Deciphering the Population Characteristics of Leiqiong Cattle Using Whole-Genome Sequencing Data
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection and Genome Sequencing
2.3. Read Mapping and SNP Calling
2.4. Genome-Wide Patterns of Genetic Diversity and Divergence
2.5. Phylogenetic and Population Structure Analyses
2.6. Selective Sweep Analysis
2.7. Functional Enrichment Analyses
3. Results
3.1. Whole-Genome Sequencing and Genetic Variation
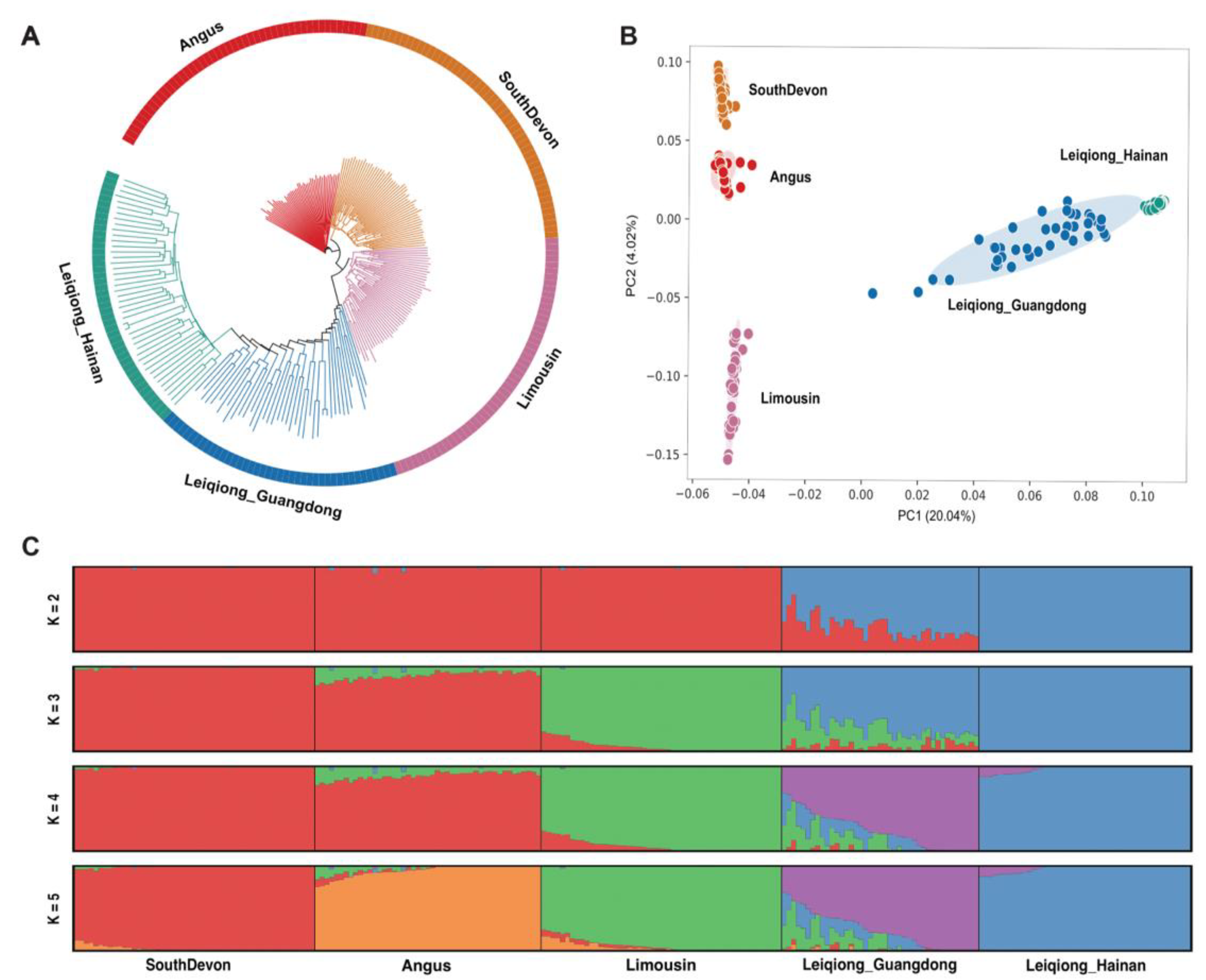
3.2. Population Structure and Relationships
3.3. Patterns of Genomic Variation
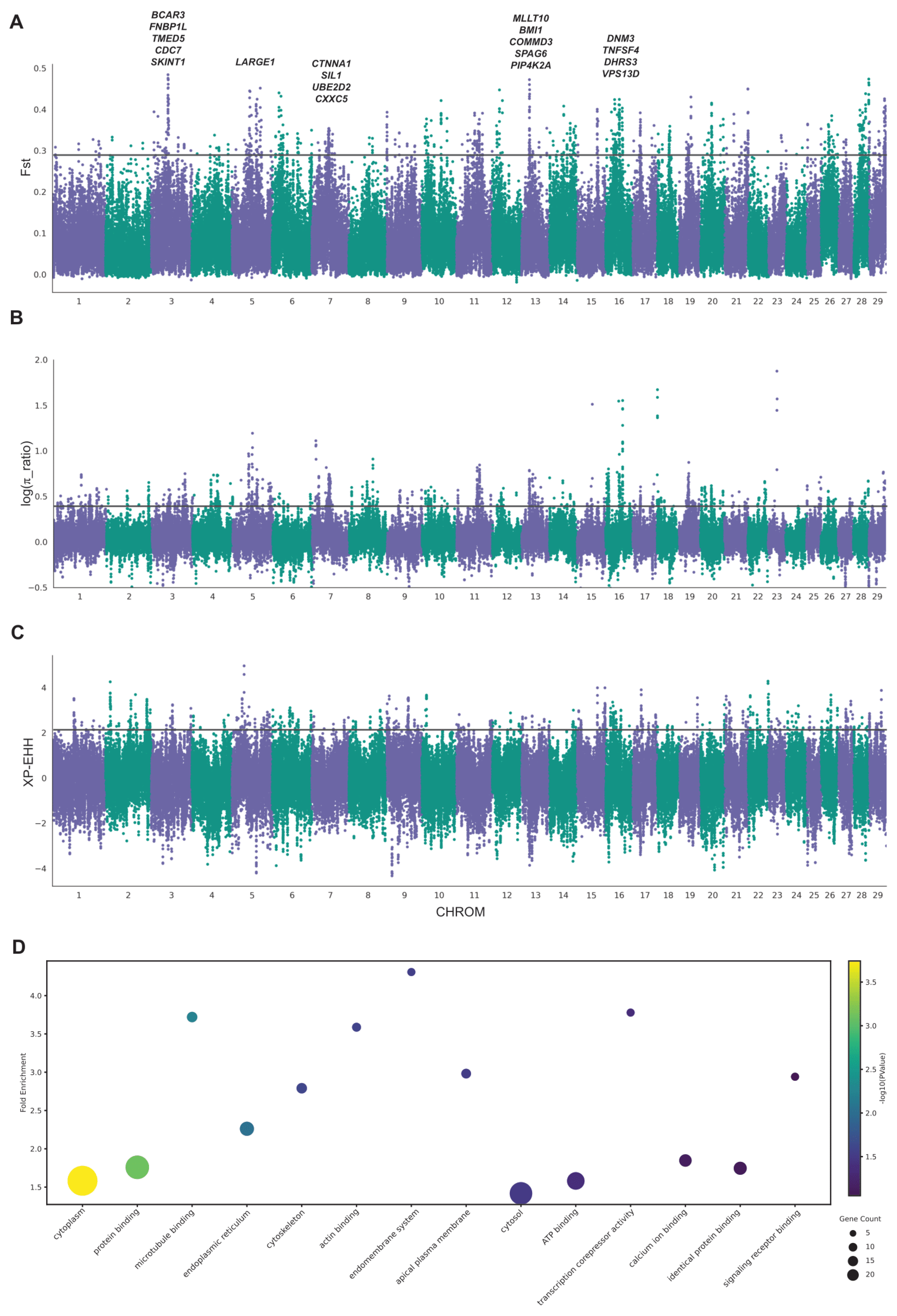
3.4. Genome-Wide Selective Sweep Between the Subgroup of Leiqiong Cattle
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hesse, B. The first steps of animal domestication. J. Ethnobiol. 2006, 26, 171–174. [Google Scholar]
- Felius, M.; Koolmees, P.A.; Theunissen, B.; Consortium, E.C.G.D.; Lenstra, J.A. On the Breeds of Cattle—Historic and Current Classifications. Diversity 2011, 3, 660–692. [Google Scholar] [CrossRef]
- Gao, Y.; Gautier, M.; Ding, X.; Zhang, H.; Wang, Y.; Wang, X.; Faruque, M.O.; Li, J.; Ye, S.; Gou, X.; et al. Species composition and environmental adaptation of indigenous Chinese cattle. Sci. Rep. 2017, 7, 16196. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.; Yao, T.; Chen, Z.; Huangfu, R.; Cheng, H.; Ma, W.; Qi, X.; Li, F.; Chen, N.; Lei, C. Genomic characterization of dryland adaptation in endangered Anxi cattle in China. Anim. Genet. 2024, 55, 352–361. [Google Scholar] [CrossRef]
- Lu, X.; Arbab, A.A.I.; Zhang, Z.; Fan, Y.; Han, Z.; Gao, Q.; Sun, Y.; Yang, Z. Comparative Transcriptomic Analysis of the Pituitary Gland between Cattle Breeds Differing in Growth: Yunling Cattle and Leiqiong Cattle. Animals 2020, 10, 1271. [Google Scholar] [CrossRef]
- Luo, J.; Liu, Q.; Yang, Z.; Kong, L.; Li, B.; Liu, Y.; Liu, D.; Li, Y. Comparison of production performance among F1 hybrids of Leiqiong yellow cattle and different beef breeds. J. South China Agric. Univ. 2020, 41, 10–17. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, L.; Yang, L.; Zhao, G.; Li, J.; Liu, D.; Li, Y. Discovery of Genomic Characteristics and Selection Signatures in Southern Chinese Local Cattle. Front. Genet. 2020, 11, 533052. [Google Scholar] [CrossRef]
- Ma, J.; Gao, X.; Li, J.; Gao, H.; Wang, Z.; Zhang, L.; Xu, L.; Gao, H.; Li, H.; Wang, Y.; et al. Assessing the Genetic Background and Selection Signatures of Huaxi Cattle Using High-Density SNP Array. Animals 2021, 11, 3469. [Google Scholar] [CrossRef]
- Wang, X.; Ju, Z.; Jiang, Q.; Zhong, J.; Liu, C.; Wang, J.; Hoff, J.L.; Schnabel, R.D.; Zhao, H.; Gao, Y.; et al. Introgression, admixture, and selection facilitate genetic adaptation to high-altitude environments in cattle. Genomics 2021, 113, 1491–1503. [Google Scholar] [CrossRef]
- Wei, C.; Wang, H.; Liu, G.; Wu, M.; Cao, J.; Liu, Z.; Liu, R.; Zhao, F.; Zhang, L.; Lu, J.; et al. Genome-wide analysis reveals population structure and selection in Chinese indigenous sheep breeds. BMC Genom. 2015, 16, 194. [Google Scholar] [CrossRef]
- Nielsen, R.; Williamson, S.; Kim, Y.; Hubisz, M.J.; Clark, A.G.; Bustamante, C. Genomic scans for selective sweeps using SNP data. Genome Res. 2005, 15, 1566–1575. [Google Scholar] [CrossRef] [PubMed]
- Sabeti, P.C.; Varilly, P.; Fry, B.; Lohmueller, J.; Hostetter, E.; Cotsapas, C.; Xie, X.; Byrne, E.H.; Mccarroll, S.A.; Gaudet, R.; et al. Genome-wide detection and characterization of positive selection in human populations. Nature 2007, 449, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Zhang, F.; Li, S.; Luo, X.; Peng, L.; Dong, Z.; Pausch, H.; Leonard, A.S.; Crysnanto, D.; Wang, S.; et al. Structural variation and introgression from wild populations in East Asian cattle genomes confer adaptation to local environment. Genome Biol. 2023, 24, 211. [Google Scholar] [CrossRef]
- Li, X.; Yang, J.; Shen, M.; Xie, X.L.; Liu, G.J.; Xu, Y.X.; Lv, F.H.; Yang, H.; Yang, Y.L.; Liu, C.B.; et al. Whole-genome resequencing of wild and domestic sheep identifies genes associated with morphological and agronomic traits. Nat. Commun. 2020, 11, 2815. [Google Scholar] [CrossRef]
- Jin, M.; Wang, H.; Liu, G.; Lu, J.; Yuan, Z.; Li, T.; Liu, E.; Lu, Z.; Du, L.; Wei, C. Whole-genome resequencing of Chinese indigenous sheep provides insight into the genetic basis underlying climate adaptation. Genet. Sel. Evol. 2024, 56, 26. [Google Scholar] [CrossRef]
- Cai, Y.; Fu, W.; Cai, D.; Heller, R.; Zheng, Z.; Wen, J.; Li, H.; Wang, X.; Alshawi, A.; Sun, Z.; et al. Ancient Genomes Reveal the Evolutionary History and Origin of Cashmere-Producing Goats in China. Mol. Biol. Evol. 2020, 37, 2099–2109. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, X.; Yang, Y.; Wang, L.; Xu, C.; Xu, W.; Chen, Q.; Li, M.; Lu, S. Population Structure and Selection Signatures in Chinese Indigenous Zhaotong Pigs Revealed by Whole-Genome Resequencing. Animals 2024, 14, 3129. [Google Scholar] [CrossRef]
- Xie, X.; Shi, L.; Zhong, Z.; Wang, Z.; Pan, D.; Hou, G.; Xiao, Q. Danzhou chicken: A unique genetic resource revealed by genome-wide resequencing data. Poult. Sci. 2024, 103, 103960. [Google Scholar] [CrossRef]
- Xu, N.Y.; Si, W.; Li, M.; Gong, M.; Lariviere, J.M.; Nanaei, H.A.; Bian, P.P.; Jiang, Y.; Zhao, X. Genome-wide scan for selective footprints and genes related to cold tolerance in Chantecler chickens. Zool. Res. 2021, 42, 710–720. [Google Scholar] [CrossRef]
- Zhang, S.; Yao, Z.; Li, X.; Zhang, Z.; Liu, X.; Yang, P.; Chen, N.; Xia, X.; Lyu, S.; Shi, Q.; et al. Assessing genomic diversity and signatures of selection in Pinan cattle using whole-genome sequencing data. BMC Genom. 2022, 23, 460. [Google Scholar] [CrossRef]
- Xia, X.; Zhang, S.; Zhang, H.; Zhang, Z.; Chen, N.; Li, Z.; Sun, H.; Liu, X.; Lyu, S.; Wang, X.; et al. Assessing genomic diversity and signatures of selection in Jiaxian Red cattle using whole-genome sequencing data. BMC Genom. 2021, 22, 43. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cheng, H.; Wang, S.; Luo, X.; Ma, X.; Sun, L.; Chen, N.; Zhang, J.; Qu, K.; Wang, M.; et al. Genomic Diversity and Selection Signatures for Weining Cattle on the Border of Yunnan-Guizhou. Front. Genet. 2022, 13, 848951. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Yao, Z.; Zhang, Z.; Lyu, S.; Chen, N.; Qi, X.; Liu, X.; Ma, W.; Wang, W.; Lei, C.; et al. Whole-genome sequencing reveals genomic diversity and selection signatures in Xia’nan cattle. BMC Genomics 2024, 25, 559. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Mckenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 10–11. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; Mccarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.-S.; Xu, J.-Y.; He, W.-M.; Yang, T.-L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Terhorst, J.; Kamm, J.A.; Song, Y.S. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 2017, 49, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Jivanji, S.; Harland, C.; Cole, S.; Brophy, B.; Garrick, D.; Snell, R.; Littlejohn, M.; Laible, G. The genomes of precision edited cloned calves show no evidence for off-target events or increased de novo mutagenesis. BMC Genom. 2021, 22, 457. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, M.; Pedersen, C.N. Rapid computation of distance estimators from nucleotide and amino acid alignments. In Proceedings of the 2011 ACM Symposium on Applied Computing, Taichung, Taiwan, 21–25 March 2011. [Google Scholar]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Behr, A.A.; Liu, K.Z.; Liu-Fang, G.; Nakka, P.; Ramachandran, S. pong: Fast analysis and visualization of latent clusters in population genetic data. Bioinformatics 2016, 32, 2817–2823. [Google Scholar] [CrossRef]
- Szpiech, Z.A. Selscan 2.0: Scanning for sweeps in unphased data. Bioinformatics 2024, 40, btae006. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Liao, F.; Yin, D.; Zhang, Y.; Hou, Q.; Zheng, Z.; Yang, L.; Shu, Y.; Xu, H.; Li, Y. Association Between PIP4K2A Polymorphisms and Acute Lymphoblastic Leukemia Susceptibility. Medicine 2016, 95, e3542. [Google Scholar] [CrossRef]
- Fu, W.; Wang, R.; Nanaei, H.A.; Wang, J.; Hu, D.; Jiang, Y. RGD v2.0: A major update of the ruminant functional and evolutionary genomics database. Nucleic Acids Res. 2022, 50, D1091–D1099. [Google Scholar] [CrossRef]
- Gaur, U.; Aggarwal, B.B. Regulation of proliferation, survival and apoptosis by members of the TNF superfamily. Biochem. Pharmacol. 2003, 66, 1403–1408. [Google Scholar] [CrossRef]
- Liu, C.; Sello, C.; Sun, Y.; Zhou, Y.; Lu, H.; Sui, Y.; Hu, J.; Xu, C.; Sun, Y.; Liu, J.; et al. De Novo Transcriptome Sequencing Analysis of Goose (Anser anser) Embryonic Skin and the Identification of Genes Related to Feather Follicle Morphogenesis at Three Stages of Development. Int. J. Mol. Sci. 2018, 19, 3170. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, K.; Cheng, G.; Mei, C.; Wang, H.; Zan, L. Genome-wide analysis reveals genomic diversity and signatures of selection in Qinchuan beef cattle. BMC Genom. 2024, 25, 558. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Bosse, M.; Rochus, C.M.; Groenen, M.A.M.; Crooijmans, R.P.M.A. Genomic insight into the influence of selection, crossbreeding, and geography on population structure in poultry. Genet. Sel. Evol. 2023, 55, 5. [Google Scholar] [CrossRef] [PubMed]
- Bergström, A.; McCarthy, S.A.; Hui, R.; Almarri, M.A.; Ayub, Q.; Danecek, P.; Chen, Y.; Felkel, S.; Hallast, P.; Kamm, J. Insights into human genetic variation and population history from 929 diverse genomes. Science 2020, 367, eaay5012. [Google Scholar] [CrossRef]
- Panigrahi, M.; Rajawat, D.; Nayak, S.S.; Ghildiyal, K.; Sharma, A.; Jain, K.; Lei, C.; Bhushan, B.; Mishra, B.P.; Dutt, T. Landmarks in the history of selective sweeps. Anim. Genet. 2023, 54, 667–688. [Google Scholar] [CrossRef]
- Sumita, K.; Lo, Y.H.; Takeuchi, K.; Senda, M.; Kofuji, S.; Ikeda, Y.; Terakawa, J.; Sasaki, M.; Yoshino, H.; Majd, N.; et al. The Lipid Kinase PI5P4Kbeta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis. Mol. Cell 2016, 61, 187–198. [Google Scholar] [CrossRef]
- Wang, D.G.; Paddock, M.N.; Lundquist, M.R.; Sun, J.Y.; Mashadova, O.; Amadiume, S.; Bumpus, T.W.; Hodakoski, C.; Hopkins, B.D.; Fine, M.; et al. PIP4Ks Suppress Insulin Signaling through a Catalytic-Independent Mechanism. Cell Rep. 2019, 27, 1991–2001.e5. [Google Scholar] [CrossRef]
- Hu, A.; Zhao, X.T.; Tu, H.; Xiao, T.; Fu, T.; Wang, Y.; Liu, Y.; Shi, X.J.; Luo, J.; Song, B.L. PIP4K2A regulates intracellular cholesterol transport through modulating PI(4,5)P(2) homeostasis. J. Lipid Res. 2018, 59, 507–514. [Google Scholar] [CrossRef]
- Emerling, B.M.; Hurov, J.B.; Poulogiannis, G.; Tsukazawa, K.S.; Choo-Wing, R.; Wulf, G.M.; Bell, E.L.; Shim, H.S.; Lamia, K.A.; Rameh, L.E.; et al. Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell 2013, 155, 844–857. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Li, M.; Sun, H.; Chen, Q.; Yan, D.; Dong, X.; Pan, Y.; Lu, S. Genome-wide association study reveals genetic loci and candidate genes for meat quality traits in a four-way crossbred pig population. Front. Genet. 2023, 14, 1001352. [Google Scholar] [CrossRef]
- Lee, T.; Shin, D.-H.; Cho, S.; Kang, H.S.; Kim, S.H.; Lee, H.-K.; Kim, H.; Seo, K.-S. Genome-wide Association Study of Integrated Meat Quality-related Traits of the Duroc Pig Breed. Asian Australas. J. Anim. Sci. 2014, 27, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Lau, L.Y.; Fortes, M.R.S. Proteomic Analysis of Hypothalamus and Pituitary Gland in Pre and Postpubertal Brahman Heifers. Front. Genet. 2022, 13, 935433. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Li, J.; Xiao, C.; Sun, L.; Xiang, W.; Chen, N.; Lei, C.; Lei, H.; Long, Y.; Long, T.; et al. Whole-Genome Resequencing of Xiangxi Cattle Identifies Genomic Diversity and Selection Signatures. Front. Genet. 2022, 13, 816379. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.K.; Yang, W.; Zhao, M.; Mok, C.C.; Chan, T.M.; Wong, R.W.S.; Lee, K.W.; Mok, M.Y.; Wong, S.N.; Ng, I.O.L.; et al. Association of BANK1 and TNFSF4 with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun. 2009, 10, 414–420. [Google Scholar] [CrossRef][Green Version]
- Graham, D.S.C.; Graham, R.R.; Manku, H.; Wong, A.K.; Whittaker, J.C.; Gaffney, P.M.; Moser, K.L.; Rioux, J.D.; Altshuler, D.; Behrens, T.W.; et al. Polymorphism at the TNF superfamily gene TNFSF4 confers susceptibility to systemic lupus erythematosus. Nat. Genet. 2008, 40, 83–89. [Google Scholar] [CrossRef]
- Ria, M.; Lagercrantz, J.; Samnegård, A.; Boquist, S.; Hamsten, A.; Eriksson, P. A Common Polymorphism in the Promoter Region of the TNFSF4 Gene Is Associated with Lower Allele-Specific Expression and Risk of Myocardial Infarction. PLoS ONE 2011, 6, e17652. [Google Scholar] [CrossRef]
- Wang, X.; Ria, M.; Kelmenson, P.M.; Eriksson, P.; Higgins, D.C.; Samnegård, A.; Petros, C.; Rollins, J.; Bennet, A.M.; Wiman, B.; et al. Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat. Genet. 2005, 37, 365–372. [Google Scholar] [CrossRef]
- Zhao, H.; Guo, T.; Lu, Z.; Liu, J.; Zhu, S.; Qiao, G.; Han, M.; Yuan, C.; Wang, T.; Li, F.; et al. Genome-wide association studies detects candidate genes for wool traits by re-sequencing in Chinese fine-wool sheep. BMC Genom. 2021, 22, 127. [Google Scholar] [CrossRef]
- Megdiche, S.; Mastrangelo, S.; Ben Hamouda, M.; Lenstra, J.A.; Ciani, E. A Combined Multi-Cohort Approach Reveals Novel and Known Genome-Wide Selection Signatures for Wool Traits in Merino and Merino-Derived Sheep Breeds. Front. Genet. 2019, 10, 1025. [Google Scholar] [CrossRef]
- Mota-Rojas, D.; Titto, C.G.; de Mira Geraldo, A.; Martinez-Burnes, J.; Gomez, J.; Hernandez-Avalos, I.; Casas, A.; Dominguez, A.; Jose, N.; Bertoni, A.; et al. Efficacy and Function of Feathers, Hair, and Glabrous Skin in the Thermoregulation Strategies of Domestic Animals. Animals 2021, 11, 3472. [Google Scholar] [CrossRef]
- Sejian, V.; Bhatta, R.; Gaughan, J.B.; Dunshea, F.R.; Lacetera, N. Review: Adaptation of animals to heat stress. Animal 2018, 12, s431–s444. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Zhao, Z.; Ge, F.; Yu, H.; Lyu, C.; Liu, Y.; Li, J.; Chen, Y. Deciphering the Population Characteristics of Leiqiong Cattle Using Whole-Genome Sequencing Data. Animals 2025, 15, 342. https://doi.org/10.3390/ani15030342
Guo Y, Zhao Z, Ge F, Yu H, Lyu C, Liu Y, Li J, Chen Y. Deciphering the Population Characteristics of Leiqiong Cattle Using Whole-Genome Sequencing Data. Animals. 2025; 15(3):342. https://doi.org/10.3390/ani15030342
Chicago/Turabian StyleGuo, Yingwei, Zhihui Zhao, Fei Ge, Haibin Yu, Chenxiao Lyu, Yuxin Liu, Junya Li, and Yan Chen. 2025. "Deciphering the Population Characteristics of Leiqiong Cattle Using Whole-Genome Sequencing Data" Animals 15, no. 3: 342. https://doi.org/10.3390/ani15030342
APA StyleGuo, Y., Zhao, Z., Ge, F., Yu, H., Lyu, C., Liu, Y., Li, J., & Chen, Y. (2025). Deciphering the Population Characteristics of Leiqiong Cattle Using Whole-Genome Sequencing Data. Animals, 15(3), 342. https://doi.org/10.3390/ani15030342





