11.03% | 1009/76.73% |
| OratIR40a-3 | Orat_gene21361 | 151/29.96% | 121/24.01% | 232/46.03% |
| OratIR40a-4 | Orat_gene21362 | 202/35.19% | 122/21.25% | 250/43.55% |
| OratIR40a-5 | Orat_gene21363 | 270/33.01% | 182/22.25% | 366/44.74% |
| OratIR40a-6 | Orat_gene21364 | 234/27.89% | 163/19.43% | 442/52.68% |
| OratIR40a-7 | Orat_gene21367 | 887/47.13% | 264/14.03% | 731/38.84% |
| OratIR40a-8 | Orat_gene21368 | 178/36.40% | 94/19.22% | 217/44.38% |
| OratIR68a | Orat_gene00368 | 237/31.52% | 159/21.14% | 356/47.34% |
| OratIR75-1 | Orat_gene17760 | 251/41.56% | 70/11.59% | 283/46.85% |
| OratIR75-2 | Orat_gene17762 | 337/30.44% | 195/17.62% | 575/51.94% |
| OratIR1018-1 | Orat_gene29704 | 161/33.89% | 117/24.63% | 197/41.47% |
| OratIR1018-2 | Orat_gene29705 | 179/33.90% | 117/22.16% | 232/43.94% |
| OratIR1020 | Orat_gene06678 | 168/44.80% | 73/19.47% | 134/35.73% |
| OratIR1021-1 | Orat_gene20793 | 114/27.47% | 100/24.10% | 201/48.43% |
| OratIR1021-2 | Orat_gene20797 | 137/35.96% | 75/19.69% | 169/44.36% |
| OratIR1039 | Orat_gene07724 | 237/32.33% | 138/18.83% | 358/48.84% |
| OratIR1064 | Orat_gene06913 | 207/30.67% | 131/19.41% | 337/49.93% |
| OratIR1066 | Orat_gene18414 | 241/36.13% | 151/22.64% | 275/41.23% |
| OratIR1067 | Orat_gene09232 | 239/33.19% | 166/23.06% | 315/43.75% |
| OratIR1069-1 | Orat_gene09184 | 127/18.81% | 164/24.30% | 384/56.89% |
| OratIR1069-2 | Orat_gene09189 | 254/37.74% | 122/18.13% | 297/44.13% |
| OratIR02114 | Orat_gene02114 | 237/32.07% | 155/20.97% | 347/46.96% |
| OratIR03796 | Orat_gene03796 | 151/37.47% | 74/18.36% | 178/44.17% |
| OratIR04024 | Orat_gene04024 | 182/27.87% | 156/23.89% | 315/48.24% |
| OratIR04168 | Orat_gene04168 | 413/24.91% | 345/20.81% | 900/54.28% |
| OratIR07352 | Orat_gene07352 | 265/39.85% | 118/17.74% | 282/42.41% |
| OratIR07629 | Orat_gene07629 | 261/32.95% | 174/21.97% | 357/45.08% |
| OratIR08699 | Orat_gene08699 | 156/34.98% | 81/18.16% | 209/46.86% |
| OratIR09105 | Orat_gene09105 | 345/23.99% | 304/21.14% | 789/54.87% |
| OratIR09241 | Orat_gene09241 | 261/37.77% | 122/17.66% | 308/44.57% |
| OratIR11498 | Orat_gene11498 | 189/28.55% | 128/19.34% | 345/52.11% |
| OratIR11499 | Orat_gene11499 | 132/21.67% | 155/25.45% | 322/52.87% |
| OratIR11500 | Orat_gene11500 | 173/29.27% | 119/20.14% | 299/50.59% |
| OratIR11534 | Orat_gene11534 | 446/39.47% | 182/16.11% | 502/44.42% |
| OratIR11918 | Orat_gene11918 | 260/27.99% | 157/16.90% | 512/55.11% |
| OratIR13001 | Orat_gene13001 | 221/25.00% | 193/21.83% | 470/53.17% |
| OratIR13003 | Orat_gene13003 | 267/42.18% | 111/17.54% | 255/40.28% |
| OratIR14286 | Orat_gene14286 | 204/32.13% | 140/22.05% | 291/45.83% |
| OratIR18123 | Orat_gene18123 | 166/35.02% | 117/24.68% | 191/40.30% |
| OratIR18942 | Orat_gene18942 | 146/20.56% | 169/23.80% | 395/55.63% |
| OratIR22841 | Orat_gene22841 | 213/30.00% | 161/22.68% | 336/47.32% |
| OratIR22845 | Orat_gene22845 | 216/30.42% | 158/22.25% | 336/47.32% |
| OratIR23765 | Orat_gene23765 | 114/22.71% | 131/26.10% | 257/51.20% |
| OratIR23802 | Orat_gene23802 | 114/22.71% | 132/26.29% | 256/51.00% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, W.-Q.; Ding, G.; Wang, L.-L.; Yin, C.-J.; Wu, H.-Y.; Zhang, H.-B.; Liu, Q.-N.; Jiang, S.-H.; Tang, B.-P.; Wang, G.; et al. Genome-Wide Identification and Evolutionary Analysis of Ionotropic Receptors Gene Family: Insights into Olfaction Ability Evolution and Antennal Expression Patterns in Oratosquilla oratoria. Animals 2025, 15, 852. https://doi.org/10.3390/ani15060852
Yang W-Q, Ding G, Wang L-L, Yin C-J, Wu H-Y, Zhang H-B, Liu Q-N, Jiang S-H, Tang B-P, Wang G, et al. Genome-Wide Identification and Evolutionary Analysis of Ionotropic Receptors Gene Family: Insights into Olfaction Ability Evolution and Antennal Expression Patterns in Oratosquilla oratoria. Animals. 2025; 15(6):852. https://doi.org/10.3390/ani15060852
Chicago/Turabian StyleYang, Wen-Qi, Ge Ding, Lin-Lin Wang, Chi-Jie Yin, Hai-Yue Wu, Hua-Bin Zhang, Qiu-Ning Liu, Sen-Hao Jiang, Bo-Ping Tang, Gang Wang, and et al. 2025. "Genome-Wide Identification and Evolutionary Analysis of Ionotropic Receptors Gene Family: Insights into Olfaction Ability Evolution and Antennal Expression Patterns in Oratosquilla oratoria" Animals 15, no. 6: 852. https://doi.org/10.3390/ani15060852
APA StyleYang, W.-Q., Ding, G., Wang, L.-L., Yin, C.-J., Wu, H.-Y., Zhang, H.-B., Liu, Q.-N., Jiang, S.-H., Tang, B.-P., Wang, G., & Zhang, D.-Z. (2025). Genome-Wide Identification and Evolutionary Analysis of Ionotropic Receptors Gene Family: Insights into Olfaction Ability Evolution and Antennal Expression Patterns in Oratosquilla oratoria. Animals, 15(6), 852. https://doi.org/10.3390/ani15060852







