Transport Vehicles as a Vector of Goose Parvovirus Infections (GPV)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Behboudi, S. Derzsy’s Disease. CABI Compendium. 2022. Available online: https://www.cabidigitallibrary.org/doi/abs/10.1079/cabicompendium.85757 (accessed on 21 November 2024).
- Buzała, M.; Adamski, M.; Janicki, B. Characteristics of performance traits and the quality of meat and fat in Polish oat geese. Worlds Poult. Sci. J. 2014, 70, 531–542. [Google Scholar] [CrossRef]
- Siedlecka, M.; Chmielewska-Władyka, M.; Kublicka, A.; Wieliczko, A.; Matczuk, K.A. Goose parvovirus, goose hemorrhagic polyomavirus and goose circovirus infections are prevalent in commercial geese flocks in Poland and contribute to overall health and production outcomes: A two-year observational study. BMC Vet Res. 2025, 21, 216. [Google Scholar] [CrossRef]
- Kapgate, S.S.; Kumanan, K.; Vijayarani, K.; Barbuddhe, S.B. Avian parvovirus: Classification, phylogeny, pathogenesis and diagnosis. Avian Pathol. 2018, 47, 536–545. [Google Scholar] [CrossRef]
- Glavits, R.; Zolnai, A.; Szabo, E.; Ivanics, E.; Zarka, P.; Mato, T.; Palya, V. Comparative pathological studies on domestic geese Anser anser domestica and Muscovy ducks Cairina moschata experimentally infected with parvovirus strains of goose and Muscovy duck origin. Acta Vet. Hung. 2005, 53, 73–89. [Google Scholar] [CrossRef]
- Fan, W.; Sun, Z.; Shen, T.; Xu, D.; Huang, K.; Zhou, J.; Song, S.; Yan, L. Analysis of evolutionary processes of species jump in waterfowl parvovirus. Front. Microbiol. 2017, 8, 421. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, H.; Li, J.; Liu, D.; Meng, R.; Zhang, Q.; Shaozhou, W.; Bai, X.; Zhang, T.; Liu, M.; et al. A conserved epitope mapped with a monoclonal antibody against the VP3 protein of goose parvovirus by using peptide screening and phage display approaches. PLoS ONE 2016, 11, e0147361. [Google Scholar] [CrossRef] [PubMed]
- Limn, C.K.; Yamada, T.; Nakamura, M. Detection of goose parvovirus genome by polymerase chain reaction: Distribution of goose parvovirus in muscovy ducklings. Virus Res. 1996, 1, l67–172. [Google Scholar] [CrossRef]
- Mackay, M.; Arden, K.E.; Nitsche, A. Real-time PCR in virology. Nucleic Acids Res. 2002, 30, 1292–1305. [Google Scholar] [CrossRef]
- Mengyu, T.; Fei, L.; Shun, C.; Mingshu, W.; Anchun, C. Advances in parvovirus non-structural protein NS1 induced apoptosis. Chin. J. Virol. 2015, 31, 679–684. [Google Scholar]
- Racicot, M.; Kocher, A.; Beauchamp, G.; Letellier, A.; Vaillancourt, J.P. Assessing most practical and effective protocols to sanitize hands of poultry catching crew members. Prev. Vet. Med. 2013, 111, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Zhang, W.; Wang, H.; Zhou, Y.; Shao, S. Identification of recombination between Muscovy duck parvovirus and goose parvovirus structural protein genes. Arch. Virol. 2015, 160, 2617–2621. [Google Scholar] [CrossRef]
- Stoute, S.T.; Tsai, H.; Metwally, S.A.; Cheng, A.; Guerin, J.-L.; Palya, V. Viral infections of waterfowl. In Diseases of Poultry; Wiley: New York, NY, USA, 2020; pp. 446–497. [Google Scholar]
- Jansson, D.S.; Feinstein, R.; Kardi, V.; Mató, T.; Palya, V. Epidemiologic investigation of an outbreak of Goose parvovirus infection in Sweden. Avian Dis. 2007, 51, 609–613. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A Simple Method of Estimating Fifty Percent Endpoints. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar]
- Irvine, R.; Ceeraz, V.; Cox, B.; Twomey, F.; Young, S.; Bradshaw, J.; Featherstone, C.; Holmes, J.P.; Ainsworth, H.; Jones, R. Goose parvovirus in Great Britain. Vet. Rec. 2008, 163, 461. [Google Scholar] [CrossRef]
- Kardogan, O.; Mustak, H.K.; Mustak, I.B. The first detection and characterization of goose parvovirus (GPV) in Turkey. Trop. Anim. Health Prod. 2020, 53, 36. [Google Scholar] [CrossRef]
- Daeffler, L.; Hörlein, R.; Rommelaere, J.; Nüesch, J.P.F. Modulation of minute virus of mice cytotoxic activities through site-directed mutagenesis within the NS coding region. J. Virol. 2003, 77, 12466–12478. [Google Scholar] [CrossRef]
- Caillet-Fauquet, P.; Perros, M.; Brandenburger, A.; Spegelaere, P.; Rommelaere, J. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 1990, 9, 2989–2995. [Google Scholar] [CrossRef]
- Dzieciolowski, T.; Boqvist, S.; Rydén, J.; Hansson, I. Cleaning and disinfection of transport crates for poultry–comparison of four treatments at slaughter plant. Poult. Sci. 2022, 101, 101521. [Google Scholar] [CrossRef]
- Dos Santos, V.M.; Dallago, B.S.L.; Racanicci, A.M.C.; Santana, Â.P.; Bernal, F.E.M. Effects of season and distance during transport on broiler chicken meat. Poult. Sci. 2017, 96, 4270–4279. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, V.M.; Dallago, B.S.L.; Racanicci, A.M.C.; Santana, Â.P.; Cue, R.I.; Bernal, F.E.M. Effect of transportation distances, seasons and crate microclimate on broiler chicken production losses. PLoS ONE 2020, 15, e0232004. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; et al. Transmission of antimicrobial resistance (AMR) during animal transport. EFSA J. 2022, 20, e07586. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Cheng, A.; Wang, M.; Shen, C.; Jia, R.; Chen, S.; Zhang, N. Development of TaqMan MGB fluorescent real-time PCR assay for the detection of anatid herpesvirus 1. Virol. J. 2009, 6, 71. [Google Scholar] [CrossRef]
- Herrero y Calle, M.; Cornelis, J.J.; Herold-Mende, C.; Rommelaere, J.; Schlehofer, J.R.; Geletneky, K. Parvovirus H-1 infection of human glioma cells leads to complete viral replication and efficient cell killing. Int. J. Cancer 2004, 109, 76–84. [Google Scholar] [CrossRef]
- Tatár-kis, T.; Mató, T.; Markos, B.; Palya, V. Phylogenetic analysis of Hungarian goose parvovirus isolates and vaccine strains. Avian Pathol. 2004, 33, 438–444. [Google Scholar] [CrossRef]
- Huneau-Salaün, A.; Scoizec, A.; Thomas, R.; Martenot, C.; Schmitz, A.; Pierre, I.; Allée, C.; Busson, R.; Massin, P.; Briand, F.X.; et al. Avian influenza outbreaks: Evaluating the efficacy of cleaning and disinfection of vehicles and transport crates. Poult. Sci. 2022, 101, 101569. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, R.; Nurjanah, S.; Furukawa, K.; Murai, A.; Kikusato, M.; Nochi, T.; Toyomizu, M. Heat stress causes immune abnormalities via massive damage to effect proliferation and differentiation of lymphocytes in broiler chickens. Front. Vet. Sci. 2020, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- Kraszczuk, V. Verificação do Processo de Higienização Pré-Operacional de um Abatedouro de Aves. Bachelor’s Thesis, Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil, 2010. [Google Scholar]

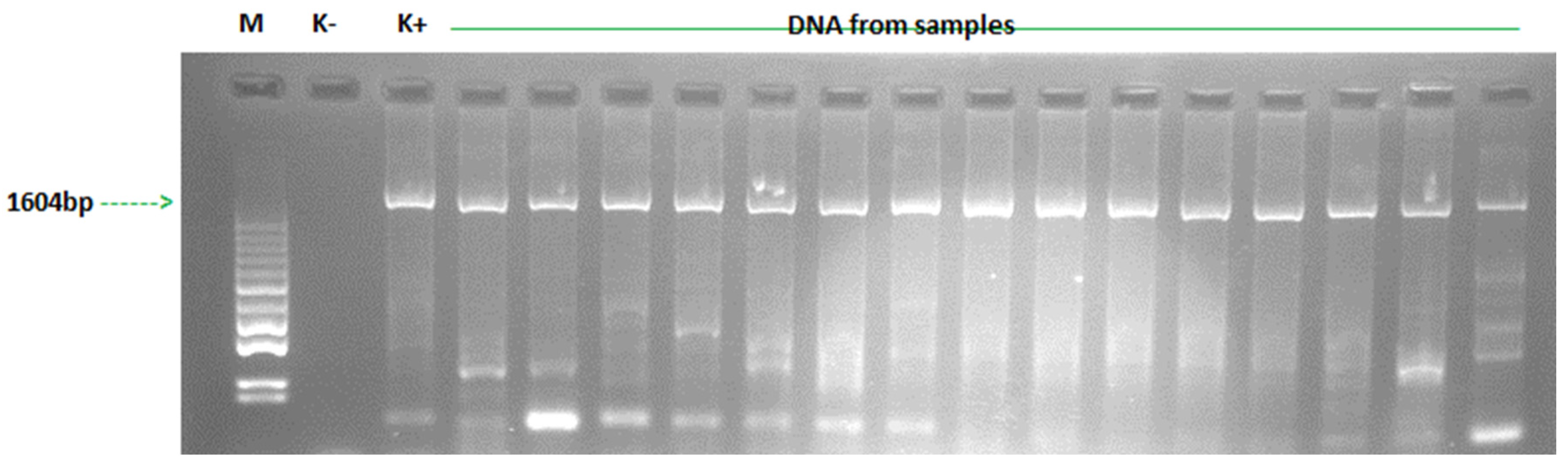
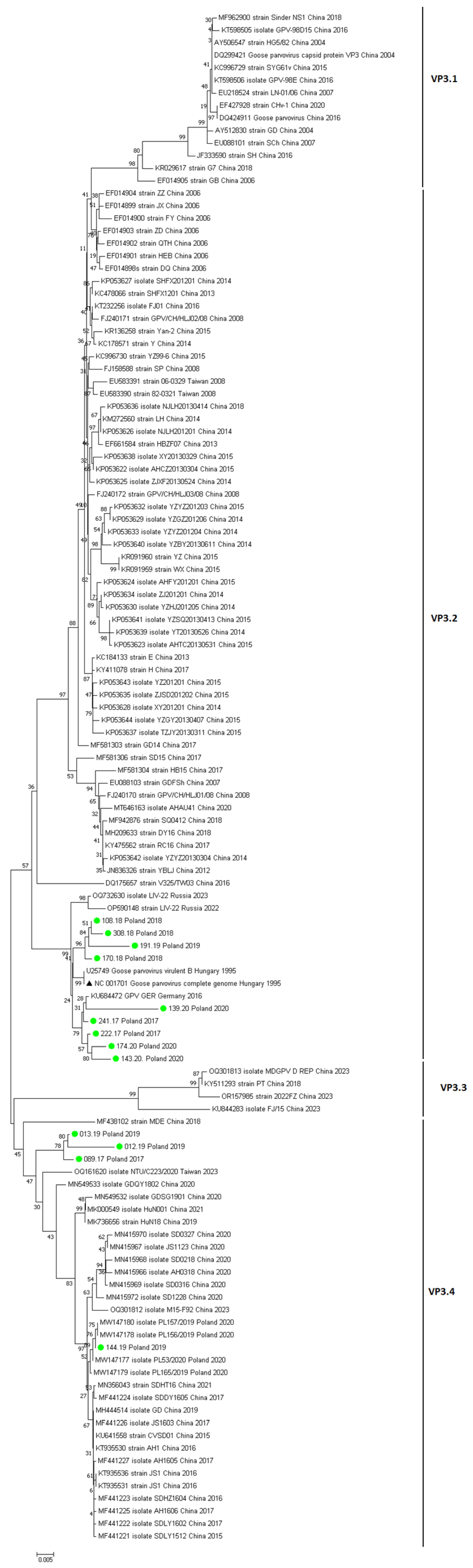
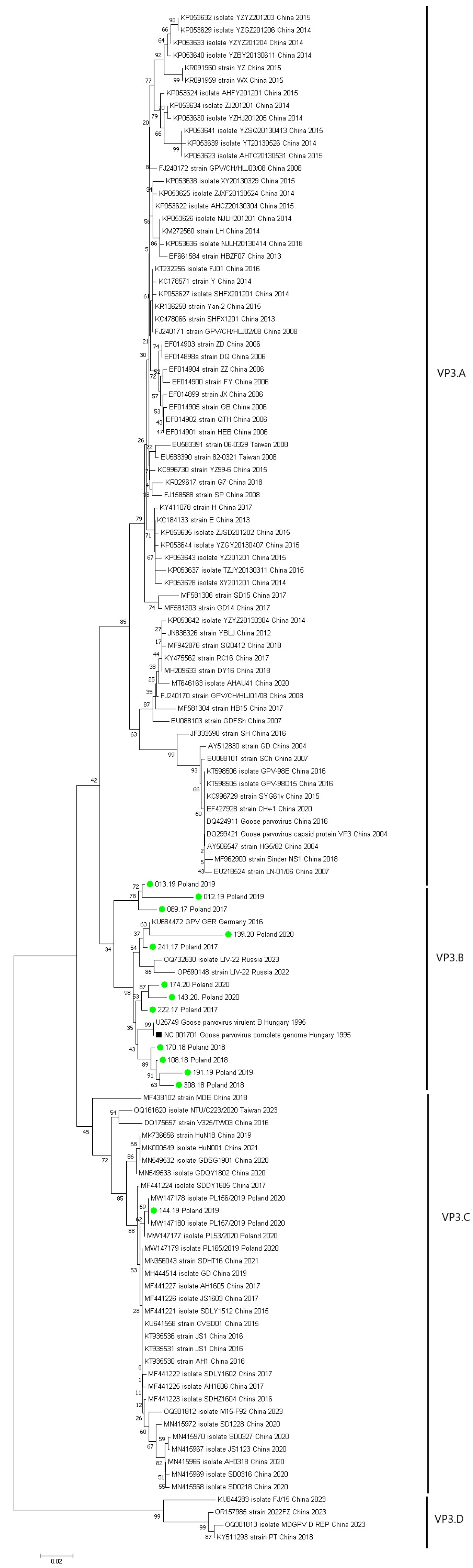
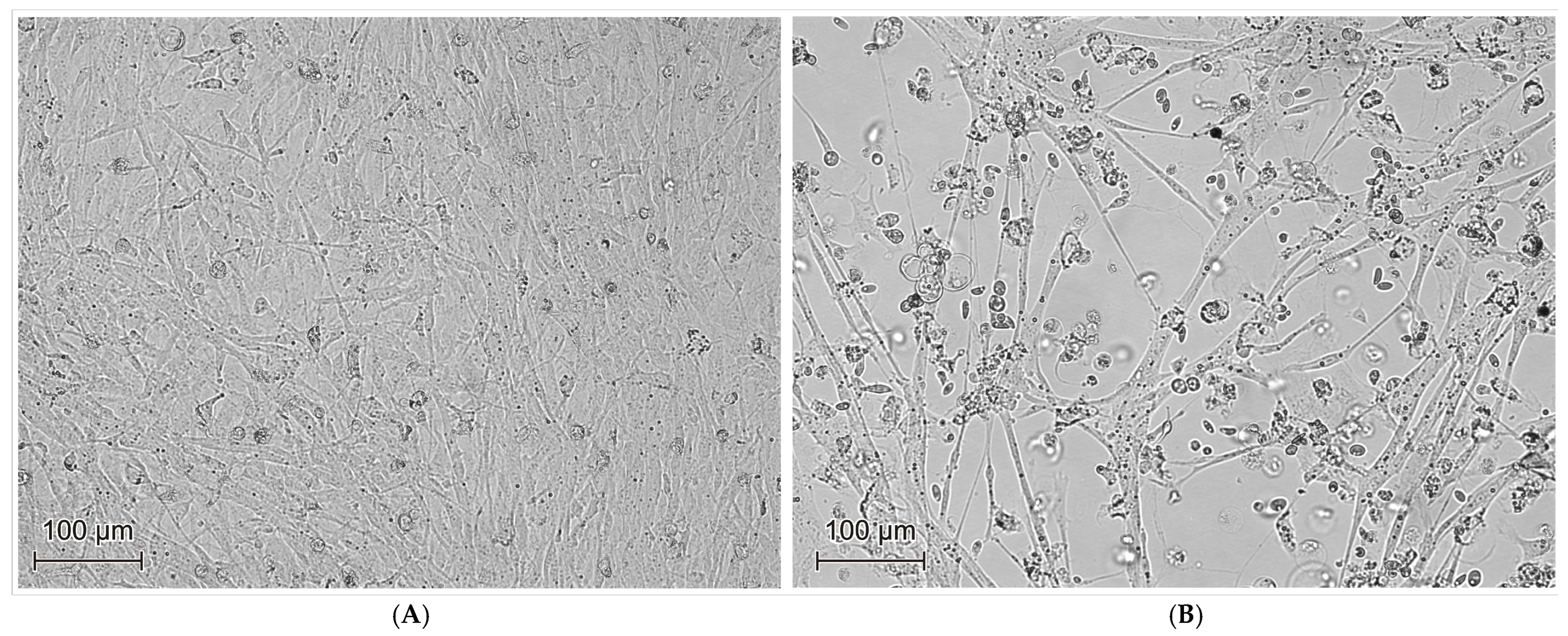
| Year | Number of Vehicles Examined | Number of Vehicles Testing Positive for GPV | Number of Sequences Obtained | Accession Number GenBANK |
|---|---|---|---|---|
| 2017 | 15 | 6 (40%) | 3 (50%) | waiting |
| 2018 | 22 | 9 (40.91%) | 3 (33.34) | waiting |
| 2019 | 17 | 5 (29.41%) | 4 (80%) | waiting |
| 2020 | 12 | 5 (41.67%) | 3 (60%) | waiting |
| Total | 66 | 25 (37.87%) | 13 |
| Lp. | Amino Acid Position | Amino Acid of Reference Strain Sequence * | Amino Acid of Field Strain | Strains with Mutation Detected |
|---|---|---|---|---|
| 1 | 20 | G | S | 139.20_Poland_2020 |
| 2 | 21 | N | H | 139.20_Poland_2020 |
| 3 | 22 | A | G | 139.20_Poland_2020 |
| 4 | 25 | N | T | 139.20_Poland_2020 |
| 5 | 26 | W | S | 308.18_Poland_2018 |
| 6 | 29 | D | E | 139.20_Poland_2020 |
| 7 | 30 | S | N | 139.20_Poland_2020 |
| 8 | 31 | Q | V | 139.20_Poland_2020 |
| 9 | 56 | K | Q | 139.20_Poland_2020 |
| 10 | 58 | I | L | 143.20_Poland_2020 |
| 11 | 59 | T | P | 143.20_Poland_2020 |
| 12 | 60 | S | R | 174.20_Poland_2020 012.19_Poland_2019 |
| G | 143.20_Poland_2020 | |||
| 13 | 61 | G | A | 143.20_Poland_2020 |
| 14 | 64 | Q | D | 012.19_Poland_2019 |
| 15 | 65 | D | H | 012.19_Poland_2019 |
| 16 | 67 | N | Y | 143.20_Poland_2020 |
| 17 | 70 | Y | F | 143.20_Poland_2020 |
| 18 | 77 | W | G | 012.19_Poland_2019 |
| 19 | 80 | F | L | 012.19_Poland_2019 |
| 20 | 83 | N | C | 012.19_Poland_2019 |
| 21 | 94 | W | G | 139.20_Poland_2020 |
| 22 | 95 | Q | L | 012.19_Poland_2019 |
| 23 | 99 | N | S | 139.20_Poland_2020 |
| 24 | 103 | G | E | 012.19_Poland_2019 |
| 25 | 104 | I | F | 012.19_Poland_2019 |
| 26 | 126 | Q | H | 139.20_Poland_2020 |
| 27 | 127 | T | A | 139.20_Poland_2020 |
| 28 | 192 | C | R | 144.19_Poland_2019 012.19_Poland_2019 013.19_Poland_2019 089.17_Poland_2017 |
| 29 | 204 | A | V | 191.19_Poland_2019 108.18_Poland_2018 170.18_Poland_2018 308.18_Poland_2018 |
| 30 | 205 | K | E | 144.19_Poland_2019 012.19_Poland_2019 013.19_Poland_2019 089.17_Poland_2017 |
| 31 | 208 | Q | R | 139.20_Poland_2020 |
| 32 | 222 | P | S | 191.19_Poland_2019 108.18_Poland_2018 170.18_Poland_2018 308.18_Poland_2018 |
| 33 | 231 | L | P | 139.20_Poland_2020 174.20_Poland_2020 143.20_Poland_2020 241.17_Poland_2017 222.17_Poland_2017 |
| 34 | 232 | R | G | 012.19_Poland_2019 |
| 35 | 235 | D | Y | 144.19_Poland_2019 012.19_Poland_2019 013.19_Poland_2019 089.17_Poland_2017 |
| 36 | 236 | E | K | 139.20_Poland_2020 |
| 37 | 253 | Q | H | 139.20_Poland_2020 |
| 38 | 260 | G | S | 144.19_Poland_2019 012.19_Poland_2019 013.19_Poland_2019 089.17_Poland_2017 |
| 39 | 261 | C | R | 139.20_Poland_2020 174.20_Poland_2020 143.20_Poland_2020 241.17_Poland_2017 222.17_Poland_2017 |
| 40 | 262 | E | K | 308.18_Poland_2018 |
| 41 | 266 | W | C | 139.20_Poland_2020 |
| 42 | 270 | P | S | 144.19_Poland_2019 013.19_Poland_2019 |
| 43 | 275 | R | G | 144.19_Poland_2019 012.19_Poland_2019 013.19_Poland_2019 089.17_Poland_2017 |
| 44 | 282 | K | E | 144.19_Poland_2019 012.19_Poland_2019 013.19_Poland_2019 089.17_Poland_2017 |
| 45 | 293 | L | Q | 139.20_Poland_2020 174.20_Poland_2020 143.20_Poland_2020 |
| P | 191.19_Poland_2019 108.18_Poland_2018 170.18_Poland_2018 308.18_Poland_2018 | |||
| 46 | 308 | E | K | 139.20_Poland_2020 174.20_Poland_2020 143.20_Poland_2020 144.19_Poland_2019 012.19_Poland_2019 013.19_Poland_2019 089.17_Poland_2017 241.17_Poland_2017 222.17_Poland_2017 |
| 47 | 309 | R | G | 144.19_Poland_2019 012.19_Poland_2019 013.19_Poland_2019 089.17_Poland_2017 |
| 48 | 311 | T | A | all sequences |
| 49 | 323 | L | S | 139.20_Poland_2020 174.20_Poland_2020 143.20_Poland_2020 144.19_Poland_2019 012.19_Poland_2019 013.19_Poland_2019 089.17_Poland_2017 241.17_Poland_2017 222.17_Poland_2017 |
| 50 | 326 | R | M | 012.19_Poland_2019 |
| 51 | 332 | S | P | 144.19_Poland_2019 012.19_Poland_2019 013.19_Poland_2019 089.17_Poland_2017 |
| 52 | 333 | S | G | 139.20_Poland_2020 174.20_Poland_2020 143.20_Poland_2020 222.17_Poland_2017 |
| 53 | 335 | K | E | 144.19_Poland_2019 012.19_Poland_2019 013.19_Poland_2019 089.17_Poland_2017 |
| 54 | 347 | R | W | all sequences |
| 55 | 351 | S | R | 139.20_Poland_2020 |
| 56 | 352 | R | W | 191.19_Poland_2019 108.18_Poland_2018 170.18_Poland_2018 308.18_Poland_2018 |
| 57 | 356 | H | Y | 144.19_Poland_2019 012.19_Poland_2019 013.19_Poland_2019 089.17_Poland_2017 |
| D | 222.17_Poland_2017 | |||
| 58 | 358 | G | V | 012.19_Poland_2019 |
| 59 | 361 | K | T | all sequences except 222.17_Poland_2017 |
| 60 | 362 | N | S | 144.19_Poland_2019 012.19_Poland_2019 013.19_Poland_2019 089.17_Poland_2017 |
| 61 | 363 | K | R | 139.20_Poland_2020 144.19_Poland_2019 013.19_Poland_2019 |
| 62 | 367 | L | P | 144.19_Poland_2019 012.19_Poland_2019 013.19_Poland_2019 089.17_Poland_2017 |
| 63 | 368 | Q | R | 144.19_Poland_2019 012.19_Poland_2019 013.19_Poland_2019 089.17_Poland_2017 |
| 64 | 370 | E | Q | 191.19_Poland_2019 108.18_Poland_2018 170.18_Poland_2018 308.18_Poland_2018 |
| 65 | 382 | M | I | 139.20_Poland_2020 |
| 66 | 396 | M | K | 139.20_Poland_2020 241.17_Poland_2017 |
| R | 144.19_Poland_2019 | |||
| 67 | 397 | F | S | 191.19_Poland_2019 308.18_Poland_2018 |
| 68 | 405 | F | C | 144.19_Poland_2019 |
| 69 | 408 | T | A | 241.17_Poland_2017 |
| 70 | 409 | G | E | 144.19_Poland_2019 |
| 71 | 426 | V | A | 139.20_Poland_2020 191.19_Poland_2019 108.18_Poland_2018 308.18_Poland_2018 |
| 72 | 432 | R | Q | 144.19_Poland_2019 |
| 73 | 441 | I | Q | 174.20_Poland_2020 |
| 74 | 442 | H | N | 174.20_Poland_2020 |
| 75 | 444 | R | H | 144.19_Poland_2019 089.17_Poland_2017 |
| 76 | 446 | C | S | 144.19_Poland_2019 |
| 77 | 453 | C | Y | 144.19_Poland_2019 089.17_Poland_2017 |
| 78 | 461 | T | M | 144.19_Poland_2019 |
| 79 | 467 | I | T | 144.19_Poland_2019 |
| 80 | 471 | P | L | 139.20_Poland_2020 144.19_Poland_2019 |
| 81 | 472 | S | N | 144.19_Poland_2019 |
| 82 | 475 | R | Q | 144.19_Poland_2019 |
| 83 | 481 | R | K | 144.19_Poland_2019 |
| 84 | 508 | Q | L | 144.19_Poland_2019 089.17_Poland_2017 |
| 85 | 509 | A | V | 144.19_Poland_2019 089.17_Poland_2017 |
| 86 | 512 | L | K | 191.19_Poland_2019 |
| 87 | 518 | V | L | 191.19_Poland_2019 |
| 88 | 523 | I | T | 191.19_Poland_2019 |
| 89 | 529 | D | A | 191.19_Poland_2019 |
| 90 | 530 | I | R | 191.19_Poland_2019 |
| 91 | 535 | C | R | 191.19_Poland_2019 |
| L | 308.18_Poland_2018 |
| Lp. | Inoculum | Cytopathic Effect (CPE) at First Passage | Cytopathic Effect (CPE) at Third Passage | Presence of GPV Genetic Material After the Third Passage |
|---|---|---|---|---|
| 1 | 089/17 | negative | negative | negative |
| 2 | 222/17 | positive | positive | positive |
| 3 | 241/17 | positive | positive | positive |
| 4 | 108/18 | negative | positive | positive |
| 5 | 170/18 | negative | positive | positive |
| 6 | 308/18 | positive | positive | positive |
| 7 | 012/19 | negative | negative | negative |
| 8 | 013/19 | negative | negative | negative |
| 9 | 144/19 | positive | positive | positive |
| 10 | 191/19 | positive | positive | positive |
| 11 | 139/20 | negative | negative | negative |
| 12 | 143/20 | negative | positive | positive |
| 13 | 174/20 | negative | positive | positive |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozdruń, W.; Olszowiec, P.; Piekarska, K.; Niczyporuk, J.S. Transport Vehicles as a Vector of Goose Parvovirus Infections (GPV). Animals 2025, 15, 3572. https://doi.org/10.3390/ani15243572
Kozdruń W, Olszowiec P, Piekarska K, Niczyporuk JS. Transport Vehicles as a Vector of Goose Parvovirus Infections (GPV). Animals. 2025; 15(24):3572. https://doi.org/10.3390/ani15243572
Chicago/Turabian StyleKozdruń, Wojciech, Paweł Olszowiec, Karolina Piekarska, and Jowita S. Niczyporuk. 2025. "Transport Vehicles as a Vector of Goose Parvovirus Infections (GPV)" Animals 15, no. 24: 3572. https://doi.org/10.3390/ani15243572
APA StyleKozdruń, W., Olszowiec, P., Piekarska, K., & Niczyporuk, J. S. (2025). Transport Vehicles as a Vector of Goose Parvovirus Infections (GPV). Animals, 15(24), 3572. https://doi.org/10.3390/ani15243572






