De Novo Assembly, Characterization and Comparative Transcriptome Analysis of the Mature Male and Female Gonads in Acrossocheilus parallens
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Extraction and Library Construction
2.3. Library Sequencing, De Novo Assembly and Annotation
2.4. Identification of Differentially Expressed Genes (DESs) and Enrichment Analysis
2.5. Validation of DEGs Using Quantitative Real-Time PCR (qRT-PCR)
3. Results
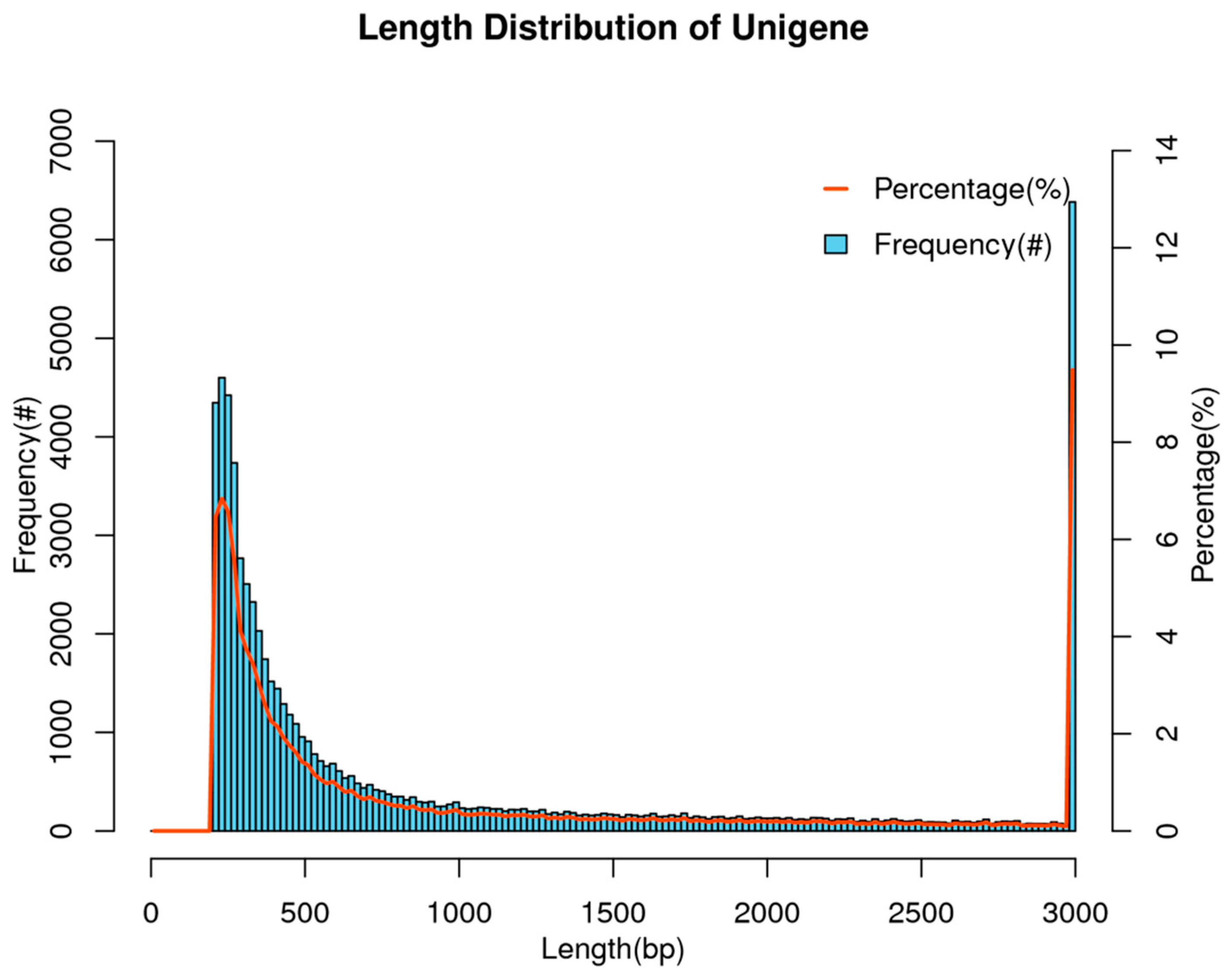
3.1. Overview of Transcriptome Assembly Results
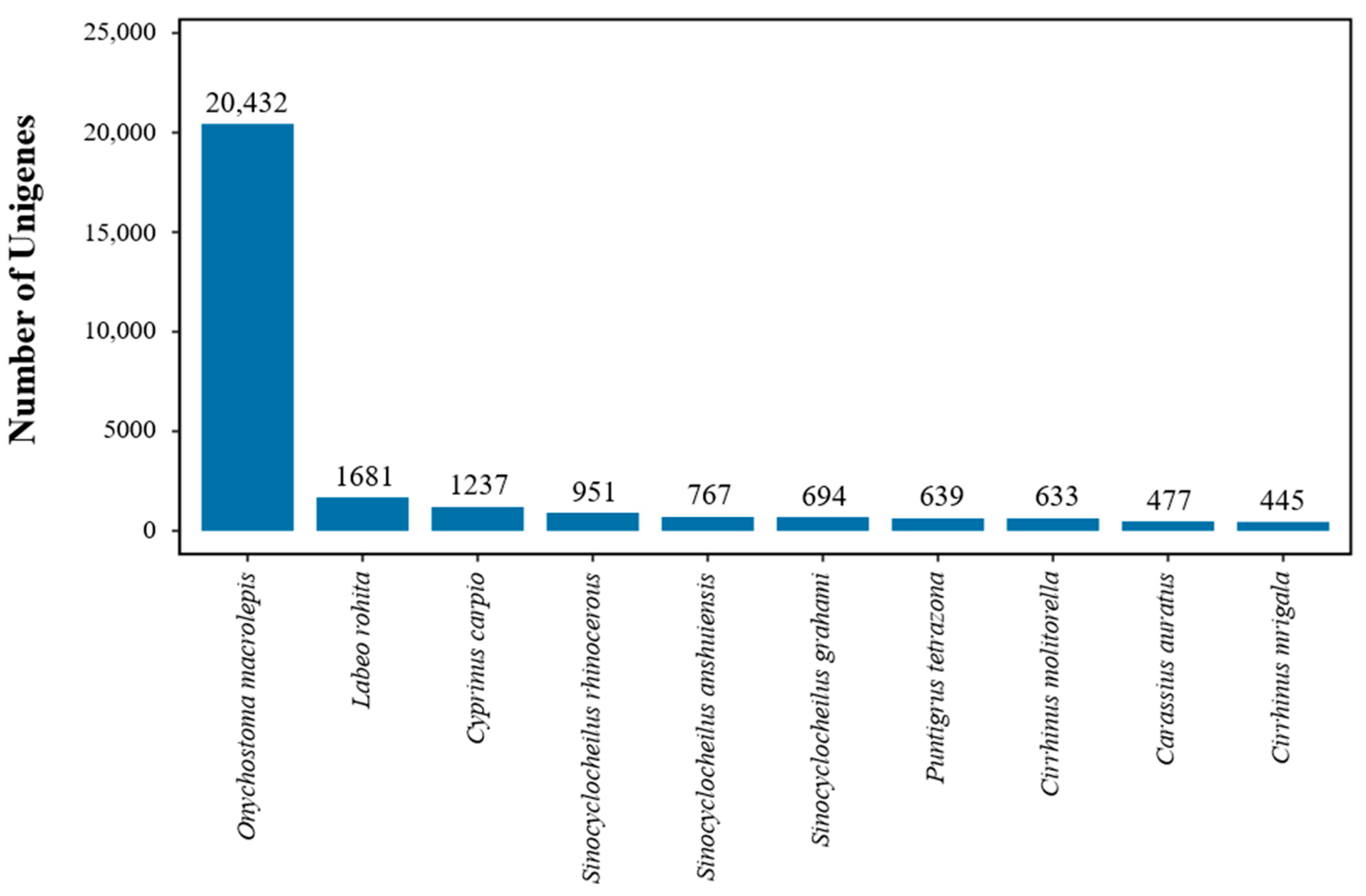
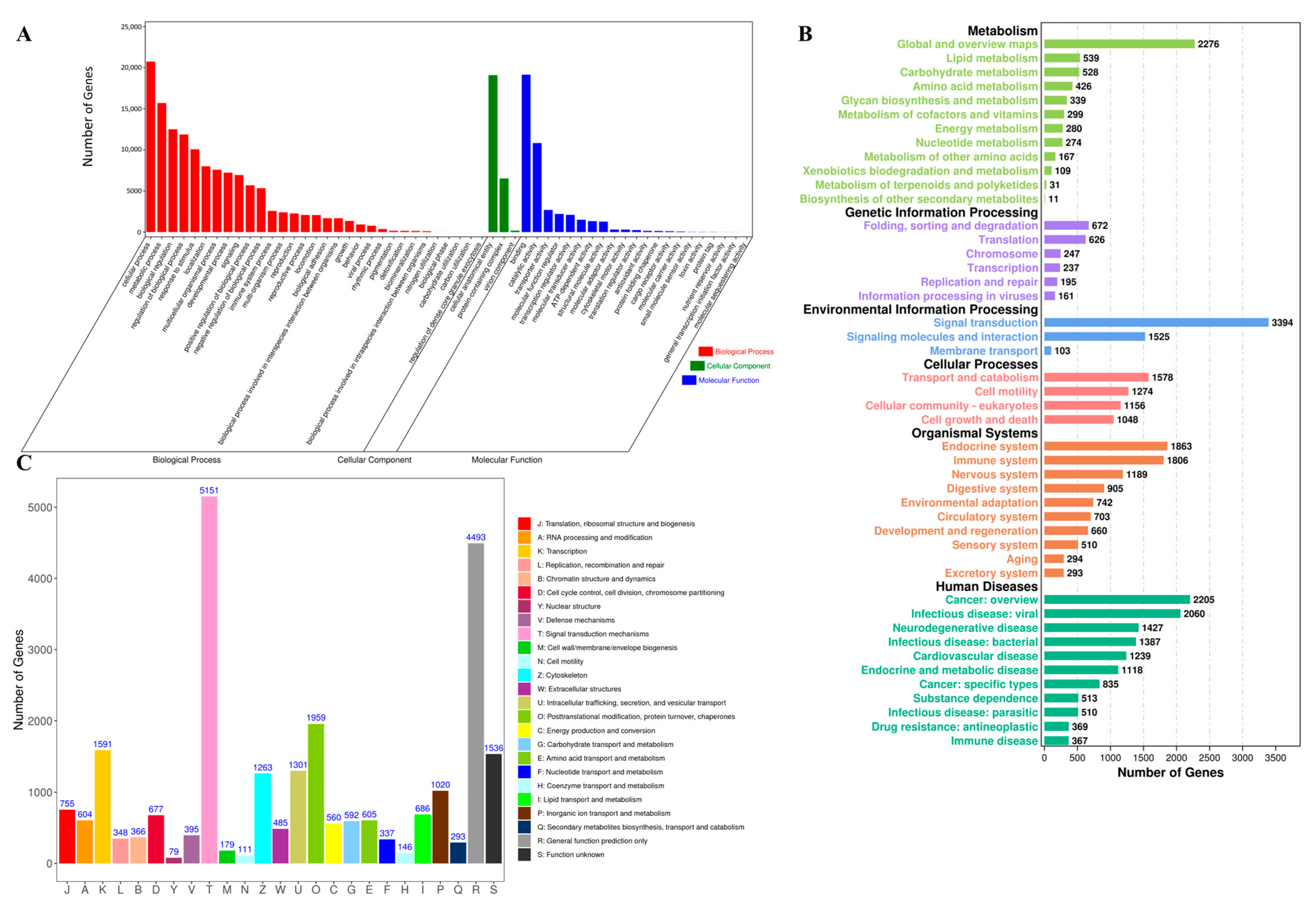
3.2. Unigene Annotation
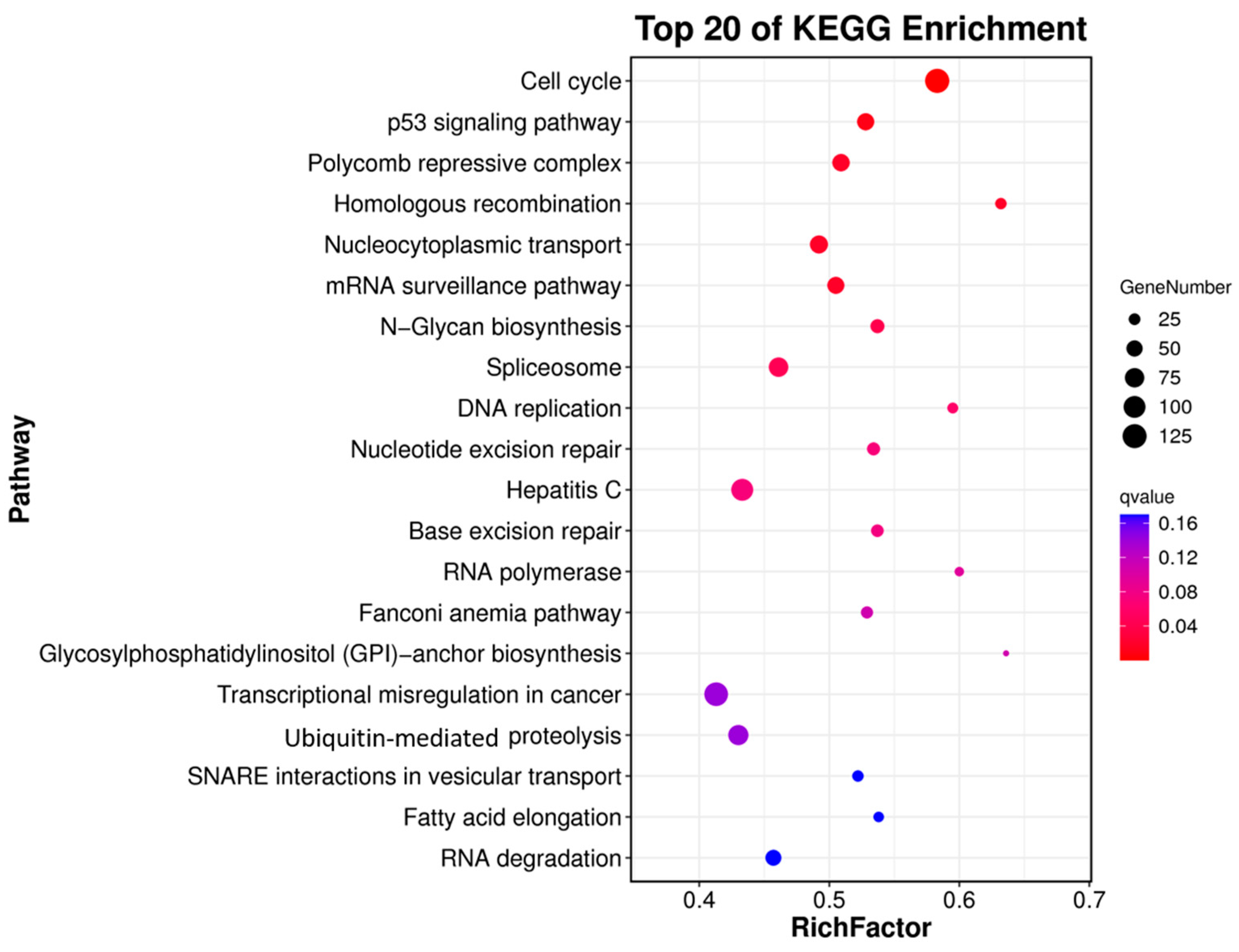
3.3. Differential Gene Expression Analysis
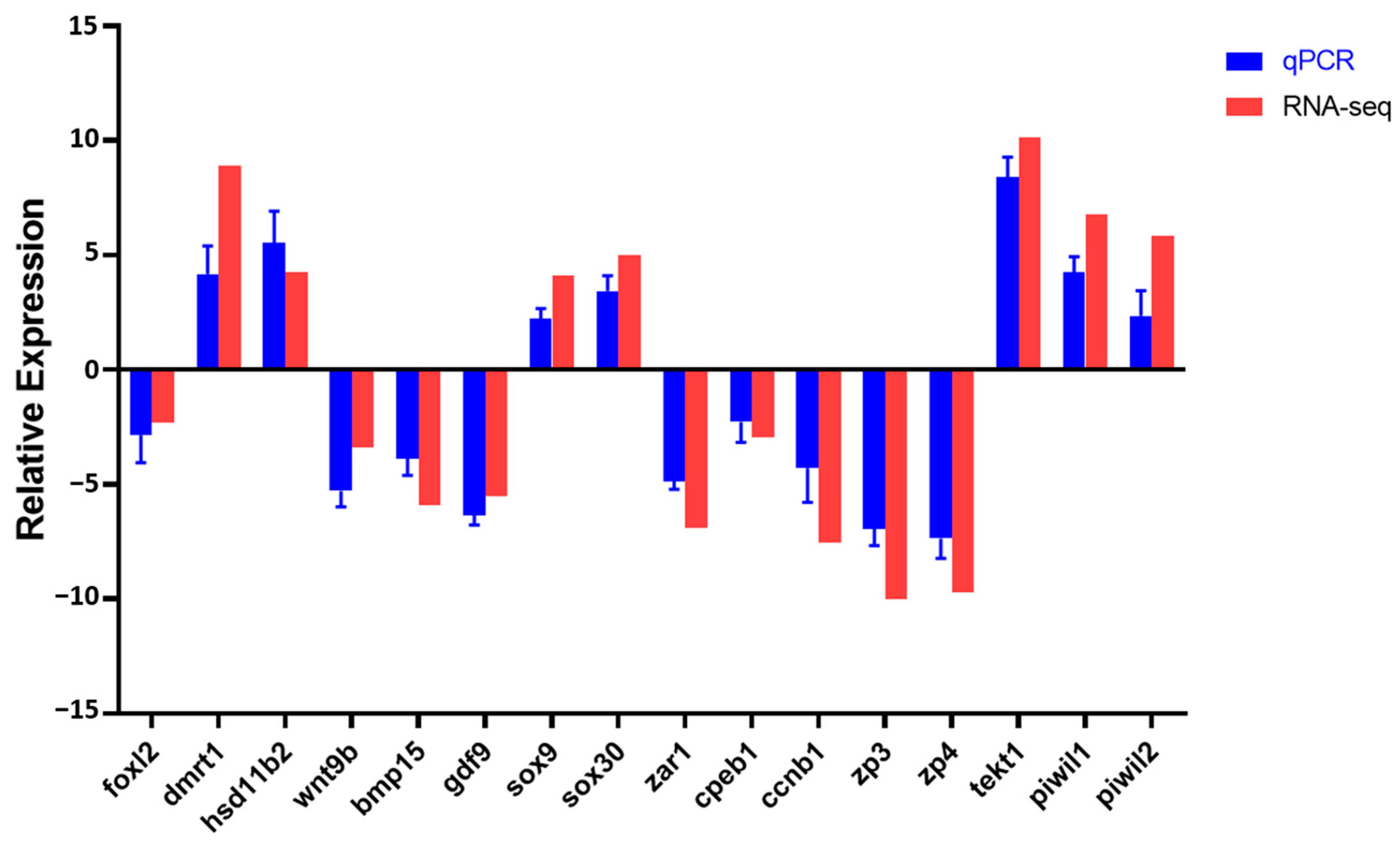
3.4. Validation of Transcriptomic Data by qRT-PCR
4. Discussion
4.1. DEGs Involved in Steroidogenesis Pathway
4.2. DEGs Involved in Gonad Differentiation and Development
4.3. DEGs Involved in Gametogenesis and Gamete Maturation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, M.L.; Zhou, H.M.; Wu, Z.Q.; Ouyang, S.; Chen, C.Q. Freshwater fish species richness and conservation of mountain streams in the Jinggangshan National Nature Reserve, China. Eco Mont-J. Prot. Mt. Areas Res. 2014, 6, 37–42. [Google Scholar] [CrossRef][Green Version]
- Li, Q.; Xu, X.L.; Huang, J.R. Length-weight relationships of 16 fish species from the Liuxihe national aquatic germplasm resources conservation area, Guangdong, China. J. Appl. Ichthyol. 2014, 30, 434–435. [Google Scholar] [CrossRef]
- Hu, M.L.; Wu, Z.Q.; Liu, Y.L. The fish fauna of mountain streams in the Guanshan National Nature Reserve, Jiangxi, China. Environ. Biol. Fishes 2009, 86, 23–27. [Google Scholar] [CrossRef]
- Zhang, C.P.; Lin, J.F.; Lai, X.X.; Zhang, M.Q.; Yuan, L.M.; Qin, W.J.; Shu, H. Genetic diversity and karyotype analysis of Acrossocheilus parallens in the Beijiang river. J. Guangzhou Univ. (Nat. Sci. Ed.) 2023, 22, 78–85, (In Chinese with English Abstract). [Google Scholar]
- Qin, Z.Q.; Lin, J.B.; Liang, P.; Qiu, M.L. Evaluation of Nutritional Components and Nutritive Quality in the Muscle of Acrossocheilus parallens. Chin. Agric. Sci. Bull. 2021, 37, 111–116, (In Chinese with English Abstract). [Google Scholar]
- Lan, Z.J.; Li, Q.; Zhao, J.; Zhong, L.M. Age and Growth of Acrossocheilus parallens in the Beijiang River. Chin. J. Zool. 2015, 50, 518–528, (In Chinese with English Abstract). [Google Scholar]
- Han, C.; Li, Q.; Xu, J.Q.; Li, X.F.; Huang, J.R. Characteristics and phylogenetic studies of Acrossocheilus parallens (Cypriniformes, Barbinae) complete mitochondrial genome. Mitochondrial DNA Part A 2016, 27, 4708–4709. [Google Scholar] [CrossRef]
- Xie, X.Y.; Huang, G.F.; Li, Y.T.; Zhang, Y.T.; Chen, S.X. Complete mitochondrial genome of Acrossocheilus parallens (Cypriniformes, Barbinae). Mitochondrial DNA Part A 2016, 27, 3339–3340. [Google Scholar] [CrossRef]
- Craves, J.A.M. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annu. Rev. Genet. 2008, 42, 565–586. [Google Scholar] [CrossRef]
- Heule, C.; Salzburger, W.; Böhne, A. Genetics of sexual development: An evolutionary playground for fish. Genetics 2014, 196, 579–591. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Nagahama, Y.; Nakamura, M. Diversity and plasticity of sex determination and differentiation in fishes. Sex. Dev. 2013, 7, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.H.; Wang, Y.W.; Hsu, C.W.; Chung, B.C. Zebrafish Foxl2l functions in proliferating germ cells for female meiotic entry. Dev. Biol. 2025, 517, 91–99. [Google Scholar] [CrossRef]
- Hu, Y.C.; Tan, R.H.; Li, Y.; Shu, T.T.; Yang, G.; Chu, Z.; Wang, H.R.; Liu, X.Q.; Zhu, X.; Wang, B.Z.; et al. Molecular cloning, expression and functional analysis of foxl2 from Chinese sturgeon (Acipenser sinensis) in relation to sex differentiation. Front. Mar. Sci. 2024, 11, 1506932. [Google Scholar] [CrossRef]
- Hattori, R.S.; Strüssmann, C.A.; Fernandino, J.I.; Somoza, G.M. Genotypic sex determination in teleosts: Insights from the testis-determining amhy gene. Gen. Comp. Endocrinol. 2013, 192, 55–59. [Google Scholar] [CrossRef]
- Sheng, Y.; Chen, B.; Zhang, L.; Luo, M.J.; Cheng, H.H.; Zhou, R.J. Identification of Dmrt genes and their up-regulation during gonad transformation in the swamp eel (Monopterus albus). Mol. Biol. Rep. 2014, 41, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Herpin, A.; Schartl, M. Dmrt1 genes at the crossroads: A widespread and central class of sexual development factors in fish. FEBS J. 2011, 278, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.J.; Liu, L.; Guo, L.Q.; Yu, H.S.; Cheng, H.; Huang, X.; Tiersch, T.R.; Berta, P. Similar gene structure of two Sox9a genes and their expression patterns during gonadal differentiation in a teleost fish, rice field eel (Monopterus albus). Mol. Reprod. Dev. 2003, 66, 211–217. [Google Scholar] [CrossRef]
- Yan, Y.-L.; Batzel, P.; Titus, T.; Sydes, J.; Desvignes, T.; BreMiller, R.; Draper, B.; Postlethwait, J.H. A hormone that lost its receptor: Anti-Mullerian hormone (AMH) in zebrafish gonad development and sex determination. Genetics 2019, 213, 529–553. [Google Scholar] [CrossRef]
- Liu, S.Y.; Han, C.; Huang, J.J.; Li, M.H.; Yang, J.Y.; Li, G.F.; Lin, H.R.; Li, S.S.; Zhang, Y. Genome-wide identification, evolution and expression of TGF-β signaling pathway members in mandarin fish (Siniperca chuatsi). Int. J. Biol. Macromol. 2023, 253, 126949. [Google Scholar] [CrossRef]
- Sreenivasan, R.; Jiang, J.H.; Wang, X.G.; Bártfai, R.; Kwan, H.Y.; Christoffels, A.; Orbán, L. Gonad differentiation in zebrafish is regulated by the canonical Wnt signaling pathway. Biol. Reprod. 2014, 90, 45. [Google Scholar] [CrossRef]
- Huang, X.L.; Huang, Z.; Li, Q.; Li, W.J.; Han, C.; Yang, Y.K.; Lin, H.Z.; Wu, Q.E.; Zhou, Y.B. De novo assembly, characterization, and comparative transcriptome analysis of mature male and female gonads of rabbitfish (Siganus oramin) (Bloch & Schneider, 1801). Animals 2024, 14, 1346. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Huang, W.W.; Peng, S.H.; Zhou, J.W.; Zhan, H.W.; Zhang, Y.Y.; Li, W.J.; Gong, J.; Li, Q. De novo assembly, characterization and comparative transcriptome analysis of the mature gonads in Spinibarbus hollandi. Animals 2023, 13, 166. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Y.; Han, C.; Zhang, Y. De novo assembly, characterization and comparative transcriptome analysis of gonads reveals sex-biased genes in Coreoperca whiteheadi. Comp. Biochem. Physiol. D-Genom. Proteom. 2023, 47, 101115. [Google Scholar] [CrossRef]
- Liu, S.Y.; Lian, Y.Y.; Song, Y.K.; Chen, Q.H.; Huang, J.R. De novo assembly, characterization and comparative transcriptome analysis of the gonads of jade perch (Scortum barcoo). Animals 2023, 13, 2254. [Google Scholar] [CrossRef]
- Shen, X.Y.; Yanez, J.M.; Gomes, G.B.; Poon, Z.W.J.; Foster, D.; Alarcon, J.F.; Domingos, J.A. Comparative gonad transcriptome analysis in cobia (Rachycentron canadum). Front. Genet. 2023, 14, 1128943. [Google Scholar] [CrossRef]
- Shen, F.F.; Long, Y.; Li, F.Y.; Ge, G.D.; Song, G.L.; Li, Q.; Qiao, Z.G.; Cui, Z.B. De novo transcriptome assembly and sex-biased gene expression in the gonads of Amur catfish (Silurus asotus). Genomics 2020, 112, 2603–2614. [Google Scholar] [CrossRef]
- Yang, W.; Chen, H.P.; Cui, X.F.; Zhang, K.W.; Jiang, D.N.; Deng, S.P.; Zhu, C.H.; Li, G.L. Sequencing, de novo assembly and characterization of the spotted scat Scatophagus argus (Linnaeus 1766) transcriptome for discovery of reproduction related genes and SSRs. J. Oceanol. Limnol. 2018, 36, 1329–1341. [Google Scholar] [CrossRef]
- Yue, H.M.; Li, C.J.; Du, H.; Zhang, S.H.; Wei, Q.W. Sequencing and De novo assembly of the gonadal transcriptome of the endangered Chinese sturgeon (Acipenser sinensis). PLoS ONE 2015, 10, e0127332. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644-U130. [Google Scholar] [CrossRef]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357-U121. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Anders, S.; McCarthy, D.J.; Chen, Y.S.; Okoniewski, M.; Smyth, G.K.; Huber, W.; Robinson, M.D. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013, 8, 1765–1786. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Baroiller, J.F.; Guiguen, Y.; Fostier, A. Endocrine and environmental aspects of sex differentiation in fish. Cell. Mol. Life Sci. 1999, 55, 910–931. [Google Scholar] [CrossRef]
- Göppert, C.; Harris, R.M.; Theis, A.; Boila, A.; Hohl, S.; Rüegg, A.; Hofmann, H.A.; Salzburger, W.; Böhne, A. Inhibition of aromatase induces partial sex change in a cichlid fish: Distinct functions for sex steroids in brains and gonads. Sex. Dev. 2016, 10, 97–110. [Google Scholar] [CrossRef]
- Sun, L.N.; Jiang, X.L.; Xie, Q.P.; Yuan, J.; Huang, B.F.; Tao, W.J.; Zhou, L.Y.; Nagahama, Y.; Wang, D.S. Transdifferentiation of differentiated ovary into functional testis by long-term treatment of aromatase inhibitor in Nile tilapia. Endocrinology 2014, 155, 1476–1488. [Google Scholar] [CrossRef]
- Wang, D.S.; Kobayashi, T.; Zhou, L.Y.; Paul-Prasanth, B.; Ijiri, S.; Sakai, F.; Okubo, K.; Morohashi, K.I.; Nagahama, Y. Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with Ad4 binding protein/steroidogenic factor 1. Mol. Endocrinol. 2007, 21, 712–725. [Google Scholar] [CrossRef]
- Loffler, K.A.; Zarkower, D.; Koopman, P. Etiology of ovarian failure in blepharophimosis ptosis epicanthus inversus syndrome: FOXL2 is a conserved, early-acting gene in vertebrate ovarian development. Endocrinology 2003, 144, 3237–3243. [Google Scholar] [CrossRef]
- Wang, D.S.; Zhou, L.Y.; Kobayashi, T.; Matsuda, M.; Shibata, Y.; Sakai, F.; Nagahama, Y. Doublesex- and mab-3-related transcription factor-1 repression of aromatase transcription, a possible mechanism favoring the male pathway in tilapia. Endocrinology 2010, 151, 1331–1340. [Google Scholar] [CrossRef]
- Li, M.H.; Yang, H.H.; Li, M.R.; Sun, Y.L.; Jiang, X.L.; Xie, Q.P.; Wang, T.R.; Shi, H.J.; Sun, L.N.; Zhou, L.Y.; et al. Antagonistic roles of Dmrt1 and Foxl2 in sex differentiation via estrogen production in tilapia as demonstrated by TALENs. Endocrinology 2013, 154, 4814–4825. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhu, H.; Dong, Y.; Dong, T.; Tian, Z.H.; Hu, H.X. Identification and dimorphic expression of sex-related genes during gonadal differentiation in sterlet Acipenser ruthenus, a primitive fish species. Aquaculture 2019, 500, 178–187. [Google Scholar] [CrossRef]
- Wang, W.; Zhu, H.; Dong, Y.; Tian, Z.H.; Dong, T.; Hu, H.X.; Niu, C.J. Dimorphic expression of sex-related genes in different gonadal development stages of sterlet, Acipenser ruthenus, a primitive fish species. Fish Physiol. Biochem. 2017, 43, 1557–1569. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, X.L.; Wei, X.K.; Li, Y.; Liu, W.; Gan, G.C.; Xiao, L.L.; Wang, X.Y.; Luo, H. Gonadal transcriptome analysis of paradise fish Macropodus opercularis to reveal sex-related genes. Comp. Biochem. Physiol. D-Genom. Proteom. 2023, 48, 101125. [Google Scholar] [CrossRef]
- Xu, Y.; Zhong, Z.W.; Feng, Y.; Zhang, Z.Y.; Ao, L.L.; Liu, H.W.; Wang, Y.L.; Jiang, Y.H. Expression pattern analysis of anti-Mullerian hormone in testis development of pearlscale angelfish (Centropyge vrolikii). J. Fish Biol. 2023, 102, 1067–1078. [Google Scholar] [CrossRef]
- Hu, Y.C.; Wang, B.Z.; Du, H.J. A review on sox genes in fish. Rev. Aquac. 2021, 13, 1986–2003. [Google Scholar] [CrossRef]
- Shi, R.; Li, X.H.; Xu, X.W.; Chen, Z.F.; Zhu, Y.; Wang, N. Genome-wide analysis of BMP/GDF family and DAP-seq of YY1 suggest their roles in Cynoglossus semilaevis sexual size dimorphism. Int. J. Biol. Macromol. 2023, 253, 127201. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, J.Y.; Ma, Z.H.; Pu, H.Q.; Zhang, T.; Guo, J.Y.; Luo, Z.P.; Chen, H.P.; Liang, W.M.; Liang, Z.F.; et al. Molecular and cellular regulation on sexual fate and gonadal development in hermaphrodite yellowfin seabream Acanthopagrus latus. Aquac. Rep. 2024, 34, 101913. [Google Scholar] [CrossRef]
- Liu, Y.L.; Kossack, M.E.; McFaul, M.E.; Christensen, L.N.; Siebert, S.; Wyatt, S.R.; Kamei, C.N.; Horst, S.; Arroyo, N.; Drummond, I.A.; et al. Single-cell transcriptome reveals insights into the development and function of the zebrafish ovary. eLife 2022, 11, e76014. [Google Scholar] [CrossRef]
- Clelland, E.; Kohli, G.; Campbell, R.K.; Sharma, S.; Shimasaki, S.; Peng, C. Bone morphogenetic protein-15 in the zebrafish ovary: Complementary deoxyribonucleic acid cloning, genomic organization, tissue distribution, and role in oocyte maturation. Endocrinology 2006, 147, 201–209. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, C.J.; Lawrence, S.; Smith, P.; Juengel, J.L.; McNatty, K.P. Active immunization against the proregions of GDF9 or BMP15 alters ovulation rate and litter size in mice. Reproduction 2012, 143, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Li, X.H.; Cheng, P.; Yang, Q.; Chen, Z.F.; Chen, S.L.; Wang, N. Characterization of growth differentiation factor 9 and bone morphogenetic factor 15 in Chinese tongue sole (Cynoglossus semilaevis): Sex-biased expression pattern and promoter regulation. Theriogenology 2022, 182, 119–128. [Google Scholar] [CrossRef]
- Yadav, H.; Lal, B. Cellular localization and seasonal variation in BMP15 expression in ovary of the catfish Clarias batrachus and its role in ovarian steroidogenesis. Theriogenology 2019, 129, 14–22. [Google Scholar] [CrossRef]
- Wu, K.; Zhai, Y.; Qin, M.M.; Zhao, C.; Ai, N.A.; He, J.G.; Ge, W. Genetic evidence for differential functions of figla and nobox in zebrafish ovarian differentiation and folliculogenesis. Commun. Biol. 2023, 6, 1185. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Li, X.Y.; Li, M.H.; Tang, Y.H.; Wei, J.; Wang, D.S. Dmrt1 directly regulates the transcription of the testis-biased Sox9b gene in Nile tilapia (Oreochromis niloticus). Gene 2019, 687, 109–115. [Google Scholar] [CrossRef]
- Barrionuevo, F.; Scherer, G. SOX E genes: SOX9 and SOX8 in mammalian testis development. Int. J. Biochem. Cell Biol. 2010, 42, 433–436. [Google Scholar] [CrossRef]
- Anitha, A.; Senthilkumaran, B. Role of sox30 in regulating testicular steroidogenesis of common carp. J. Steroid Biochem. Mol. Biol. 2020, 204, 105769. [Google Scholar] [CrossRef]
- Liang, B.; Jerry, D.R.; Shen, X.Y.; Koh, J.; Terence, C.; Nayfa, M.G.; Nguyen, V.; Loo, G.; Vij, S.; Domingos, J.A. Transcriptomic analysis of gonads in Malabar red snapper (Lutjanus malabaricus) reveals genes associated with gonad development. Aquaculture 2024, 592, 741258. [Google Scholar] [CrossRef]
- Tseng, P.W.; Wu, G.C.; Kuo, W.L.; Tseng, Y.C.; Chang, C.F. The ovarian transcriptome at the early stage of testis removal-induced male-to-female sex change in the protandrous black porgy Acanthopagrus schlegelii. Front. Genet. 2022, 13, 816955. [Google Scholar] [CrossRef]
- Zhu, Q.Y.; Han, C.; Liu, S.Y.; Ouyang, H.F.; Liu, D.R.; Zhang, Z.W.; Huang, J.J.; Han, L.Q.; Li, S.S.; Li, G.F.; et al. Development and gene expression analysis of gonad during 17α-methyltestosterone-induced sex reversal in mandarin fish (Siniperca chuatsi). Aquac. Rep. 2022, 23, 101049. [Google Scholar] [CrossRef]
- Amos, L.A. The tektin family of microtubule-stabilizing proteins. Genome Biol. 2008, 9, 229. [Google Scholar] [CrossRef]
- Tao, M.; Zhou, Y.; Li, S.N.; Zhong, H.; Hu, H.; Yuan, L.J.; Luo, M.; Chen, J.; Ren, L.; Luo, J.; et al. MicroRNA alternations in the testes related to the sterility of triploid fish. Mar. Biotechnol. 2018, 20, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Wang, D.; Li, X.R.; Zhao, C.; Wang, T.; Qian, X.M.; Yin, S.W. Differential expression of two Piwil orthologs during embryonic and gonadal development in pufferfish, Takifugu fasciatus. Comp. Biochem. Physiol. B-Biochem. Mol. Biol. 2018, 219, 44–51. [Google Scholar] [CrossRef]
- Ni, F.F.; Yu, H.Y.; Liu, Y.Z.; Meng, L.H.; Yan, W.J.; Zhang, Q.Q.; Yu, H.Y.; Wang, X.B. Roles of piwil1 gene in gonad development and gametogenesis in Japanese flounder, Paralichthys olivaceus. Gene 2019, 701, 104–112. [Google Scholar] [CrossRef]
- Zong, W.Y.; Wang, Y.P.; Zhang, L.Q.; Lu, W.; Li, W.G.; Wang, F.C.; Cheng, J. DNA methylation mediates sperm quality via piwil1 and piwil2 regulation in Japanese flounder (Paralichthys olivaceus). Int. J. Mol. Sci. 2024, 25, 5935. [Google Scholar] [CrossRef]
- Wu, X.M.; Wang, P.; Brown, C.A.; Zilinski, C.A.; Matzuk, M.M. Zygote arrest 1 (Zar1) is an evolutionarily conserved gene expressed in vertebrate ovaries. Biol. Reprod. 2003, 69, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Zhang, S.Y.; Ma, Z.S.; Qiang, J.; Wei, J.; Sun, L.N.; Kocher, T.D.; Wang, D.S.; Tao, W.J. Disruption of Zar1 leads to arrested oogenesis by regulating polyadenylation via Cpeb1 in tilapia (Oreochromis niloticus). Int. J. Biol. Macromol. 2024, 260, 129632. [Google Scholar] [CrossRef]
- Weill, L.; Belloc, E.; Bava, F.A.; Méndez, R. Translational control by changes in poly(A) tail length: Recycling mRNAs. Nat. Struct. Mol. Biol. 2012, 19, 577–585. [Google Scholar] [CrossRef]
- Richter, J.D. CPEB: A life in translation. Trends Biochem. Sci. 2007, 32, 279–285. [Google Scholar] [CrossRef]
- Cheng, J.M.; Liu, Y.X. Knockout of cyclin B1 in granulosa cells causes female subfertility. Cell Cycle 2022, 21, 1867–1878. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Cai, R.; Shi, W.H.; Zhang, H.; Liu, Z.; Xie, F.F.; Chen, Y.H.; Hong, Q. Comparative transcriptome analysis of ovaries and testes reveals sex-biased genes and pathways in zebrafish. Gene 2024, 901, 148176. [Google Scholar] [CrossRef] [PubMed]
- Wassarman, P.M. Sperm receptors and fertilization in mammals. Mt. Sinai J. Med. 2002, 69, 148–155. [Google Scholar] [PubMed]
- Litscher, E.S.; Wassarman, P.M. The Fish Egg’s Zona Pellucida. In Extracellular Matrix and Egg Coats; Litscher, E.S., Wassarman, P.M., Eds.; Current Topics in Developmental Biology; Academic Press: New York, NY, USA, 2018; Volume 130, pp. 275–305. [Google Scholar]

| Gene | Sequence (5′–3′) | |
|---|---|---|
| Forward Primer | Reverse Primer | |
| β-actin | GTGTTGGCATACAGGTCCTTACG | ACGGACAGGTCATCACCATTG |
| foxl2 | AGGGTTGGCAGAACAGTATCAGG | GAAATGCGTCGGTGGAGGTC |
| dmrt1 | AACCCAAAGCAGCAGTTTTCTC | CGACAGAGAAGGTTCCCGAC |
| hsd11b2 | AGACAGGCTAAAGGCCGGA | TGACGAAGTGTGTTGGTAAGAAGAT |
| wnt9b | CTCTGAGGGAATCTGTCCGC | GCGGTCTCTTTAAAGCCTCG |
| bmp15 | TCCCAACCTCAAGTGACCTTC | ACGTGACTCTTGCCTCACAG |
| gdf9 | CGAGCAAAACCGAGAGTTCTT | ATAGCAGAGCGATGTGAAGGG |
| sox9 | AGGTCAGAGCTCCGGCTTGTACT | TGTGATTGGGTTGGGGAATGG |
| sox30 | CCTTCTGGAGCAGAAAGTGAG | GTTGCTAGCATTAGGGTTGGC |
| zar1 | CGTGAAGGAACCGCTTAGTC | TCTGCTCCAAAAACTGGAACC |
| cpeb1 | CCTGGGACATCACTGAAGCTGG | TTGGGCATATTACCTTTCGGAGG |
| ccnb1 | TTTACAACAGCTCGAGGTTGCG | GCATGAGCCAGTCGATGGTG |
| zp3 | GCCTTCAGGTTCCACCAGGAC | GCCACCCATCTGTTGGTTTCG |
| zp4 | TTGAGTGGTTGCTGGTGGTCC | CAGGTGTTTAGCGCTTTGTGG |
| tekt1 | GGACAAATTTCAGGCCGAGC | AGGAGTCACTGCAGATCCAAG |
| piwil1 | GCCATGTCTGAAGAAGCGATG | ATTAGCTGGCGAGTGTGGAC |
| piwil2 | GCATCGTGACACTTTGCATTG | ACCCTGATCTTCTGCTGACTG |
| Sample | Number of Reads | Total Base | GC Content (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|
| Ovary-1 | 41,216,910 | 6,083,067,389 | 45.86 | 97.02 | 92.49 |
| Ovary-2 | 41,264,316 | 6,077,643,211 | 46.57 | 97.04 | 92.61 |
| Ovary-3 | 40,229,586 | 5,882,642,501 | 45.89 | 96.92 | 92.42 |
| Testis-1 | 41,869,770 | 6,171,999,520 | 45.77 | 97.14 | 92.75 |
| Testis-2 | 37,750,646 | 5,572,829,473 | 48.31 | 97.34 | 93.15 |
| Testis-3 | 40,272,500 | 5,922,275,444 | 47.78 | 98.12 | 94.94 |
| Mean | 40,433,955 | 5,951,742,923 | 46.70 | 97.26 | 93.06 |
| Total | 242,603,728 | 35,710,457,538 |
| Database | Number |
|---|---|
| Assembly | |
| Gene number (#) | 67,251 |
| Total length (nt) | 73,634,351 |
| Average length (nt) | 1094 |
| Max length (nt) | 18,169 |
| Min length (nt) | 201 |
| N50 (nt) | 2416 |
| GC | 44.19% |
| Annotation | |
| Total number of annotated unigenes | 34,069 |
| Unigenes matched against Nr | 33,686 |
| Unigenes matched against UniProt | 23,221 |
| Unigenes matched against KEGG | 32,251 |
| Unigenes matched against KOG | 18,137 |
| Unigene ID | log2FC | p-Value | FDR | Nr Annotation | Gene Name |
|---|---|---|---|---|---|
| Unigene0065503 | 17.164 | 3.71 × 10−26 | 2.59 × 10−24 | tektin-4 | tekt4 |
| Unigene0020072 | 11.090 | 2.55 × 10−3 | 1.39 × 10−2 | forkhead box protein G1 | foxg1 |
| Unigene0020990 | 11.048 | 5.63 × 10−4 | 3.58 × 10−3 | paired box protein Pax-3a isoform X3 | pax3-a |
| Unigene0039747 | 10.974 | 9.04 × 10−3 | 4.19 × 10−2 | paired box protein Pax-2-A isoform X8 | pax2a |
| Unigene0068389 | 10.156 | 1.21 × 10−106 | 2.88 × 10−103 | tektin-1-like | tekt1 |
| Unigene0063623 | 10.008 | 7.00 × 10−15 | 2.09 × 10−13 | cytochrome P450 11B, mitochondrial | cyp11b |
| Unigene0012670 | 9.514 | 2.01 × 10−84 | 1.49 × 10−81 | forkhead box protein J1-A | foxj1a |
| Unigene0031953 | 8.892 | 7.47 × 10−77 | 4.06 × 10−74 | Double sex- and mab-3-related transcription factor 1 | dmrt1 |
| Unigene0042464 | 8.568 | 3.09 × 10−10 | 5.55 × 10−9 | fibroblast growth factor 13b isoform X1 | fgf13 |
| Unigene0015615 | 8.523 | 3.27 × 10−9 | 5.11 × 10−8 | bone morphogenetic protein 8A-like | bmp8a |
| Unigene0010324 | 8.393 | 8.15 × 10−82 | 5.29 × 10−79 | sperm-associated antigen 16 protein | spag16 |
| Unigene0039593 | 8.384 | 2.76 × 10−89 | 2.64 × 10−86 | sperm-associated antigen 17 isoform X4 | spag17 |
| Unigene0043996 | 7.447 | 1.37 × 10−3 | 7.97 × 10−3 | cytochrome P450 2K6-like | cyp2k6 |
| Unigene0001198 | 6.693 | 1.01 × 10−15 | 3.25 × 10−14 | protein Wnt-5a | wnt5a |
| Unigene0015091 | 6.776 | 1.25 × 10−97 | 1.90 × 10−94 | piwi-like protein 1 | piwil1 |
| Unigene0001625 | 6.622 | 5.61 × 10−17 | 2.03 × 10−15 | doublesex- and mab-3-related transcription factor A1-like | dmrta2 |
| Unigene0033990 | 6.423 | 3.45 × 10−5 | 2.84 × 10−4 | forkhead box protein P3 isoform X2 | foxp3 |
| Unigene0040562 | 6.174 | 1.60 × 10−28 | 1.29 × 10−26 | protein Wnt-7b isoform X1 | wnt7b |
| Unigene0005830 | 5.838 | 4.69 × 10−48 | 9.45 × 10−46 | piwi-like protein 2 | piwil2 |
| Unigene0009058 | 5.561 | 4.38 × 10−6 | 4.25 × 10−5 | fibroblast growth factor 14 | fgf14 |
| Unigene0039956 | 5.468 | 5.70 × 10−12 | 1.26 × 10−10 | paired box protein Pax-8 isoform X1 | pax8 |
| Unigene0052496 | 5.011 | 1.43 × 10−45 | 2.58 × 10−43 | transcription factor SOX-30-like isoform X2 | sox30 |
| Unigene0071169 | 4.442 | 1.22 × 10−3 | 7.19 × 10−3 | transcription factor Sox-14 | sox14 |
| Unigene0017663 | 4.373 | 1.39 × 10−4 | 1.01 × 10−3 | paired box protein Pax-3b isoform X1 | pax3a |
| Unigene0064185 | 4.236 | 1.50 × 10−22 | 8.30 × 10−21 | 11-beta-hydroxysteroid dehydrogenase type 2 | hsd11b2 |
| Unigene0001099 | 4.191 | 1.68 × 10−13 | 4.37 × 10−12 | insulin-like growth factor-binding protein 3 | igfbp3 |
| Unigene0014008 | 4.190 | 1.12 × 10−5 | 1.01 × 10−4 | bone morphogenetic protein 3 | bmp3 |
| Unigene0071287 | 4.147 | 2.06 × 10−26 | 1.46 × 10−24 | mothers against decapentaplegic homolog 5 | smad5 |
| Unigene0012314 | 4.110 | 7.88 × 10−13 | 1.91 × 10−11 | transcription factor Sox-9-like | sox9 |
| Unigene0030575 | 4.093 | 4.18 × 10−11 | 8.35 × 10−10 | stAR-related lipid transfer protein 4 isoform X1 | stard4 |
| Unigene0063224 | 4.029 | 1.42 × 10−16 | 4.95 × 10−15 | steroid hormone receptor ERR2 isoform X2 | esrrb |
| Unigene0044603 | 3.879 | 9.20 × 10−3 | 4.26 × 10−2 | fibroblast growth factor receptor 3 isoform X3 | fgfr3 |
| Unigene0066364 | 3.518 | 6.33 × 10−9 | 9.56 × 10−8 | cytochrome P450 4B1 | cyp4b1 |
| Unigene0040138 | 3.446 | 4.82 × 10−22 | 2.55 × 10−20 | forkhead box protein M1 isoform X2 | foxm1 |
| Unigene0035507 | 3.068 | 1.51 × 10−4 | 1.09 × 10−3 | forkhead box protein L1 | foxl1 |
| Unigene0018532 | 2.972 | 9.64 × 10−11 | 1.85 × 10−9 | growth/differentiation factor 10 | gdf10 |
| Unigene0069601 | 2.704 | 2.64 × 10−6 | 2.66 × 10−5 | fibroblast growth factor receptor 2 | fgfr2 |
| Unigene0002103 | 2.687 | 3.68 × 10−10 | 6.52 × 10−9 | stAR-related lipid transfer protein 9 isoform X1 | stard9 |
| Unigene0010222 | 2.671 | 1.04 × 10−6 | 1.12 × 10−5 | fibroblast growth factor receptor-like 1 | fgfrl1 |
| Unigene0016545 | 2.461 | 4.25 × 10−5 | 3.43 × 10−4 | cytochrome P450 4F3 | cyp4f3 |
| Unigene0040707 | 2.413 | 5.64 × 10−3 | 2.78 × 10−2 | steroidogenic acute regulatory protein, mitochondrial | star |
| Unigene0057715 | 2.304 | 1.13 × 10−4 | 8.41 × 10−4 | mothers against decapentaplegic homolog 4 isoform X2 | smad4 |
| Unigene0015555 | 2.257 | 4.05 × 10−9 | 6.25 × 10−8 | forkhead box protein O4 | foxo4 |
| Unigene0058422 | 2.240 | 8.76 × 10−6 | 8.06 × 10−5 | forkhead box protein J2 | foxj2 |
| Unigene0001059 | 2.139 | 4.00 × 10−4 | 2.64 × 10−3 | estrogen-related receptor gamma a isoform X1 | esrrg |
| Unigene0065036 | 2.127 | 5.45 × 10−3 | 2.69 × 10−2 | cytochrome P450 7A1 | cyp7a1 |
| Unigene0049698 | −1.662 | 7.54 × 10−3 | 3.59 × 10−3 | protein Wnt-11 | wnt11 |
| Unigene0063444 | −2.332 | 2.18 × 10−5 | 1.87 × 10−4 | forkhead box protein L2a isoform X1 | foxl2 |
| Unigene0002571 | −2.346 | 1.62 × 10−6 | 1.69 × 10−5 | transcription factor SOX-4b | sox4 |
| Unigene0004856 | −2.461 | 2.53 × 10−12 | 5.81 × 10−11 | very-long-chain 3-oxoacyl-CoA reductase-B | hsd17b12b |
| Unigene0001279 | −2.512 | 9.74 × 10−6 | 8.89 × 10−5 | androgen receptor isoform X2 | ar |
| Unigene0016850 | −2.522 | 4.59 × 10−13 | 1.14 × 10−11 | forkhead box protein O3a | foxo3a |
| Unigene0058062 | −2.648 | 1.48 × 10−8 | 2.13 × 10−7 | mothers against decapentaplegic homolog 6a | smad6a |
| Unigene0020261 | −2.913 | 1.21 × 10−12 | 2.88 × 10−11 | 3 beta-hydroxysteroid dehydrogenase type 7 | hsd3b7 |
| Unigene0064533 | −2.940 | 8.16 × 10−17 | 2.92 × 10−15 | cytoplasmic polyadenylation element-binding protein 1 isoform X1 | cpeb1 |
| Unigene0005781 | −3.083 | 1.81 × 10−17 | 6.85 × 10−16 | cytochrome P450 2J4 | cyp2j4 |
| Unigene0063752 | −3.147 | 3.80 × 10−21 | 1.91 × 10−19 | fibroblast growth factor receptor 1-A isoform X2 | fgfr1a |
| Unigene0057571 | −3.391 | 1.76 × 10−8 | 2.50 × 10−7 | protein Wnt-9b | wnt9b |
| Unigene0008152 | −3.417 | 3.87 × 10−7 | 4.46 × 10−6 | paired box protein Pax-1a | pax1 |
| Unigene0058533 | −3.553 | 1.26 × 10−14 | 3.69 × 10−13 | cytochrome P450 2F2-like isoform X1 | cyp2f2 |
| Unigene0021056 | −3.578 | 5.23 × 10−8 | 6.91 × 10−7 | insulin-like growth factor-binding protein 5a | igfbp5 |
| Unigene0066573 | −3.620 | 1.29 × 10−7 | 1.60 × 10−6 | forkhead box protein H1 isoform X1 | foxh1 |
| Unigene0065136 | −3.686 | 7.71 × 10−13 | 1.87 × 10−11 | paired box protein Pax-6 isoform X1 | pax6a |
| Unigene0004558 | −3.928 | 2.86 × 10−16 | 9.70 × 10−15 | transcription factor Sox-3 isoform X1 | sox3 |
| Unigene0035649 | −4.122 | 2.35 × 10−3 | 1.29 × 10−2 | cytochrome P450 aromatase | cyp19a1 |
| Unigene0049839 | −4.226 | 2.37 × 10−24 | 1.47 × 10−22 | paired box protein Pax-1 | pax1 |
| Unigene0020541 | −4.330 | 1.22 × 10−36 | 1.45 × 10−34 | sperm-associated antigen 7 homolog isoform X1 | spag7 |
| Unigene0009593 | −4.460 | 2.95 × 10−46 | 5.42 × 10−44 | transcription factor SOX-11b | sox11b |
| Unigene0067666 | −4.855 | 1.00 × 10−40 | 1.44 × 10−38 | cytochrome P450 2J2 isoform X3 | cyp2j2 |
| Unigene0057503 | −4.861 | 1.74 × 10−9 | 2.82 × 10−8 | forkhead box protein Q1b | foxq1b |
| Unigene0057502 | −5.034 | 3.95 × 10−56 | 1.05 × 10−53 | insulin-like growth factor 2a | igf2 |
| Unigene0036758 | −5.432 | 3.50 × 10−33 | 3.58 × 10−31 | transcription factor Sox-21-A | sox21a |
| Unigene0003785 | −5.515 | 1.82 × 10−36 | 2.16 × 10−34 | growth/differentiation factor 9 | gdf9 |
| Unigene0055178 | −5.932 | 3.52 × 10−75 | 1.77 × 10−72 | bone morphogenetic protein 15 | bmp15 |
| Unigene0004792 | −6.643 | 1.53 × 10−36 | 1.81 × 10−34 | bone morphogenetic protein 2 | bmp2 |
| Unigene0001965 | −6.895 | 2.29 × 10−89 | 2.21 × 10−86 | zygote arrest protein 1 | zar1 |
| Unigene0062685 | −7.559 | 7.88 × 10−43 | 1.24 × 10−40 | G2/mitotic-specific cyclin-B1-like isoform X1 | ccnb1 |
| Unigene0065315 | −7.795 | 1.24 × 10−33 | 1.29 × 10−31 | protein Wnt-8a ORF1 isoform X1 | wnt8a |
| Unigene0063202 | −9.710 | 7.99 × 10−90 | 7.97 × 10−87 | zona pellucida sperm-binding protein 4-like | zp4 |
| Unigene0039351 | −10.013 | 9.72 × 10−98 | 1.51 × 10−94 | zona pellucida sperm-binding protein 3-like | zp3 |
| Unigene0011219 | −10.483 | 7.43 × 10−37 | 9.01 × 10−35 | zona pellucida sperm-binding protein 4-like | zp1 |
| Unigene0061382 | −13.657 | 2.08 × 10−13 | 5.35 × 10−12 | forkhead box protein I3a | foxi3a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, W.; Liu, L.; Chen, D.; Wang, K.; Lin, S.; Chen, W.; Li, S.; Deng, B.; Li, Q.; Han, C. De Novo Assembly, Characterization and Comparative Transcriptome Analysis of the Mature Male and Female Gonads in Acrossocheilus parallens. Animals 2025, 15, 806. https://doi.org/10.3390/ani15060806
Liang W, Liu L, Chen D, Wang K, Lin S, Chen W, Li S, Deng B, Li Q, Han C. De Novo Assembly, Characterization and Comparative Transcriptome Analysis of the Mature Male and Female Gonads in Acrossocheilus parallens. Animals. 2025; 15(6):806. https://doi.org/10.3390/ani15060806
Chicago/Turabian StyleLiang, Weiqian, Lanyuan Liu, Dingxian Chen, Kaifeng Wang, Shengyue Lin, Weijian Chen, Sixun Li, Binhua Deng, Qiang Li, and Chong Han. 2025. "De Novo Assembly, Characterization and Comparative Transcriptome Analysis of the Mature Male and Female Gonads in Acrossocheilus parallens" Animals 15, no. 6: 806. https://doi.org/10.3390/ani15060806
APA StyleLiang, W., Liu, L., Chen, D., Wang, K., Lin, S., Chen, W., Li, S., Deng, B., Li, Q., & Han, C. (2025). De Novo Assembly, Characterization and Comparative Transcriptome Analysis of the Mature Male and Female Gonads in Acrossocheilus parallens. Animals, 15(6), 806. https://doi.org/10.3390/ani15060806





