Mitochondrial Genome of Grapsus albolineatus and Insights into the Phylogeny of Brachyura
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Extraction
2.2. PCR Amplification and Sequencing
2.3. Sequencing and Analysis
2.4. Phylogenetic Analysis
3. Results and Discussion
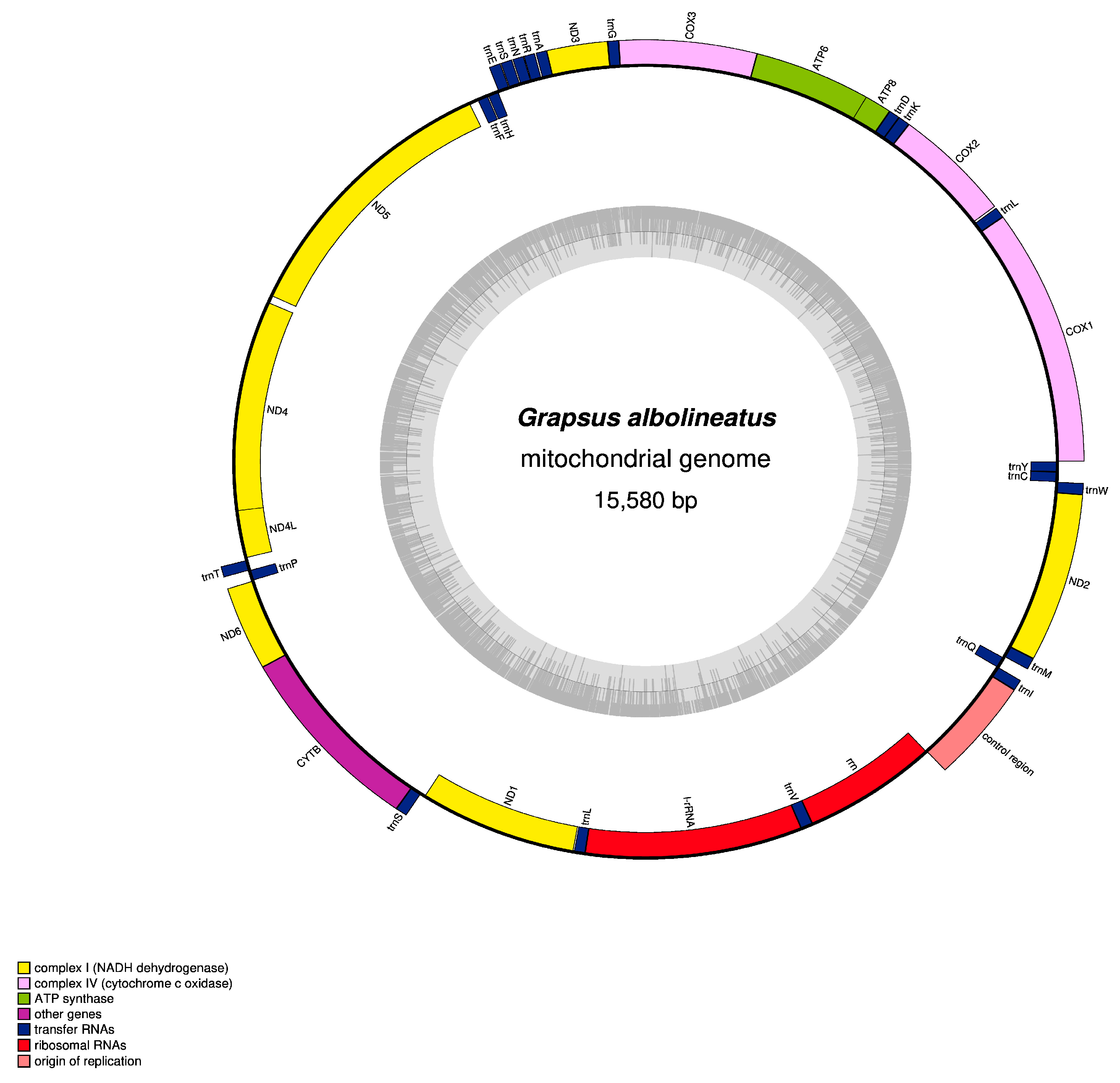
3.1. Mitogenome Organisation and Nucleotide Composition
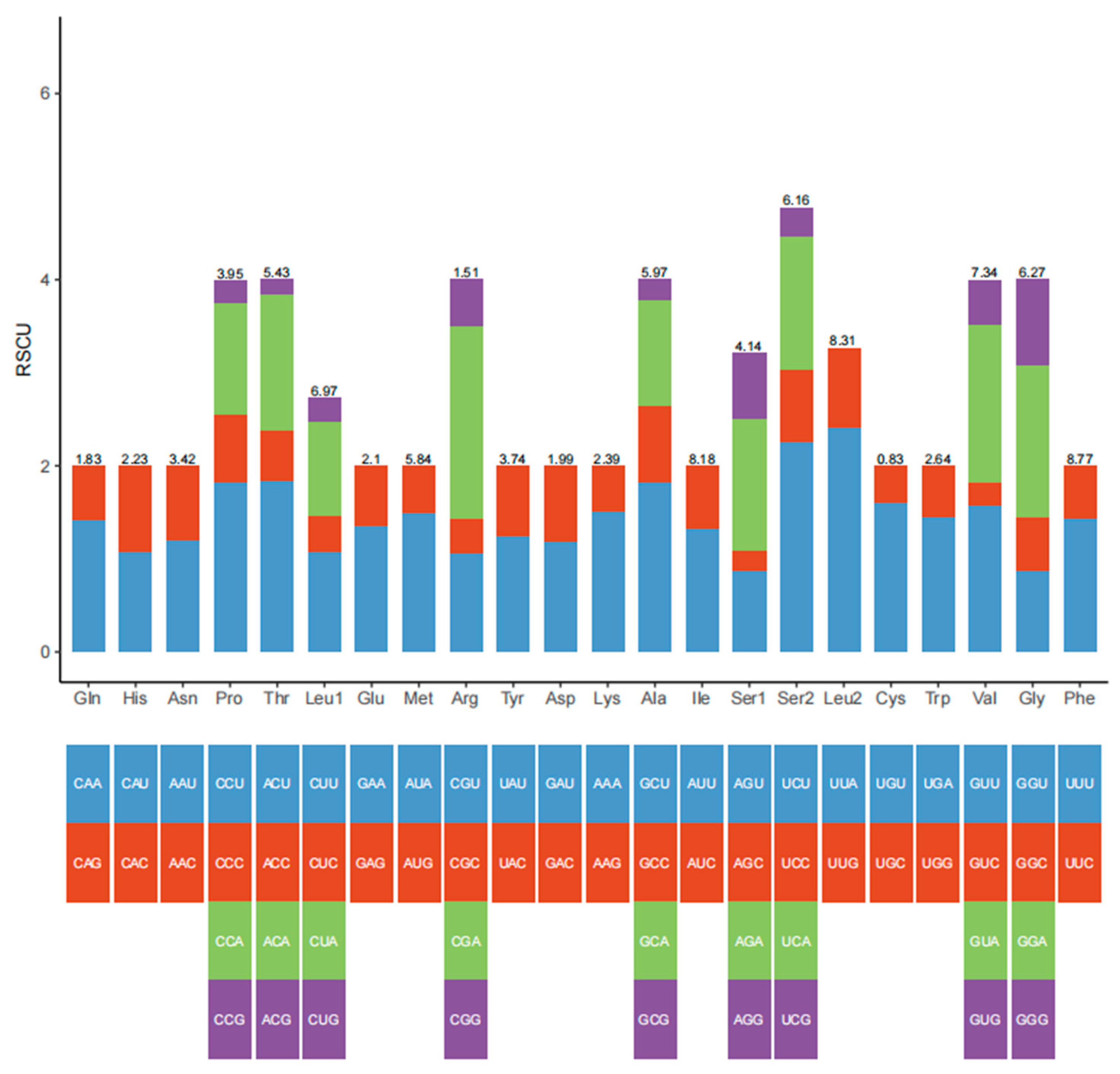
3.2. Protein-Coding Genes
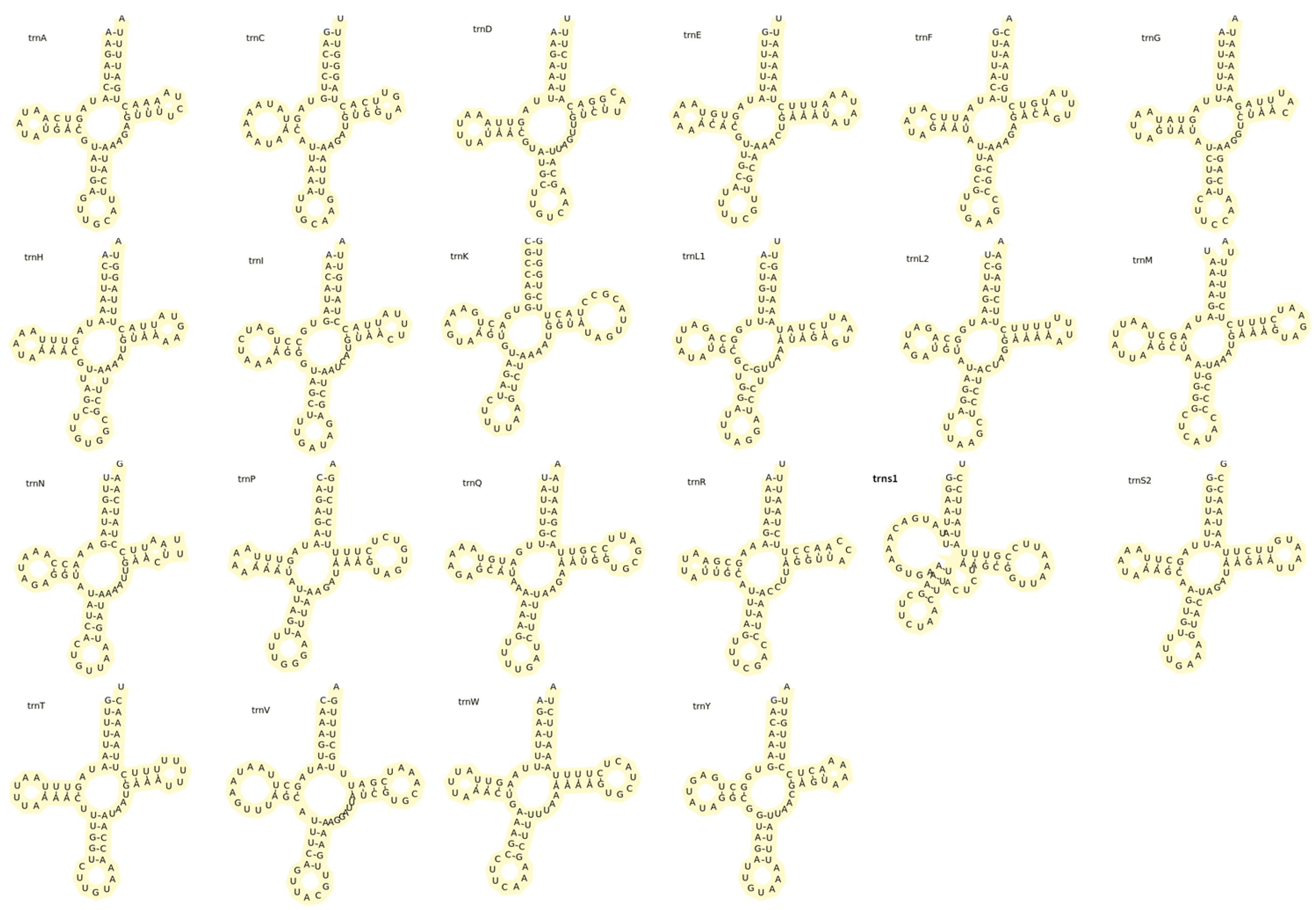
3.3. Transfer RNA and Ribosomal RNA Genes and Control Region
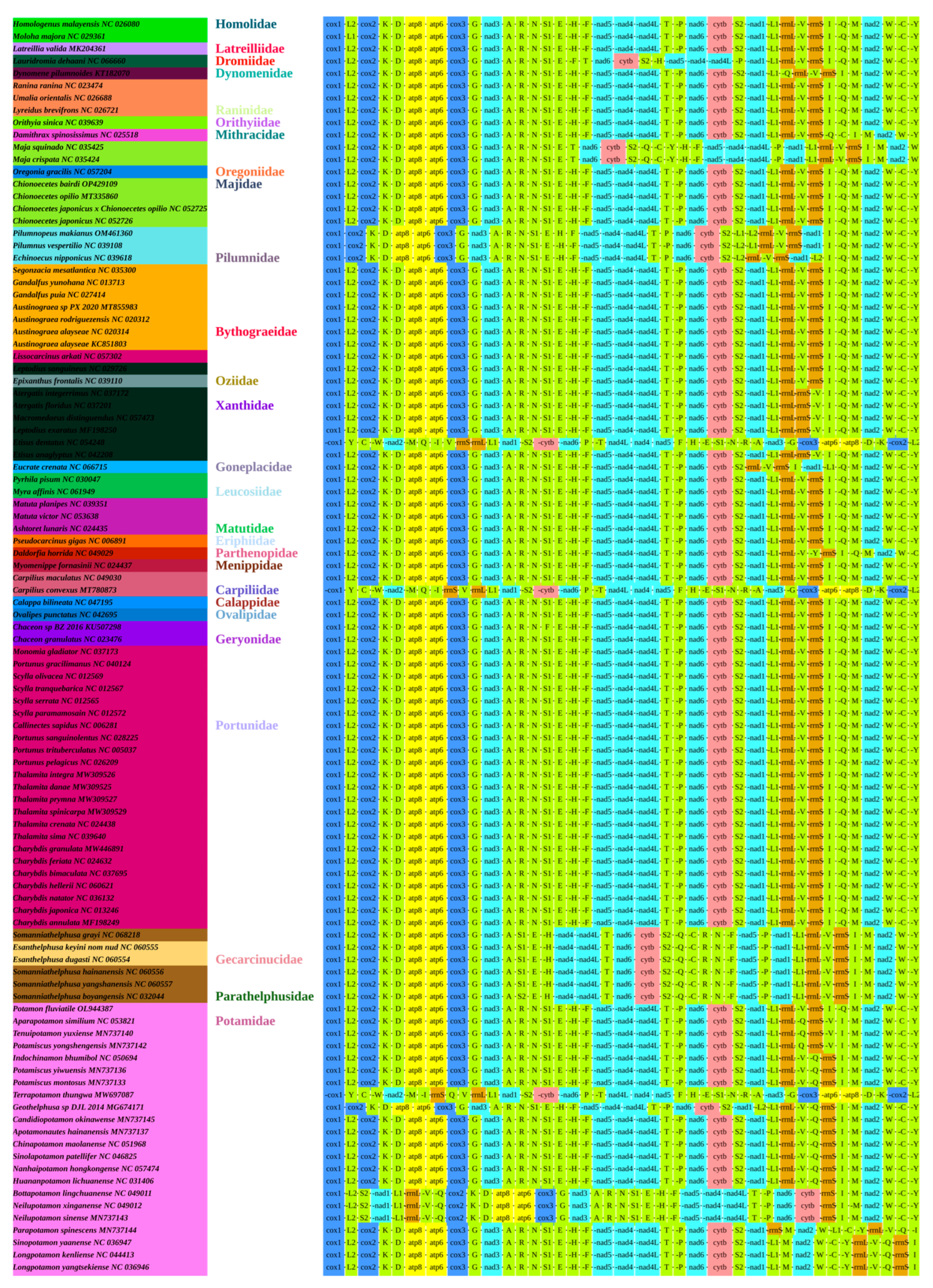
3.4. Gene Arrangement
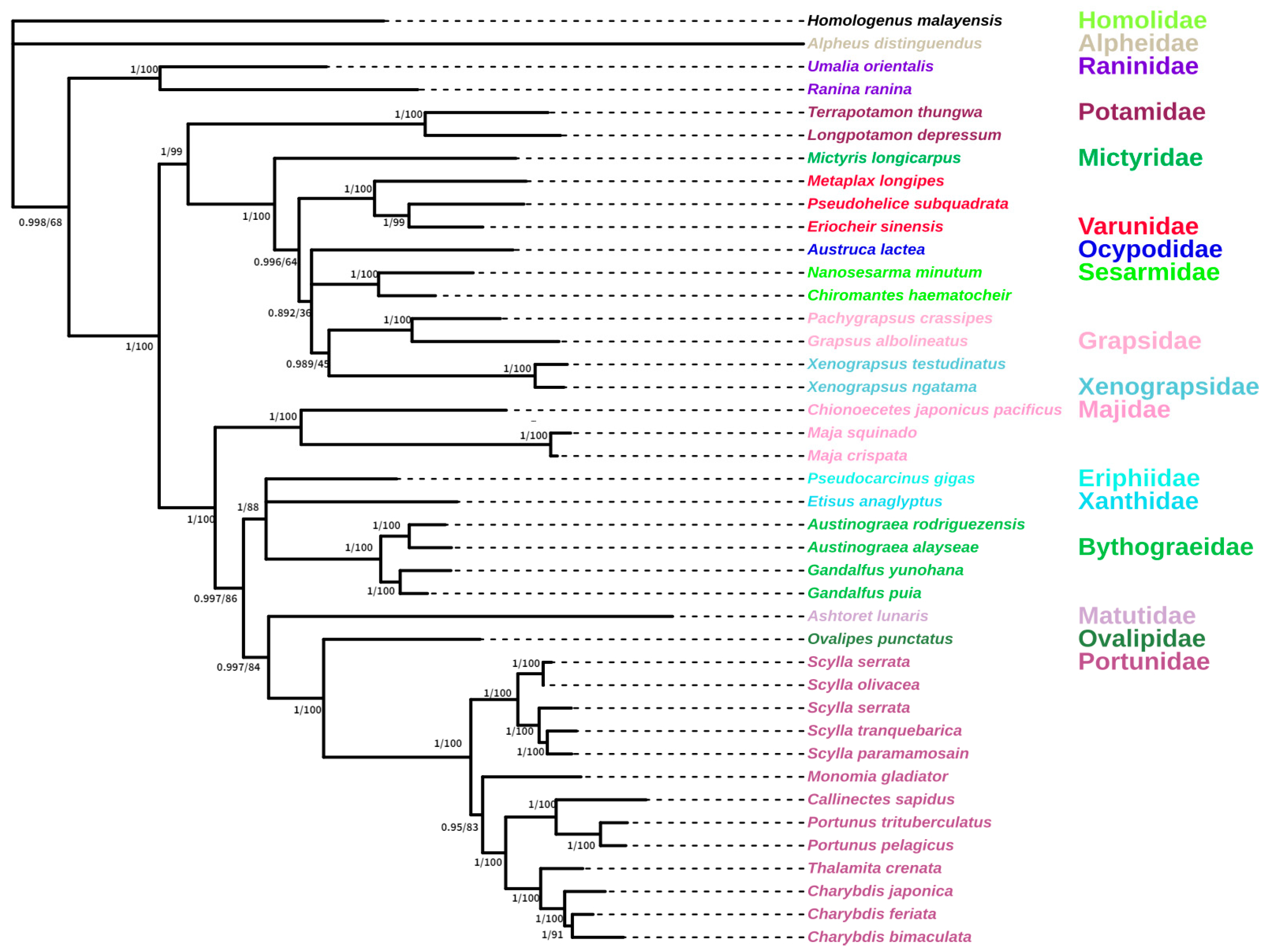
3.5. Phylogenetic Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brown, W.M.; George, M.; Wilson, A.C. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1979, 76, 1967–1971. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lu, C.; Liu, Q.; Zou, T.; Qiao, G.; Huang, X. Insights into the evolution of Aphid mitogenome features from new data and comparative analysis. Animals 2022, 12, 1970. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xin, Z.Z.; Zhu, X.Y.; Zhao, X.M.; Wang, Y.; Tang, B.P.; Zhang, H.B.; Zhang, D.Z.; Zhou, C.L.; Liu, Q.N. The complete mitochondrial genome of Euproctis similis (Lepidoptera: Noctuoidea: Erebidae) and phylogenetic analysis. Int. J. Biol. Macromol. 2017, 105, 219–227. [Google Scholar] [CrossRef]
- Guinot, D.; Tavares, M.; Castro, P. Significance of the sexual openings and supplementary structures on the phylogeny of brachyuran crabs (Crustacea, Decapoda, Brachyura), with new nomina for higher-ranked podotreme taxa. Zootaxa 2013, 3665, 1–414. [Google Scholar] [CrossRef]
- Martin, J.W.; Davis, G.E. An updated classification of the recent crustacea. Invertebr. Syst. 2001, 16, 837–838. [Google Scholar]
- Guinot, D.; Jamieson, B.G.M.; Deforges, B.R. Relationship of Homolidae and Dromiidae: Evidence from Spermatozoal Ultrastructure (Crustacea, Decapoda). Acta Zool. 1994, 75, 255–267. [Google Scholar] [CrossRef]
- Ng, P.K.L.; Guinot, D. Systema Brachyurorum: Part I. An annotated checklist of extant Brachyuran crabs of the world. Raffles Bull. Zool. 2008, 17, 1–286. [Google Scholar]
- Tsang, L.M.; Schubart, C.D.; Ahyong, S.T.; Lai, J.C.Y.; Au, E.Y.C.; Chan, T.Y.; Ng, P.K.L.; Chu, K.H. Evolutionary History of True Crabs (Crustacea: Decapoda: Brachyura) and the Origin of Freshwater Crabs. Mol. Biol. Evol. 2014, 31, 1173–1187. [Google Scholar] [CrossRef]
- Ahyong, S.T.; O’Meally, D. Phylogeny of the Decapoda reptantia: Resolution using three molecular loci and morphology. Raffles Bull. Zool. 2004, 52, 673–693. [Google Scholar]
- Karasawa, H.; Schweitzer, C.E.; Feldmann, R.M. Phylogenetic analysis and revised classification of podotrematous Brachyura (Decapoda) including extinct and extant families. J. Crustac. Biol. 2011, 31, 523–565. [Google Scholar] [CrossRef]
- Metzker, M.L. Sequencing technologies-the next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Lei, R.H.; Frasier, C.L.; Hawkins, M.T.R.; Engberg, S.E.; Bailey, C.A.; Johnson, S.E.; McLain, A.T.; Groves, C.P.; Perry, G.H.; Nash, S.D.; et al. Phylogenomic reconstruction of Sportive Lemurs (genus Lepilemur) recovered from mitogenomes with inferences for Madagascar biogeography. J. Hered. 2017, 108, 107–119. [Google Scholar] [PubMed]
- Mueller, R.L.; Macey, J.R.; Jaekel, M.; Wake, D.B.; Boore, J.L. Morphological homoplasy, life history evolution, and historical biogeography of plethodontid salamanders inferred from complete mitochondrial genomes. Proc. Natl. Acad. Sci. USA 2004, 101, 13820–13825. [Google Scholar] [CrossRef] [PubMed]
- Cerny, V.; Fernandes, V.; Costa, M.D.; Hajek, M.; Mulligan, C.J.; Pereira, L. Migration of Chadic speaking pastoralists within Africa based on population structure of Chad Basin and phylogeography of mitochondrial L3f haplogroup. BMC Evol. Biol. 2009, 9, 63. [Google Scholar] [CrossRef]
- Inak, G.; Lorenz, C.; Lisowski, P.; Zink, A.; Mlody, B.; Prigione, A. Concise review: Induced pluripotent stem cell-based drug discovery for mitochondrial disease. Stem Cells 2017, 35, 1655–1662. [Google Scholar] [CrossRef]
- Janzen, D.H.; Burns, J.M.; Cong, Q.; Hallwachs, W.; Dapkey, T.; Manjunath, R.; Hajibabaei, M.; Hebert, P.D.N.; Grishin, N.V. Nuclear genomes distinguish cryptic species suggested by their DNA barcodes and ecology. Proc. Natl. Acad. Sci. USA 2017, 114, 8313–8318. [Google Scholar] [CrossRef]
- Tang, B.P.; Liu, Y.; Xin, Z.Z.; Zhang, D.Z.; Wang, Z.F.; Zhu, X.Y.; Wang, Y.; Zhang, H.B.; Zhou, C.L.; Chai, X.Y.; et al. Characterisation of the complete mitochondrial genome of Helice wuana (Grapsoidea: Varunidae) and comparison with other Brachyuran crabs. Genomics 2018, 110, 221–230. [Google Scholar] [CrossRef]
- Boore, J.L.; Brown, W.M. Big trees from little genomes: Mitochondrial gene order as a phylogenetic tool. Curr. Opin. Genet. Dev. 1998, 8, 668–674. [Google Scholar] [CrossRef]
- Carter, J.K.; Innes, P.; Goebl, A.M.; Johnson, B.; Gebert, M.; Attia, Z.; Kane, N.C. Complete mitochondrial genomes provide current refined phylogenomic hypotheses for relationships among ten Hirundo species. Mitochondrial DNA Part B 2020, 5, 2881–2885. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, S.; Chen, J.; Chen, C.; Lin, X.; Peng, H.; Wang, X. Characterization and phylogenetic analysis of the complete mitochondrial genome sequence of Photinia serratifolia. Sci. Rep. 2023, 13, 770. [Google Scholar] [CrossRef]
- Shen, J.; Dai, A. Illustrated Fauna of China: Crustacea; Crabs; Science Press: Beijing, China, 1964; Volume 2, pp. 1–142. [Google Scholar]
- VanHook, A.M.; Patel, N.H. Crustaceans. Curr. Biol. 2008, 18, R547–R550. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P.; Teslenko, M. Draft MrBayes, Version 3.2 Manual; Tutorials and Model Summaries. 2011. Available online: https://web-genobioinfo.toulouse.inrae.fr/~formation/11_Phylogeny/doc/mb3.2_manual.pdf (accessed on 10 May 2022).
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, H.S.; Ju, S.J. Mitochondrial genome of the hydrothermal vent crab Austinograea alayseae (Crustacea: Bythograeidae): Genetic differences between individuals from Tofua Arc and Manus Basin. Mitochondrial DNA 2014, 25, 251–252. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Ma, C.; Li, X.; Xu, Z.; Feng, N.; Ma, L. The complete mitochondrial genome sequence and gene organization of the mud crab (Scylla paramamosain) with phylogenetic consideration. Gene 2013, 519, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Ma, C.; Li, C.; Lu, J.; Zou, X.; Gong, Y.; Wang, W.; Chen, W.; Ma, L.; Xia, L. First mitochondrial genome for the red crab (Charybdis feriata) with implication of phylogenomics and population genetics. Sci. Rep. 2015, 5, 11524. [Google Scholar] [CrossRef]
- Inoue, J.G.; Miya, M.; Tsukamoto, K.; Nishida, M. Complete mitochondrial DNA sequence of the Japanese sardine, Sardinops melanostictus. Fish. Sci. 2000, 66, 924–932. [Google Scholar] [CrossRef]
- Liu, Q.N.; Tang, Y.Y.; Yang, T.T.; Li, Y.T.; Yu, X.M. Phylogenetic relationships of Grapsoidea and insights into the higher phylogeny of Brachyuran. Genomics 2021, 113, 429–439. [Google Scholar] [CrossRef]
- Li, Y.T.; Xin, Z.Z.; Tang, Y.Y.; Yang, T.T.; Tang, B.P.; Sun, Y.; Zhang, D.Z.; Zhou, C.L.; Liu, Q.N.; Yu, X.M. Comparative mitochondrial genome analyses of Sesarmid and other Brachyuran crabs reveal gene rearrangements and phylogeny. Front. Genet. 2020, 11, 536640. [Google Scholar] [CrossRef]
- Ki, J.S.; Dahms, H.U.; Hwang, J.S.; Lee, J.S. The complete mitogenome of the hydrothermal vent crab Xenograpsus testudinatus (Decapoda, Brachyura) and comparison with brachyuran crabs. Comp. Biochem. Physiol. D 2009, 4, 290–299. [Google Scholar] [CrossRef]
- Boore, J.L.; Lavrov, D.V.; Brown, W.M. Gene translocation links insects and crustaceans. Nature 1998, 392, 667–668. [Google Scholar] [CrossRef]
- Shen, X.; Ren, J.; Cui, Z.; Sha, Z.; Wang, B.; Xiang, J.; Liu, B. The complete mitochondrial genomes of two common shrimps (Litopenaeus vannamei and Fenneropenaeus chinensis) and their phylogenomic considerations. Gene 2007, 403, 98–109. [Google Scholar] [CrossRef]
- Xin, Z.-Z.; Liu, Y.; Zhang, D.-Z.; Chai, X.-Y.; Wang, Z.-F.; Zhang, H.-B.; Zhou, C.-L.; Tang, B.-P.; Liu, Q.-N. Complete mitochondrial genome of Clistocoeloma sinensis (Brachyura: Grapsoidea): Gene rearrangements and higher-level phylogeny of the Brachyura. Sci. Rep. 2017, 7, 4489. [Google Scholar] [CrossRef] [PubMed]
- Jühling, F.; Pütz, J.; Bernt, M.; Donath, A.; Middendorf, M.; Florentz, C.; Stadler, P.F. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res. 2012, 40, 2833–2845. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, T.A.; Collins, T.M.; Bieler, R. Changing identities: tRNA duplication and remolding within animal mitochondrial genomes. Proc. Natl. Acad. Sci. USA 2003, 100, 15700–15705. [Google Scholar] [CrossRef]
- Sahyoun, A.H.; Hölzer, M.; Jühling, F.; Höner zu Siederdissen, C.; Al-Arab, M.; Tout, K.; Marz, M.; Middendorf, M.; Stadler, P.F.; Bernt, M. Towards a comprehensive picture of alloacceptor tRNA remolding in metazoan mitochondrial genomes. Nucleic Acids Res. 2015, 43, 8044–8056. [Google Scholar] [CrossRef]
- Du, S.; Pan, D.; Zhang, K.; Liu, C.; Yin, J.; Cumberlidge, N.; Sun, H. A novel gene order and remolded tRNAs revealed in the mitogenome of Asian gecarcinucid freshwater crabs (Brachyura, Gecarcinucidae). Gene 2022, 813, 146102. [Google Scholar] [CrossRef]
- Shi, W.; Gong, L.; Wang, S.Y.; Miao, X.G.; Kong, X.Y. Tandem Duplication and Random Loss for mitogenome rearrangement in Symphurus (Teleost: Pleuronectiformes). BMC Genom. 2015, 16, 355. [Google Scholar] [CrossRef]
- Simon, C.; Buckley, T.R.; Frati, F.; Stewart, J.B.; Beckenbach, A.T. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2016, 37, 545–579. [Google Scholar] [CrossRef]
- Komai, T.; Yamasaki, I.; Kobayashi, S.; Yamamoto, T.; Watanabe, S. Eriocheir ogasawaraensis Komai, a new species of mitten crab (Crustacea: Decapoda: Brachyura: Varunidae) from the Ogasawara Islands, Japan, with notes on the systematics of Eriocheir De Haan, 1835. Zootaxa 2006, 1168, 1–20. [Google Scholar] [CrossRef]
- Xin, Z.-Z.; Liu, Y.; Zhang, D.-Z.; Wang, Z.-F.; Zhang, H.-B.; Tang, B.-P.; Zhou, C.-L.; Chai, X.-Y.; Liu, Q.-N. Mitochondrial genome of Helice tientsinensis (Brachyura: Grapsoidea: Varunidae): Gene rearrangements and higher-level phylogeny of the Brachyura. Gene 2017, 627, 307–314. [Google Scholar] [CrossRef]
- Ingman, M.; Kaessmann, H.; Pääbo, S.; Gyllensten, U. Mitochondrial genome variation and the origin of modern humans. Nature 2000, 408, 708–713. [Google Scholar] [CrossRef]
- Tang, B.-P.; Xin, Z.-Z.; Liu, Y.; Zhang, D.-Z.; Wang, Z.-F.; Zhang, H.-B.; Chai, X.-Y.; Zhou, C.-L.; Liu, Q.-N. The complete mitochondrial genome of Sesarmops sinensis reveals gene rearrangements and phylogenetic relationships in Brachyura. PLoS ONE 2017, 12, e0179800. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.-Y.; Tang, B.-P.; Xin, Z.-Z.; Li, Y.-T.; Zha, X.-H.; Zhang, D.-Z.; Sun, Y.; Liu, Q.-N.; Ma, Y.-F. Characterization of the complete mitochondrial genome of Helice latimera and its phylogenetic implications in Brachyura. Genomics 2020, 112, 5180–5187. [Google Scholar] [CrossRef] [PubMed]
- Tsang, C.T.T.; Schubart, C.D.; Chu, K.H.; Ng, P.K.L.; Tsang, L.M. Molecular phylogeny of Thoracotremata crabs (Decapoda, Brachyura): Toward adopting monophyletic superfamilies, invasion history into terrestrial habitats and multiple origins of symbiosis. Mol. Phylogenet. Evol. 2022, 177, 107596. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.T.; Zhou, X.J.; Yang, Q.; Lu, Y.B.; Wang, J.; Zou, J.X. Adaptive evolution characteristics of mitochondrial genomes in genus Aparapotamon (Brachyura, Potamidae) of freshwater crabs. BMC Genom. 2023, 24, 193. [Google Scholar] [CrossRef]
- Ghanavi, H.R.; Rahimi, P.; Tavana, M.; Rezaei Tavabe, K.; Jouladeh-Roudbar, A.; Doadrio, I. The evolutionary journey of freshwater crabs of the genus Potamon (Decapoda: Brachyura: Potamidae). Mol. Phylogenet. Evol. 2023, 180, 107690. [Google Scholar] [CrossRef]

| Species | Family | Size (bp) | Accession No. |
|---|---|---|---|
| Grapsus albolineatus | Grapsidae | 15,580 | MF198247 |
| Chionoecetes japonicus pacificus | Majidae | 15,341 | AB735678 |
| Scylla serrata | Portunidae | 15,721 | HM590866 |
| Scylla paramamosain | Portunidae | 15,824 | JX457150 |
| Austinograea alayseae | Bythograeidae | 15,611 | KC581803 |
| Umalia orientalis | Raninidae | 15,466 | KM365084 |
| Eriocheir sinensis | Varunidae | 16,350 | KP064329 |
| Gandalfus puia | Bythograeidae | 15,548 | KR002727 |
| Maja crispata | Majidae | 16,592 | KY650651 |
| Maja squinado | Majoidea | 16,598 | KY650652 |
| Xenograpsus ngatama | Xenograpsidae | 16,106 | KY985236 |
| Ashtoret lunaris | Matutidae | 15,807 | LK391941 |
| Mictyris longicarpus | Mictyridae | 15,548 | LN611670 |
| Metaplax longipes | Varunidae | 16,424 | MF198248 |
| Charybdis bimaculata | Portunidae | 15,712 | MG787408 |
| Terrapotamon thungwa | Potamidae | 16,156 | MW697078 |
| Portunus trituberculatus | Portunidae | 16,026 | NC_005037 |
| Callinectes sapidus | Portunidae | 16,263 | NC_006281 |
| Pseudocarcinus gigas | Eriphiidae | 15,515 | NC_006891 |
| Scylla serrata | Portunidae | 15,775 | NC_012565 |
| Scylla tranquebarica | Portunidae | 15,833 | NC_012567 |
| Scylla olivacea | Portunidae | 15,723 | NC_012569 |
| Charybdis japonica | Portunidae | 15,738 | NC_013246 |
| Xenograpsus testudinatus | Xenograpsidae | 15,798 | NC_013480 |
| Gandalfus yunohana | Bythograeidae | 15,567 | NC_013713 |
| Alpheus distinguendus | Alpheidae | 15,700 | NC_014883 |
| Austinograea rodriguezensis | Bythograeidae | 15,611 | NC_020312 |
| Pachygrapsus crassipes | Grapsidae | 15,652 | NC_021754 |
| Ranina ranina | Raninidae | 15,557 | NC_023474 |
| Thalamita crenata | Portunidae | 15,787 | NC_024438 |
| Charybdis feriata | Portunidae | 15,660 | NC_024632 |
| Homologenus malayensis | Homolidae | 15,793 | NC_026080 |
| Portunus pelagicus | Portunidae | 16,157 | NC_026209 |
| Monomia gladiator | Portunidae | 15,878 | NC_037173 |
| Nanosesarma minutum | Sesarmidae | 15,637 | NC_040977 |
| Chiromantes haematocheir | Sesarmidae | 15,899 | NC_042142 |
| Etisus anaglyptus | Xanthidae | 16,435 | NC_042208 |
| Austruca lactea | Ocypodidae | 15,659 | NC_042401 |
| Pseudohelice subquadrata | Varunidae | 16,898 | NC_042685 |
| Ovalipes punctatus | Ovalipidae | 16,084 | NC_042695 |
| Longpotamon depressum | Potamidae | 16,537 | NC_057478 |
| Gene | Direction | Location | Size (bp) | Intergenic Nucleotides | Start Codon | Stop Codon |
|---|---|---|---|---|---|---|
| cox1 | F | 1–1534 | 1534 | ATG | T | |
| trnL2 | F | 1535–1599 | 65 | |||
| cox2 | F | 1610–2297 | 688 | ATG | T | |
| trnK | F | 2298–2366 | 69 | |||
| trnD | F | 2367–2430 | 64 | |||
| atp8 | F | 2431–2589 | 159 | GTG | TAA | |
| atp6 | F | 2583–3257 | 675 | −7 | ATT | TAA |
| cox3 | F | 3257–4048 | 792 | −1 | ATG | TAA |
| trnG | F | 4048–4110 | 63 | −1 | ||
| nad3 | F | 4111–4461 | 351 | ATC | TAA | |
| trnA | F | 4460–4523 | 64 | −2 | ||
| trnR | F | 4530–4593 | 64 | |||
| trnN | F | 4595–4659 | 65 | |||
| trnS1 | F | 4664–4730 | 67 | |||
| trnE | F | 4733–4800 | 68 | |||
| trnH | R | 4804–4868 | 65 | |||
| trnF | R | 4873–4937 | 65 | |||
| nad5 | R | 4990–6720 | 1731 | ATG | TAA | |
| nad4 | R | 6765–8102 | 1338 | ATG | TAG | |
| nad4L | R | 8096–8398 | 303 | −7 | ATG | TAA |
| trnT | F | 8413–8478 | 66 | |||
| trnP | R | 8479–8545 | 67 | |||
| nad6 | F | 8548–9051 | 504 | ATT | TAA | |
| cytb | F | 9051–10,185 | 1135 | −1 | ATG | T |
| trnS2 | F | 10,186–10,253 | 68 | |||
| nad1 | R | 10,281–11,246 | 966 | ATT | TAA | |
| trnL1 | R | 11,252–11,318 | 67 | |||
| rrnL | R | 11,319–12,646 | 1328 | |||
| trnV | R | 12,647–12,719 | 73 | |||
| rrnS | R | 12,720–13,546 | 827 | |||
| CR | - | 13,547–14,762 | 616 | |||
| trnI | F | 14,164–14,229 | 67 | |||
| trnQ | R | 14,227–14,297 | 71 | |||
| trnM | F | 14,305–14,375 | 71 | |||
| nad2 | F | 14,376–15,386 | 1011 | ATG | TAG | |
| trnW | F | 15,385–15,453 | 69 | −2 | ||
| trnC | R | 15,453–15,516 | 64 | −1 | ||
| trnY | R | 15,517–15,680 | 64 |
| Species | Size (bp) | A (%) | T (%) | C (%) | G (%) | A + T (%) | A + T Skew | C + G Skew |
|---|---|---|---|---|---|---|---|---|
| G. albolineatus | 15,580 | 33.4 | 34 | 20.5 | 12.1 | 67.4 | −0.01 | −0.26 |
| C. jpacificus | 15,341 | 34.6 | 37 | 17.2 | 11.1 | 71.6 | −0.033 | −0.215 |
| S. serrata | 15,721 | 33.4 | 35.8 | 19.5 | 11.3 | 69.2 | −0.034 | −0.266 |
| S. paramamosain | 15,824 | 34.9 | 38.2 | 16.8 | 10.2 | 73.1 | −0.045 | −0.247 |
| A. alayseae | 15,611 | 34.5 | 32.4 | 21.9 | 11.3 | 66.9 | 0.032 | −0.321 |
| U. orientalis | 15,466 | 33.1 | 34.9 | 20.2 | 11.8 | 68 | −0.027 | −0.262 |
| E. sinensis | 16,350 | 35.3 | 36.4 | 17.6 | 10.7 | 71.7 | −0.015 | −0.245 |
| G. puia | 15,548 | 35.1 | 34.8 | 19.8 | 10.3 | 69.9 | 0.006 | −0.313 |
| M. crispata | 16,592 | 33.6 | 36.7 | 18.6 | 11.1 | 70.3 | −0.044 | −0.25 |
| M. squinado | 16,598 | 33.7 | 37.1 | 18.2 | 11 | 70.8 | −0.047 | −0.245 |
| X. ngatama | 16,106 | 36.1 | 36.8 | 17.5 | 9.6 | 72.9 | −0.01 | −0.293 |
| A. lunaris | 15,807 | 34.8 | 35.4 | 18.7 | 11.1 | 70.2 | −0.009 | −0.256 |
| M. longicarpus | 15,548 | 32.4 | 36.6 | 19.2 | 11.8 | 69 | −0.06 | −0.236 |
| M. longipes | 16,424 | 37.5 | 34.2 | 18 | 10.4 | 71.7 | 0.046 | −0.266 |
| C. bimaculata | 15,712 | 33.9 | 37.6 | 16.9 | 11.5 | 71.5 | −0.052 | −0.192 |
| T. thungwa | 16,156 | 37.2 | 36 | 9.1 | 17.6 | 73.2 | 0.017 | 0.318 |
| P. trituberculatus | 16,026 | 33.3 | 36.9 | 18.5 | 11.3 | 70.2 | −0.051 | −0.241 |
| C. sapidus | 16,263 | 34.2 | 34.9 | 19.8 | 11.1 | 69.1 | −0.011 | −0.279 |
| P. gigas | 15,515 | 35 | 35.5 | 18.7 | 10.8 | 70.5 | −0.006 | −0.268 |
| S. serrata | 15,775 | 34.6 | 38 | 17.1 | 10.4 | 72.6 | −0.047 | −0.242 |
| S. tranquebarica | 15,833 | 35 | 38.7 | 16.5 | 9.7 | 73.7 | −0.05 | −0.258 |
| S. olivacea | 15,723 | 33.5 | 35.9 | 19.4 | 11.2 | 69.4 | −0.035 | −0.267 |
| C. japonica | 15,738 | 33.8 | 35.4 | 18.9 | 11.9 | 69.2 | −0.024 | −0.228 |
| X. testudinatus | 15,798 | 36.7 | 37.2 | 16.8 | 9.3 | 73.9 | −0.007 | −0.286 |
| G. yunohana | 15,567 | 34.3 | 35.7 | 19.3 | 10.8 | 70 | −0.019 | −0.281 |
| A. distinguendus | 15,700 | 32.3 | 27.9 | 25.5 | 14.4 | 60.2 | 0.073 | −0.278 |
| A. rodriguezensis | 15,611 | 35.3 | 33.5 | 20.9 | 10.3 | 68.8 | 0.025 | −0.341 |
| P. crassipes | 15,652 | 30.5 | 35.8 | 21 | 12.7 | 66.3 | −0.08 | −0.245 |
| R. ranina | 15,557 | 30.3 | 36.3 | 21.1 | 12.2 | 66.6 | −0.09 | −0.266 |
| T. crenata | 15,787 | 34.4 | 35.3 | 18.8 | 11.5 | 69.7 | −0.013 | −0.24 |
| C. feriata | 15,660 | 34.1 | 36.1 | 18.6 | 11.3 | 70.2 | −0.028 | −0.246 |
| H. malayensis | 15,793 | 37.3 | 34.4 | 18.3 | 10 | 71.7 | 0.04 | −0.292 |
| P. pelagicus | 16,157 | 33.7 | 35 | 19.1 | 12.2 | 68.7 | −0.019 | −0.219 |
| M. gladiator | 15,878 | 33.3 | 35.7 | 19.2 | 11.8 | 69 | −0.034 | −0.242 |
| N. minutum | 15,637 | 38 | 39.7 | 13.4 | 8.9 | 77.7 | −0.022 | −0.201 |
| C. haematocheir | 15,899 | 37.3 | 38.3 | 15 | 9.4 | 75.6 | −0.013 | −0.226 |
| E. anaglyptus | 16,435 | 33.2 | 34.8 | 21 | 11.1 | 68 | −0.023 | −0.309 |
| A. lactea | 15,659 | 34.8 | 34.6 | 18.5 | 12 | 69.4 | 0.003 | −0.214 |
| P. subquadrata | 16,898 | 34.2 | 33.5 | 21.7 | 10.5 | 67.7 | 0.01 | −0.347 |
| O. punctatus | 16,084 | 32.6 | 35.5 | 19.4 | 12.5 | 68.1 | −0.042 | −0.218 |
| L. depressum | 16,537 | 35.4 | 37.9 | 17.3 | 9.3 | 73.3 | −0.034 | −0.302 |
| Codon | Count | RSCU | Codon | Count | RSCU | Codon | Count | RSCU | Codon | Count | RSCU |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UUU (F) | 233 | 1.43 | UCU (S) | 108 | 2.26 | UAU (Y) | 87 | 1.25 | UGU (C) | 25 | 1.61 |
| UUC (F) | 93 | 0.57 | UCC (S) | 37 | 0.77 | UAC (Y) | 52 | 0.75 | UGC (C) | 6 | 0.39 |
| UUA (L) | 229 | 2.42 | UCA (S) | 69 | 1.44 | UAA (*) | 8 | 1.6 | UGA (W) | 71 | 1.45 |
| UUG (L) | 80 | 0.85 | UCG (S) | 15 | 0.31 | UAG (*) | 2 | 0.4 | UGG (W) | 27 | 0.55 |
| CUU (L) | 102 | 1.08 | CCU (P) | 67 | 1.82 | CAU (H) | 45 | 1.08 | CGU (R) | 15 | 1.07 |
| CUC (L) | 36 | 0.38 | CCC (P) | 27 | 0.73 | CAC (H) | 38 | 0.92 | CGC (R) | 5 | 0.36 |
| CUA (L) | 96 | 1.01 | CCA (P) | 44 | 1.2 | CAA (Q) | 48 | 1.41 | CGA (R) | 29 | 2.07 |
| CUG (L) | 25 | 0.26 | CCG (P) | 9 | 0.24 | CAG (Q) | 20 | 0.59 | CGG (R) | 7 | 0.5 |
| AUU (I) | 201 | 1.32 | ACU (T) | 93 | 1.84 | AAU (N) | 76 | 1.2 | AGU (S) | 42 | 0.88 |
| AUC (I) | 103 | 0.68 | ACC (T) | 28 | 0.55 | AAC (N) | 51 | 0.8 | AGC (S) | 10 | 0.21 |
| AUA (M) | 163 | 1.5 | ACA (T) | 73 | 1.45 | AAA (K) | 67 | 1.51 | AGA (S) | 68 | 1.42 |
| AUG (M) | 54 | 0.5 | ACG (T) | 8 | 0.16 | AAG (K) | 22 | 0.49 | AGG (S) | 34 | 0.71 |
| GUU (V) | 107 | 1.57 | GCU (A) | 101 | 1.82 | GAU (D) | 44 | 1.19 | GGU (G) | 51 | 0.88 |
| GUC (V) | 17 | 0.25 | GCC (A) | 46 | 0.83 | GAC (D) | 30 | 0.81 | GGC (G) | 33 | 0.57 |
| GUA (V) | 116 | 1.7 | GCA (A) | 63 | 1.14 | GAA (E) | 53 | 1.36 | GGA (G) | 95 | 1.63 |
| GUG (V) | 33 | 0.48 | GCG (A) | 12 | 0.22 | GAG (E) | 25 | 0.64 | GGG (G) | 54 | 0.93 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Tang, S.; Chen, Y.; Liu, Q.; Tang, B. Mitochondrial Genome of Grapsus albolineatus and Insights into the Phylogeny of Brachyura. Animals 2025, 15, 679. https://doi.org/10.3390/ani15050679
Zhang X, Tang S, Chen Y, Liu Q, Tang B. Mitochondrial Genome of Grapsus albolineatus and Insights into the Phylogeny of Brachyura. Animals. 2025; 15(5):679. https://doi.org/10.3390/ani15050679
Chicago/Turabian StyleZhang, Xue, Sheng Tang, Yaohui Chen, Qiuning Liu, and Boping Tang. 2025. "Mitochondrial Genome of Grapsus albolineatus and Insights into the Phylogeny of Brachyura" Animals 15, no. 5: 679. https://doi.org/10.3390/ani15050679
APA StyleZhang, X., Tang, S., Chen, Y., Liu, Q., & Tang, B. (2025). Mitochondrial Genome of Grapsus albolineatus and Insights into the Phylogeny of Brachyura. Animals, 15(5), 679. https://doi.org/10.3390/ani15050679






