Metabolomic Insights into Energy Utilization Strategies of Asiatic Toads (Bufo gargarizans) During Hibernation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Metabolite Extraction
2.3. GC-MS and LC-MS Analysis
2.4. Data Processing and Analysis
3. Results
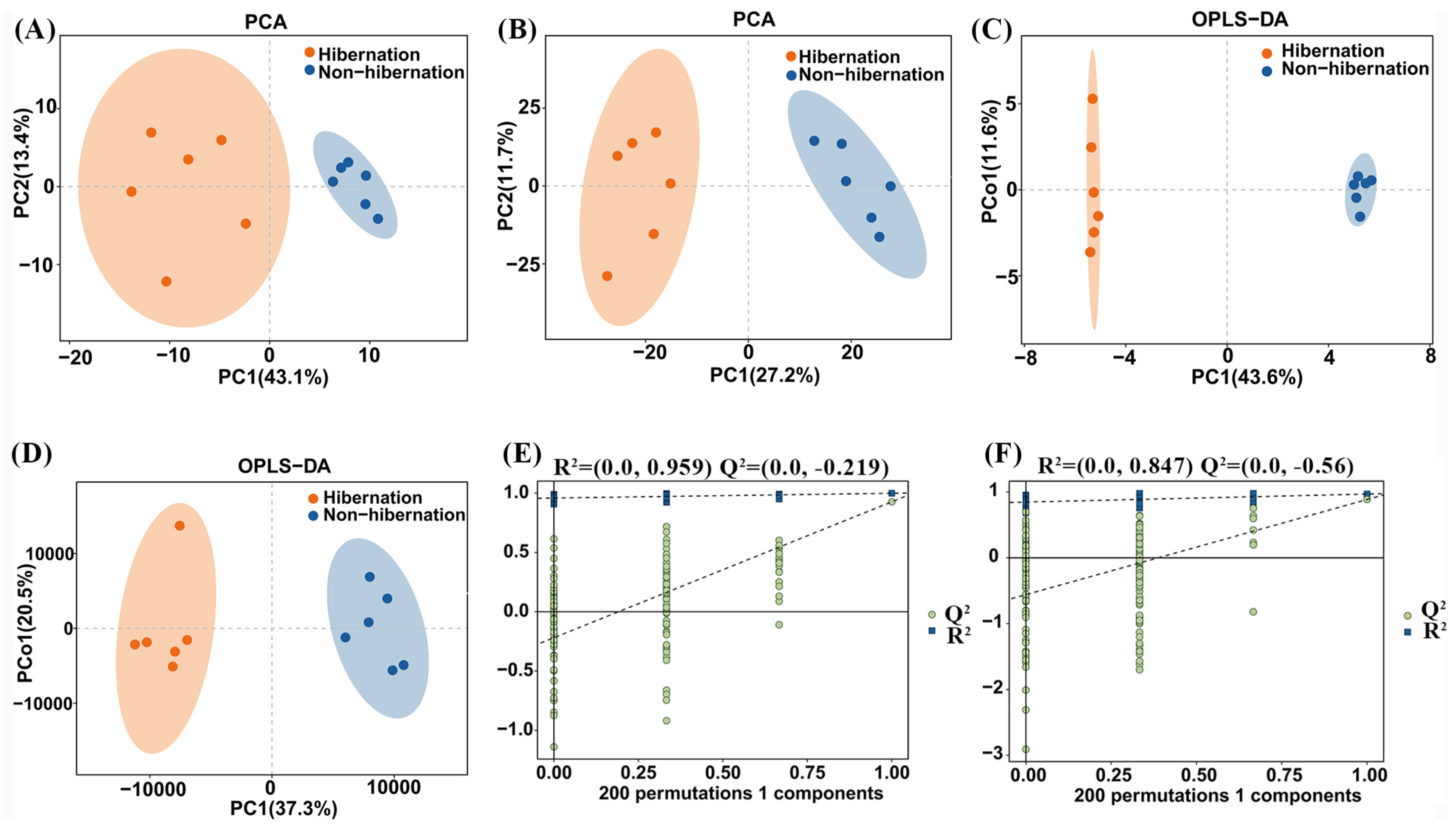
3.1. Characteristics of Hibernation and Non-Hibernation Groups
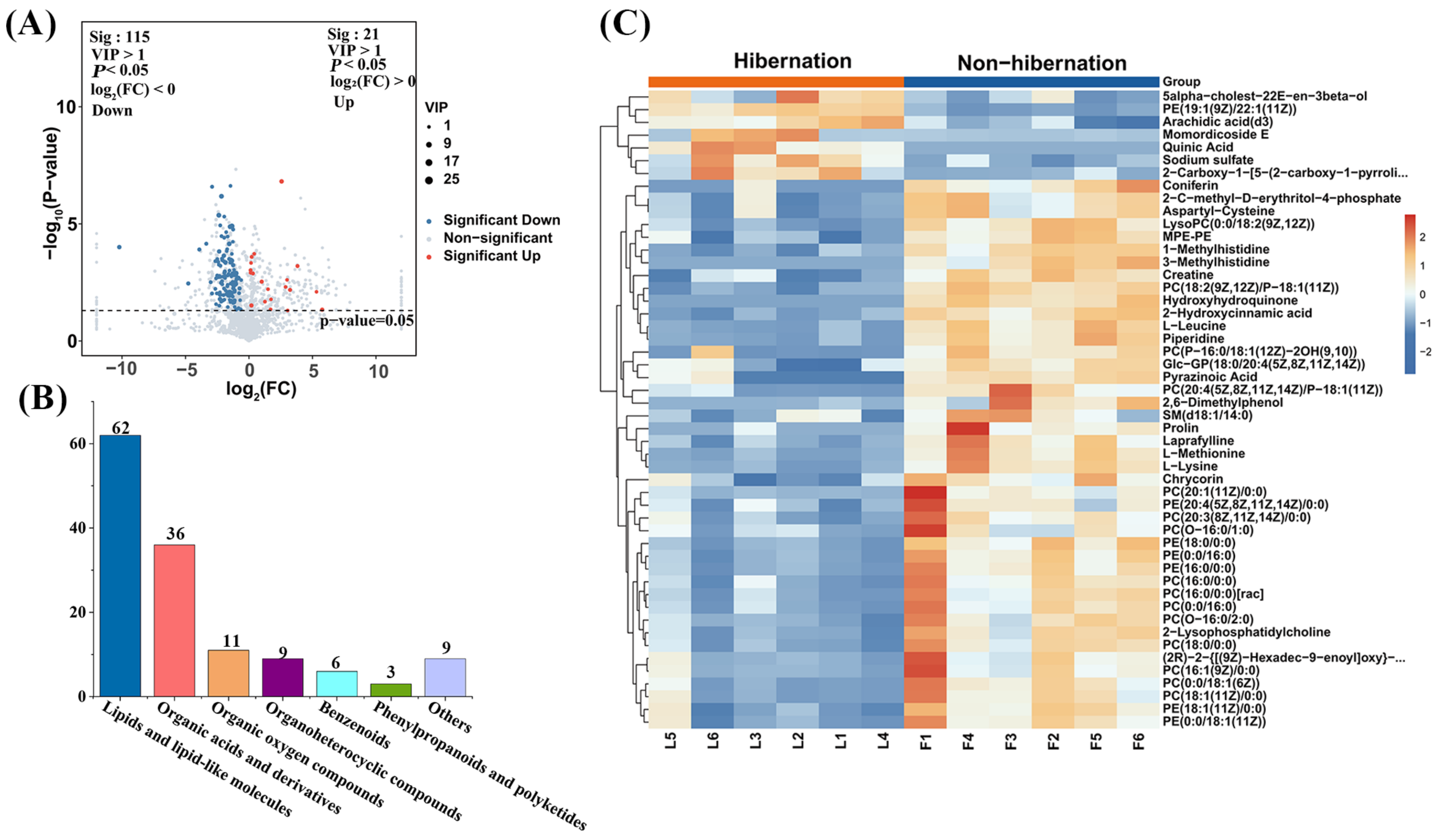
3.2. Differential Metabolite Profiles Between Hibernation and Non-Hibernation Toads
3.3. KEGG Analysis of Metabolic Pathway Differences
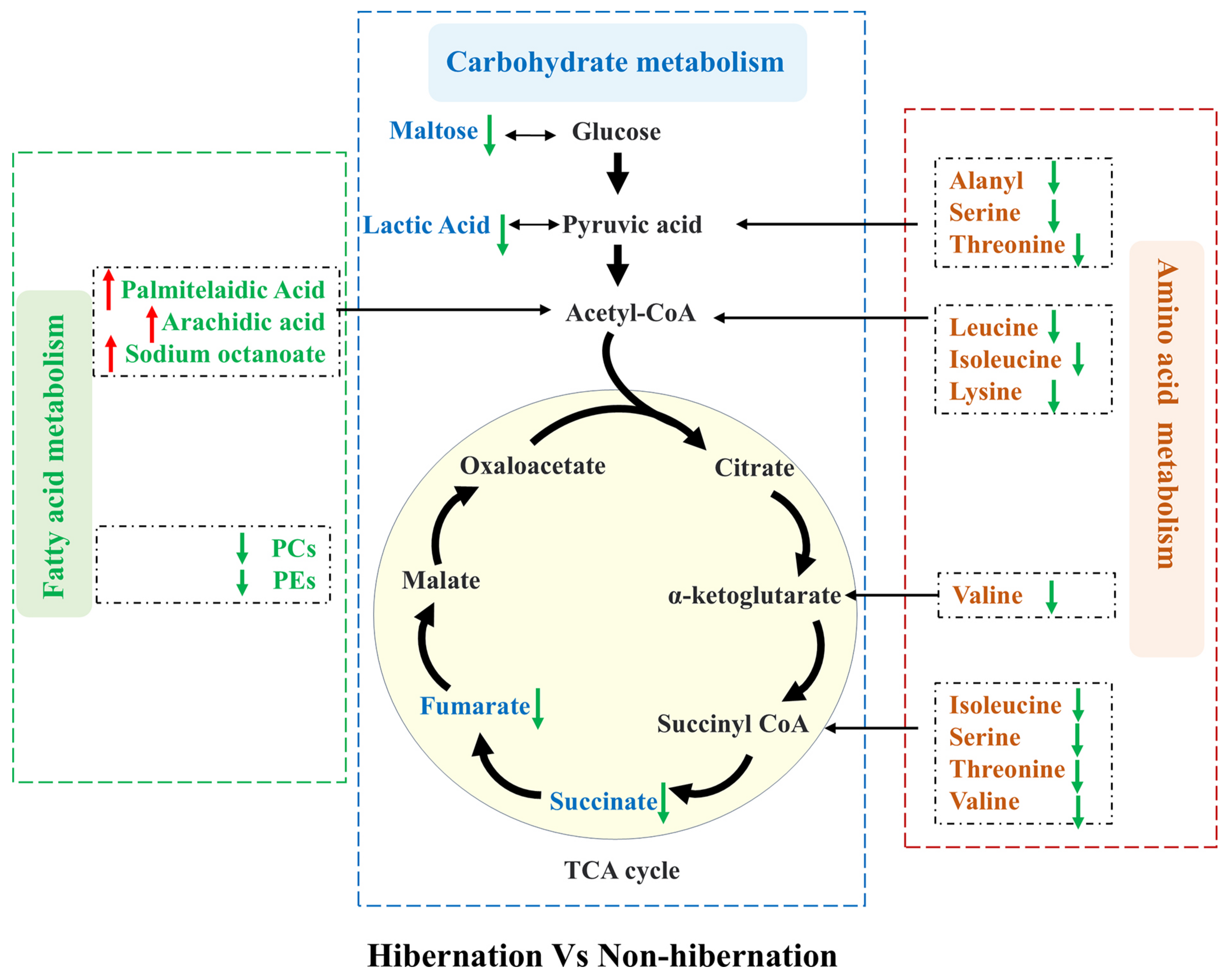
3.4. Metabolic Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DEMs | Differentially expressed metabolites; |
| PEs | Phosphatidylethanolamines; |
| PCs | Phosphatidylcholines; |
| LC-MS | Liquid chromatography–mass spectrometry; |
| GC-MS | Gas chromatography–mass spectrometry; |
| PCA | Principal component analysis; |
| OPLS-DA | Orthogonal partial least squares discriminant analysis; |
| KEGG | Kyoto encyclopedia of genes and genomes. |
References
- Mohr, S.M.; Bagriantsev, S.N.; Gracheva, E.O. Cellular, molecular, and physiological adaptations of hibernation: The solution to environmental challenges. Annu. Rev. Cell Dev. Biol. 2020, 36, 315–338. [Google Scholar] [CrossRef] [PubMed]
- Junkins, M.S.; Bagriantsev, S.N.; Gracheva, E.O. Towards understanding the neural origins of hibernation. J. Exp. Biol. 2022, 225, jeb229542. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.C.; Ultsch, G.R. Physiology of hibernation under the ice by turtles and frogs. J. Exp. Zool. Part A Ecol. Genet. Physiol. 2010, 313A, 311–327. [Google Scholar] [CrossRef] [PubMed]
- Klug, B.J.; Brigham, R.M. Changes to metabolism and cell physiology that enable mammalian hibernation. Springer Sci. Rev. 2015, 3, 39–56. [Google Scholar] [CrossRef]
- Borah, B.K.; Renthlei, Z.; Trivedi, A.K. Hypothalamus but not liver retains daily expression of clock genes during hibernation in terai tree frog (Polypedates teraiensis). Chronobiol. Int. 2020, 37, 485–492. [Google Scholar] [CrossRef]
- Wiebler, J.M.; Kohl, K.D.; Lee, R.E., Jr.; Costanzo, J.P. Urea hydrolysis by gut bacteria in a hibernating frog: Evidence for urea-nitrogen recycling in Amphibia. R. Soc. B-Biol. Sci. 2018, 285, 20180241. [Google Scholar] [CrossRef]
- Xie, Z.; Ahmad, I.M.; Zuo, L.; Xiao, F.; Wang, Y.; Li, D. Hibernation with rhythmicity: The circadian clock and hormonal adaptations of the hibernating asiatic toads (Bufo gargarizans). Integr. Zool. 2022, 17, 656–669. [Google Scholar] [CrossRef]
- Niu, Y.; Zhang, X.; Men, S.; Xu, T.; Zhang, H.; Li, X.; Storey, K.B.; Chen, Q. Effects of hibernation on two important contractile tissues in Tibetan frogs, Nanorana parkeri: A perspective from transcriptomics and metabolomics approaches. BMC Genom. 2024, 25, 454. [Google Scholar] [CrossRef]
- Sahoo, D.D.; Patnaik, P. Oxidative stress markers and antioxidant defense in hibernating common Asian toads, Duttaphrynus melanostictus. Arch. Biol. Sci. 2020, 72, 23–30. [Google Scholar] [CrossRef]
- Patnaik, P.; Sahoo, D.D. Variations in oxidative stress and antioxidant defense level during different phases of hibernation in common Asian toad, Duttaphrynus melanostictus. Biol. Open 2021, 10, bio058567. [Google Scholar] [CrossRef]
- Feder, M.E.; Burggren, W.W. Environmental Physiology of the Amphibians; University of Chicago Press: Chicago, IL, USA, 1992. [Google Scholar]
- St-Pierre, J.; Boutilier, R.G. Aerobic capacity of frog skeletal muscle during hibernation. Physiol. Biochem. Zool. 2001, 74, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Lannoo, M. Amphibian Declines: The Conservation Status of United States Species; University of California Press: Berkeley, CA, USA, 2005. [Google Scholar]
- Jiang, S.; Claussen, D.L. A bioenergetic budget for overwintering newts (Notophthalmus viridescens) from southern Ohio: Their fat reserves and aerobic metabolic rates in water. Comp. Biochem. Physiol. A Physiol. 1992, 101, 743–750. [Google Scholar] [CrossRef]
- Koskela, P.; Pasanen, S. The wintering of the common frog, Rana temporaria l., in northern Finland. Aquilo (Zool.) 1974, 15, 1–17. [Google Scholar]
- Costanzo, J.P.; do Amaral, M.C.F.; Rosendale, A.J.; Lee, R.E., Jr. Hibernation physiology, freezing adaptation and extreme freeze tolerance in a northern population of the wood frog. J. Exp. Biol. 2013, 216, 3461–3473. [Google Scholar] [CrossRef]
- Smith, C.L. Seasonal changes in blood sugar, fat body, liver glycogen, and gonads in the common frog, Rana temporaria. J. Exp. Biol. 1950, 26, 412–429. [Google Scholar] [CrossRef]
- Saoi, M.; Britz-McKibbin, P. New advances in tissue metabolomics: A review. Metabolites 2021, 11, 672. [Google Scholar] [CrossRef]
- Cao, H.; Shi, Y.; Wang, J.; Niu, Z.; Wei, L.; Tian, H.; Yu, F.; Gao, L. The intestinal microbiota and metabolic profiles of Strauchbufo raddei underwent adaptive changes during hibernation. Integr. Zool. 2024, 19, 612–630. [Google Scholar] [CrossRef]
- Yan, C.; Ma, H.; Yang, Y.; Mi, Z. Sexual dimorphism in the limb bones of asiatic toad (Bufo gargarizans) in relation to sexual selection. Animals 2023, 13, 2638. [Google Scholar] [CrossRef]
- Yan, C.; Ma, H.; Yang, Y.; Mi, Z. Metabolic adaption of flexor carpi radialis to amplexus behavior in asiatic toads (Bufo gargarizans). Int. J. Mol. Sci. 2023, 24, 10174. [Google Scholar] [CrossRef]
- Zeki, Ö.C.; Eylem, C.C.; Reçber, T.; Kır, S.; Nemutlu, E. Integration of GC-MS and LC-MS for untargeted metabolomics profiling. J. Pharm. Biomed. Anal. 2020, 190, 113509. [Google Scholar] [CrossRef]
- Chen, K.; Li, E.; Xu, C.; Wang, X.; Li, H.; Qin, J.G.; Chen, L. Growth and metabolomic responses of pacific white shrimp (Litopenaeus vannamei) to different dietary fatty acid sources and salinity levels. Aquaculture 2019, 499, 329–340. [Google Scholar] [CrossRef]
- Wu, Y.; Yuan, C.; Han, P.; Guo, J.; Wang, Y.; Chen, C.; Huang, C.; Zheng, K.; Qi, Y.; Li, J.; et al. Discovery of potential biomarkers for osteoporosis using LC/GC-MS metabolomic methods. Front. Endocrinol. 2023, 14, 1332216. [Google Scholar]
- Storey, K.B.; Storey, J.M. Metabolic rate depression: The biochemistry of mammalian hibernation. Adv. Clin. Chem. 2010, 52, 77–108. [Google Scholar] [PubMed]
- Staples, J.F. Metabolic suppression in mammalian hibernation: The role of mitochondria. J. Exp. Biol. 2014, 217, 2032–2036. [Google Scholar] [CrossRef] [PubMed]
- Finn, P.F.; Dice, J.F. Proteolytic and lipolytic responses to starvation. Nutrition 2006, 22, 830–844. [Google Scholar] [CrossRef]
- Dawaliby, R.; Trubbia, C.; Delporte, C.; Noyon, C.; Ruysschaert, J.M.; Van Antwerpen, P.; Govaerts, C. Phosphatidylethanolamine is a key regulator of membrane fluidity in eukaryotic cells. J. Biol. Chem. 2016, 291, 3658–3667. [Google Scholar] [CrossRef]
- Van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta. Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef]
- Li, Z.; Agellon, L.B.; Allen, T.M.; Umeda, M.; Jewell, L.; Mason, A.; Vance, D.E. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006, 3, 321–331. [Google Scholar] [CrossRef]
- Kabine, M.; Clemencet, M.C.; Bride, J.; El Kebbaj, M.S.; Latruffe, N.; Cherkaoui-Malki, M. Changes of peroxisomal fatty acid metabolism during cold acclimatization in hibernating jerboa (Jaculus orientalis). Biochimie 2003, 85, 707–714. [Google Scholar] [CrossRef]
- Olsen, L.; Thum, E.; Rohner, N. Lipid metabolism in adaptation to extreme nutritional challenges. Dev. Cell 2021, 56, 1417–1429. [Google Scholar] [CrossRef]
- Rigoulet, M.; Bouchez, C.L.; Paumard, P.; Ransac, S.; Cuvellier, S.; Duvezin-Caubet, S.; Mazat, J.P.; Devin, A. Cell energy metabolism: An update. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148276. [Google Scholar] [CrossRef] [PubMed]
- Horman, S.; Hussain, N.; Dilworth, S.M.; Storey, K.B.; Rider, M.H. Evaluation of the role of AMP-activated protein kinase and its downstream targets in mammalian hibernation. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2005, 142, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Staples, J.F.; Buck, L.T. Matching cellular metabolic supply and demand in energy-stressed animals. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2009, 153, 95–105. [Google Scholar] [CrossRef]
- Neinast, M.; Murashige, D.; Arany, Z. Branched chain amino acids. Annu. Rev. Physiol. 2019, 81, 139–164. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.W.; Whaley, S.G.; Rock, C.O. Branched-chain amino acid metabolism controls membrane phospholipid structure in staphylococcus aureus. J. Biol. Chem. 2021, 297, 101255. [Google Scholar] [CrossRef] [PubMed]
- Halama, A.; Horsch, M.; Kastenmuller, G.; Moller, G.; Kumar, P.; Prehn, C.; Laumen, H.; Hauner, H.; Hrabe de Angelis, M.; Beckers, J.; et al. Metabolic switch during adipogenesis: From branched chain amino acid catabolism to lipid synthesis. Arch. Biochem. Biophys. 2016, 589, 93–107. [Google Scholar] [CrossRef]
- Niu, Y.; Chen, Q.; Storey, K.B.; Teng, L.; Li, X.; Xu, T.; Zhang, H. Physiological ecology of winter hibernation by the high-altitude frog Nanorana parkeri. Physiol. Biochem. Zool. 2022, 95, 201–211. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR signaling in growth, metabolism, and disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Kim, J.; Guan, K.L. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 2019, 21, 63–71. [Google Scholar] [CrossRef]
- Bender, D.A. The aromatic amino acids: Phenylalanine, tyrosine and tryptophan. In Amino Acid Metabolism, 3rd ed.; John Wiley & Sons: Chichester, UK, 2012; pp. 323–376. [Google Scholar]
- Hui, S.; Ghergurovich, J.M.; Morscher, R.J.; Jang, C.; Teng, X.; Lu, W.; Esparza, L.A.; Reya, T.; Le, Z.; Yanxiang Guo, J.; et al. Glucose feeds the TCA cycle via circulating lactate. Nature 2017, 551, 115–118. [Google Scholar] [CrossRef]
- Wu, G. Amino acids: Metabolism, functions, and nutrition. Amino Acids 2009, 37, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Al-Attar, R.; Storey, K.B. Suspended in time: Molecular responses to hibernation also promote longevity. Exp. Gerontol. 2020, 134, 110889. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, H.; Yan, C.; Mi, Z. Metabolomic Insights into Energy Utilization Strategies of Asiatic Toads (Bufo gargarizans) During Hibernation. Animals 2025, 15, 403. https://doi.org/10.3390/ani15030403
Ma H, Yan C, Mi Z. Metabolomic Insights into Energy Utilization Strategies of Asiatic Toads (Bufo gargarizans) During Hibernation. Animals. 2025; 15(3):403. https://doi.org/10.3390/ani15030403
Chicago/Turabian StyleMa, Hui, Chengzhi Yan, and Zhiping Mi. 2025. "Metabolomic Insights into Energy Utilization Strategies of Asiatic Toads (Bufo gargarizans) During Hibernation" Animals 15, no. 3: 403. https://doi.org/10.3390/ani15030403
APA StyleMa, H., Yan, C., & Mi, Z. (2025). Metabolomic Insights into Energy Utilization Strategies of Asiatic Toads (Bufo gargarizans) During Hibernation. Animals, 15(3), 403. https://doi.org/10.3390/ani15030403




