Effects of MSTN Gene Knockout on Growth Performance and Muscle Transcriptome in Chinese Merino Sheep (Xinjiang Type)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Housing Conditions
2.2. Growth Performance Measurement
2.3. Blood Sample Collection and Analysis
2.4. Muscle Tissue RNA Extraction, Transcriptome Library Construction, and Sequencing
2.5. Identification of Differentially Expressed Genes and Functional Enrichment Analysis
2.6. Protein–Protein Interaction Network Construction
2.7. Real-Time Quantitative PCR (RT-qPCR) Validation
3. Results
3.1. Effects of MSTN Knockout on Body Weight and Body Measurements
3.2. Effects of MSTN Gene Editing on Hematological and Serum Biochemical Parameters in Sheep
3.3. Statistical Analysis of Sequencing Data
3.4. Sample Correlation Analysis
3.5. Differential Gene Screening
3.6. GO and KEGG Functional Enrichment of Differentially Expressed Genes
3.7. Protein–Protein Interaction Network Construction and Analysis
3.8. RT-qPCR Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MSTN | Myostatin |
| DEGs | Differentially expressed genes |
| PPI | Protein–protein interaction |
| GDF8 | Growth Differentiation Factor 8 |
| TGF-β | Transforming growth factor-beta |
| MT | MSTN gene-edited sheep |
| WT | Wild-type control sheep |
| PCR | Polymerase chain reaction |
| TMR | Total mixed ration |
| PCA | Principal component analysis |
| RT-qPCR | Real-time quantitative PCR |
References
- Chen, M.M.; Zhao, Y.P.; Zhao, Y.; Deng, S.L. Regulation of myostatin on the growth and development of skeletal muscle. Front. Cell Dev. Biol. 2021, 9, 785712. [Google Scholar] [CrossRef]
- McPherron, A.; Lawler, A.; Lee, S.J. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef]
- Luo, S.; Liu, Y.; Bu, L.; Wang, D.; Wen, Z.; Yang, Y.; Xu, Y.; Wu, D.; Li, G.; Yang, L. The Impact of MSTN Gene Editing on Meat Quality and Metabolomics: A Comparative Study Among Three Breeds of MSTN-Edited and Non-Edited Cattle. Animals 2025, 15, 47. [Google Scholar] [CrossRef]
- He, Z.; Zhang, T.; Jiang, L.; Zhou, M.; Wu, D.; Mei, J.; Cheng, Y. Use of CRISPR/Cas9 technology efficiently targeted goat myostatin through zygotes microinjection resulting in double-muscled phenotype in goats. Biosci. Rep. 2018, 38, BSR20180742. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.-X.; Zhan, Q.-M.; Wei, Y.-Y.; Yan, A.-F.; Feng, J.; Liu, L.; Lu, S.-S.; Tang, D.-S. CRISPR/Cas9-mediated MSTN disruption accelerates the growth of Chinese Bama pigs. Reprod. Domest. Anim. 2020, 55, 1314–1327. [Google Scholar] [CrossRef]
- Zhang, T.; Lu, Y.; Song, S.; Lu, R.; Zhou, M.; He, Z.; Yuan, T.; Yan, K.; Cheng, Y. ‘Double-muscling’ and pelvic tilt phenomena in rabbits with the cystine-knot motif deficiency of myostatin on exon 3. Biosci. Rep. 2019, 39, BSR20190207. [Google Scholar] [CrossRef]
- Ledesma, A.V.; Van Eenennaam, A.L. Global status of gene-edited animals for agricultural applications. Vet. J. 2024, 305, 106142. [Google Scholar] [CrossRef]
- Bi, Y.; Hua, Z.; Liu, X.; Hua, W.; Ren, H.; Xiao, H.; Zhang, L.; Li, L.; Wang, Z.; Laible, G.; et al. Isozygous and selectable marker-free MSTN knockout cloned pigs generated by the combined use of CRISPR/Cas9 and Cre/LoxP. Sci. Rep. 2016, 6, 31729. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-M.; Zhao, Y.; Xu, X.-L.; Zhang, X.-S.; Zhang, J.-L.; Wu, S.-J.; Liu, Z.-M.; Yuan, Y.-M.; Guo, X.-F.; Qi, S.-Y.; et al. A MSTN Del73C mutation with FGF5 knockout sheep by CRISPR/Cas9 promotes skeletal muscle myofiber hyperplasia. eLife 2024, 12, RP86827. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.-L.; Li, Z.-Y.; Zou, Y.-J.; Hao, H.-Y.; Hu, J.-X.; Li, N.; Li, Q.-Y. Generation of pigs with a Belgian Blue mutation in MSTN using CRISPR/Cpf1-assisted ssODN-mediated homologous recombination. J. Integr. Agric. 2019, 18, 1329–1336. [Google Scholar] [CrossRef]
- Dilger, A.C.; Chen, X.; Honegger, L.T.; Marron, B.M.; Beever, J.E. The potential for gene-editing to increase muscle growth in pigs: Experiences with editing myostatin. CABI Agric. Biosci. 2022, 3, 106. [Google Scholar] [CrossRef]
- Maynard, L.H.; Humbert, O.; Peterson, C.W.; Kiem, H.-P. Genome editing in large animal models. Mol. Ther. 2021, 29, 2921–2934. [Google Scholar] [CrossRef]
- Tian, Y.; An, J.; Zhang, X.; Di, J.; He, J.; Yasen, A.; Ma, Y.; Sailikehan, G.; Huang, X.; Tian, K. Genome-Wide Scan for Copy Number Variations in Chinese Merino Sheep Based on Ovine High-Density 600K SNP Arrays. Animals 2024, 14, 2897. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Duan, Y.; Zhao, S.; Xu, N.; Zhao, Y. Caprine and Ovine Genomic Selection—Progress and Application. Animals 2024, 14, 2659. [Google Scholar] [CrossRef]
- Duan, Z.; Liang, Y.; Sun, J.; Zheng, H.; Lin, T.; Luo, P.; Wang, M.; Liu, R.; Chen, Y.; Guo, S.; et al. An engineered Cas12i nuclease that is an efficient genome editing tool in animals and plants. Innovation 2024, 5, 100564. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.H.; Wang, J.Q.; Zhao, H.R.; Tan, X.; Yan, S.H.; Zhang, H.Y.; Wang, T.F.; Tang, X.C. Molecular breeding of pigs in the genome editing era. Genet. Sel. Evol. 2025, 57, 12. [Google Scholar] [CrossRef]
- Zhou, S.W.; Kalds, P.; Luo, Q.; Sun, K.X.; Zhao, X.E.; Gao, Y.W.; Cai, B.; Huang, S.H.; Kou, Q.F.; Petersen, B.; et al. Optimized Cas9:sgRNA delivery efficiently generates biallelic MSTN knockout sheep without affecting meat quality. BMC Genom. 2022, 23, 348. [Google Scholar] [CrossRef]
- Gu, R.; Wang, H.; Meng, C.; Gui, H.; Li, Y.; Chen, F.; Zhang, C.; Zhang, H.; Ding, Q.; Zhang, J.; et al. Efficient and Specific Generation of MSTN-Edited Hu Sheep Using C-CRISPR. Genes 2023, 14, 1216. [Google Scholar] [CrossRef]
- Baig, M.H.; Ahmad, K.; Moon, J.S.; Park, S.Y.; Lim, J.H.; Chun, H.J.; Qadri, A.F.; Hwang, Y.C.; Jan, A.F.; Ahmad, S.S.; et al. Myostatin and its regulation: A comprehensive review of myostatin inhibiting strategies. Front. Physiol. 2022, 13, 876078. [Google Scholar] [CrossRef] [PubMed]
- Sheng, H.; Guo, Y.; Zhang, L.; Zhang, J.; Miao, M.; Tan, H.; Hu, D.; Li, X.; Ding, X.; Li, G.; et al. Proteomic Studies on the Mechanism of Myostatin Regulating Cattle Skeletal Muscle Development. Front. Genet. 2021, 12, 752129. [Google Scholar] [CrossRef]
- Hai, C.; Liu, X.; Bai, C.; Su, G.; Yang, L.; Li, G. Loss of Myostatin Shapes the Transcriptomic and Epigenetic Landscapes Across Multiple Muscle Types in Cattle. Curr. Issues Mol. Biol. 2025, 47, 431. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, N.; Zhang, Y.; Liu, J.; Li, J.; Gao, S.; Wang, Y.; Yang, L. Generation of Gene-Modified Goats Targeting MSTN and FGF5 via Zygote Injection of CRISPR/Cas9 System. Sci. Rep. 2015, 5, 13878. [Google Scholar] [CrossRef]
- Gao, L.; Liu, Y.; Zhang, Y.; Li, J.; Wang, X. Mstn Knockdown Decreases the Trans-differentiation from Myocytes to Adipocytes by Reducing Jmjd3 Expression via the SMAD2/SMAD3 Complex. Biosci. Biotechnol. Biochem. 2019, 83, 2090–2096. [Google Scholar] [CrossRef]
- Malgwi, I.; D’Andrea, M.; Schiavon, S.; Badiani, A. Genes Related to Fat Metabolism in Pigs and Intramuscular Fat Content of Pork: A Focus on Nutrigenetics and Nutrigenomics. Animals 2022, 12, 150. [Google Scholar] [CrossRef]
- Bannister, M.L.; Zhou, H.; Dulhunty, A.F. Malignant hyperthermia mutation sites in the Leu2442-Pro2477 (DP4) region of RyR1 are clustered in a structurally and functionally definable area. Biochem. J. 2007, 403, 519–528. [Google Scholar] [CrossRef]
- Brosnan, J.T.; Brosnan, M.E. Glutamate, at the interface between amino acid and carbohydrate metabolism. J. Nutr. 2000, 130, 988S–990S. [Google Scholar] [CrossRef]
- Degens, H.; Qiao, L.; Yang, Y. Myostatin Knockout Mice Have Larger Muscle Fibers With Normal Function and Morphology. Muscle Nerve 2025, 61, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Ponnampalam, E.N.; Mann, N.J.; Norng, S.; Bekhit, A.E.D.; Trotter, M.D.; Hopkins, D.L. Sensory and Flavor Chemistry Characteristics of Australian Beef: Influence of Intramuscular Fat, Feed, and Breed. J. Agric. Food Chem. 2016, 64, 3425–3435. [Google Scholar] [CrossRef]
- Pei, Y.; Fan, Z.; Song, Y.; Chen, C.; Mu, Y.; Li, B.; Feng, Z.; Li, H.; Li, K. Viscera characteristics of MSTN-edited heterozygous pigs. Front. Genet. 2022, 13, 764965. [Google Scholar] [CrossRef]
- Wu, D.; Gu, M.; Wei, Z.; Bai, C.; Su, G.; Liu, X.; Zhao, Y.; Yang, L.; Li, G. Myostatin Knockout Regulates Bile Acid Metabolism by Promoting Bile Acid Synthesis in Cattle. Animals 2022, 12, 205. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.B.; Han, S.; Yin, X.J.; Liu, H.; Wang, J.; Xuan, M.; Hao, C.; Wang, D.; Liu, Y.; Chang, S.; et al. Fecal transplant from myostatin deletion pigs positively impacts the gut-muscle axis. eLife 2023, 12, e81858. [Google Scholar] [CrossRef]
- Chen, M.; Zhao, Y.; Li, Y.; Chen, T.; Zhou, W.; Zhang, X.; Deng, S.; Liu, Z.; Qi, S.; Wang, L.; et al. Reproduction and viscera organ characteristics of MSTN and FGF5 dual-gene knockout sheep. Front. Vet. Sci. 2023, 10, 1119312. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Wang, S.; Di, A.; Wu, D.; Hai, C.; Liu, X.; Bai, C.; Su, G.; Yang, L.; Li, G. Combined Transcriptome and Metabolome Analysis of Smooth Muscle of Myostatin Knockout Cattle. Int. J. Mol. Sci. 2023, 24, 8120. [Google Scholar] [CrossRef]
- Welle, S.; Bhatt, K.; Pinkert, C.A. Myofibrillar protein synthesis in myostatin-deficient mice. Am. J. Physiol. Endocrinol. Metab. 2005, 288, E1088–E1094. [Google Scholar] [CrossRef] [PubMed]
- Qiu, K.; Wang, Y.; Xu, D.; He, L.; Zhang, X.; Yan, E.; Wang, L.; Yin, J. Ryanodine receptor RyR1-mediated elevation of Ca2+ concentration is required for the late stage of myogenic differentiation and fusion. J. Anim. Sci. Biotechnol. 2022, 13, 9. [Google Scholar] [CrossRef]
- Li, X.; Zhao, X.; Qin, Z.; Li, J.; Sun, B.; Liu, L. Regulation of calcium homeostasis in endoplasmic reticulum–mitochondria crosstalk: Implications for skeletal muscle atrophy. Cell Commun. Signal. 2025, 23, 17. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.; Xiong, Q.; Shang, Y.Y.; Zheng, R.; Peng, J.; Jiang, S.W. Evidence for a new allele at the SERCA1 locus affecting pork meat quality in part through the imbalance of Ca2+ homeostasis. J. Anim. Sci. 2009, 87, 3276–3283. [Google Scholar] [CrossRef]
- Pflügers Archiv Editorial Staff. Skeletal muscle CaV1.1 channelopathies. Pflügers Arch. Eur. J. Physiol. 2020, 472, 739–754. [Google Scholar] [CrossRef]
- Nodari, A.; Scambi, I.; Peroni, D.; Calabria, E.; Benati, D.; Mannucci, S.; Manfredi, M.; Frontini, A.; Visonà, S.; Bozzato, A.; et al. Interferon regulatory factor-7 impairs cellular metabolism in aging adipose-derived stromal cells. J. Cell Sci. 2021, 134, jcs256230. [Google Scholar] [CrossRef]
- Olie, C.S.; Pinto-Fernández, A.; Damianou, A.; Vendrell, I.; Mei, H.; Hamer, B.; Wal, E.; Greef, J.C.; Raz, V.; Kessler, B.M. USP18 is an essential regulator of muscle cell differentiation and maturation. Cell Death Dis. 2023, 14, 231. [Google Scholar] [CrossRef]
- Perng, Y.-C.; Lenschow, D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018, 16, 423–439. [Google Scholar] [CrossRef]
- Han, S.-Z.; Gao, K.; Chang, S.-Y.; Choe, H.-M.; Paek, H.-J.; Quan, B.-H.; Liu, X.-Y.; Yang, L.-H.; Lv, S.-T.; Yin, X.-J.; et al. miR-455-3p Is Negatively Regulated by Myostatin in Skeletal Muscle and Promotes Myoblast Differentiation. J. Agric. Food Chem. 2023, 71, 3456–3467. [Google Scholar] [CrossRef] [PubMed]
- Nageeb, R.S.; Mohamed, N.M.; Soliman, A.S.; Ebald, A.M.; Mohammad, N.A. Serum level of myostatin and type I interferon-inducible gene expression in dermatomyositis patients and its relation to insulin resistance. Egypt J. Neurol. Psychiatry Neurosurg. 2024, 60, 136. [Google Scholar] [CrossRef]
- Jin, Z.; Choe, H.M.; Lv, S.; Chang, S.; Yin, X. Esophageal striated muscle hypertrophy and muscle fiber type transformation in MSTN knockout pigs. Transgenic Res. 2022, 31, 341–349. [Google Scholar] [CrossRef]
- Sammut, M.J.; Thorne, B.R.; Melling, C.W.J. Skeletal muscle growth to combat diabetes and obesity: The potential role of muscle-secreted factors. Obesity 2025, 33, 435–451. [Google Scholar] [CrossRef]
- Gim, G.-M.; Kwon, D.-H.; Eom, K.-H.; Moon, J.H. Production of MSTN-Mutated Cattle Without Exogenous Gene Integration Using CRISPR-Cas9. Biotechnol. J. 2021, 17, 2100198. [Google Scholar] [CrossRef]
- Wang, Y.; Li, N.; Lv, Z.; Zhang, Y.; Cao, J.; Liu, B.; Wang, X.; Han, Y.; Bai, Y.; Li, J.; et al. Generation and Evaluation of Myostatin Knock-Out Rabbits and Goats Using CRISPR/Cas9 System. Sci. Rep. 2016, 6, 29855. [Google Scholar] [CrossRef]
- Perea, A.; Pérez, E.; Rodríguez, A.; García-García, R.; Fernández, A.; de la Fuente, J.; Silvestre, M.A.; González, S. Efficient Generation of Myostatin Knock-Out Sheep Using CRISPR/Cas9 Technology and Microinjection into Zygotes. PLoS ONE 2015, 10, e0136690. [Google Scholar] [CrossRef]
- Guo, X.; Li, N.; Liu, G.; Zhang, Y.; Wang, Y.; Lv, Z.; Cao, J.; Liu, B.; Han, Y.; Bai, Y.; et al. Myostatin Regulates Fatty Acid Desaturation and Fat Deposition Through MEF2C/miR222/SCD5 Cascade in Pigs. Commun. Biol. 2020, 3, 612. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, C.; Kong, Y.; Li, F.; Yue, X. Effects of intramuscular fat on meat quality and its regulation mechanism in Tan sheep. Front. Nutr. 2022, 9, 908355. [Google Scholar] [CrossRef]
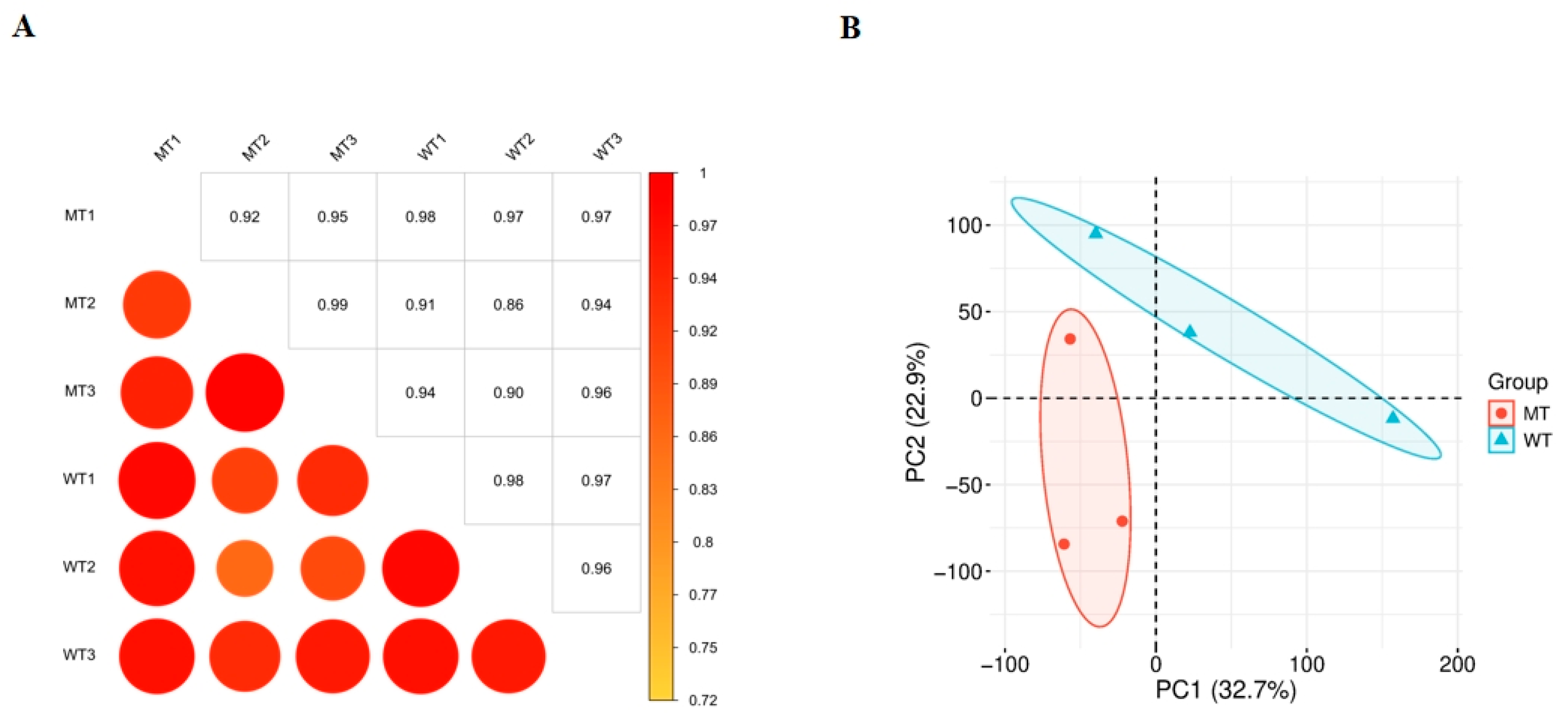
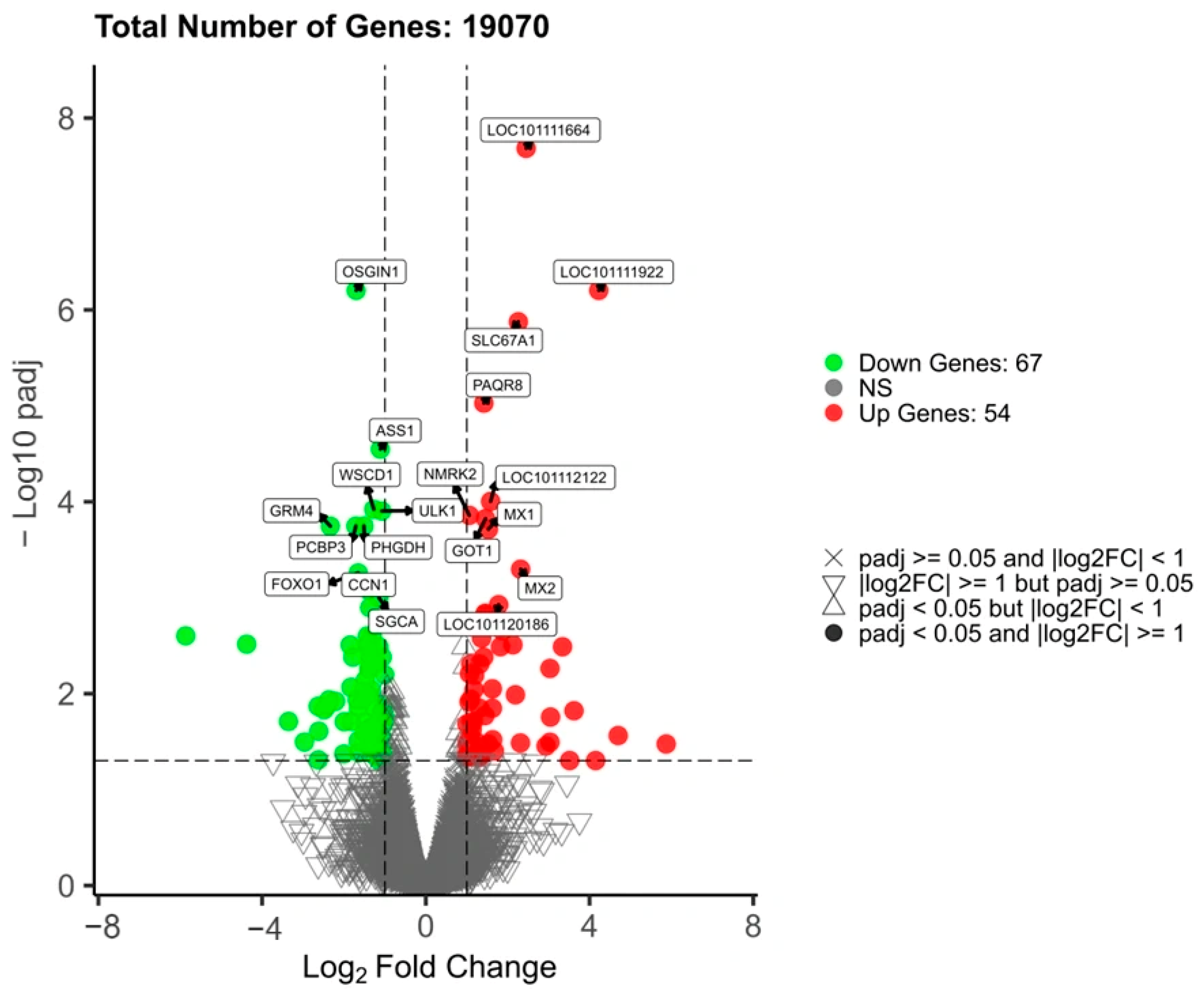
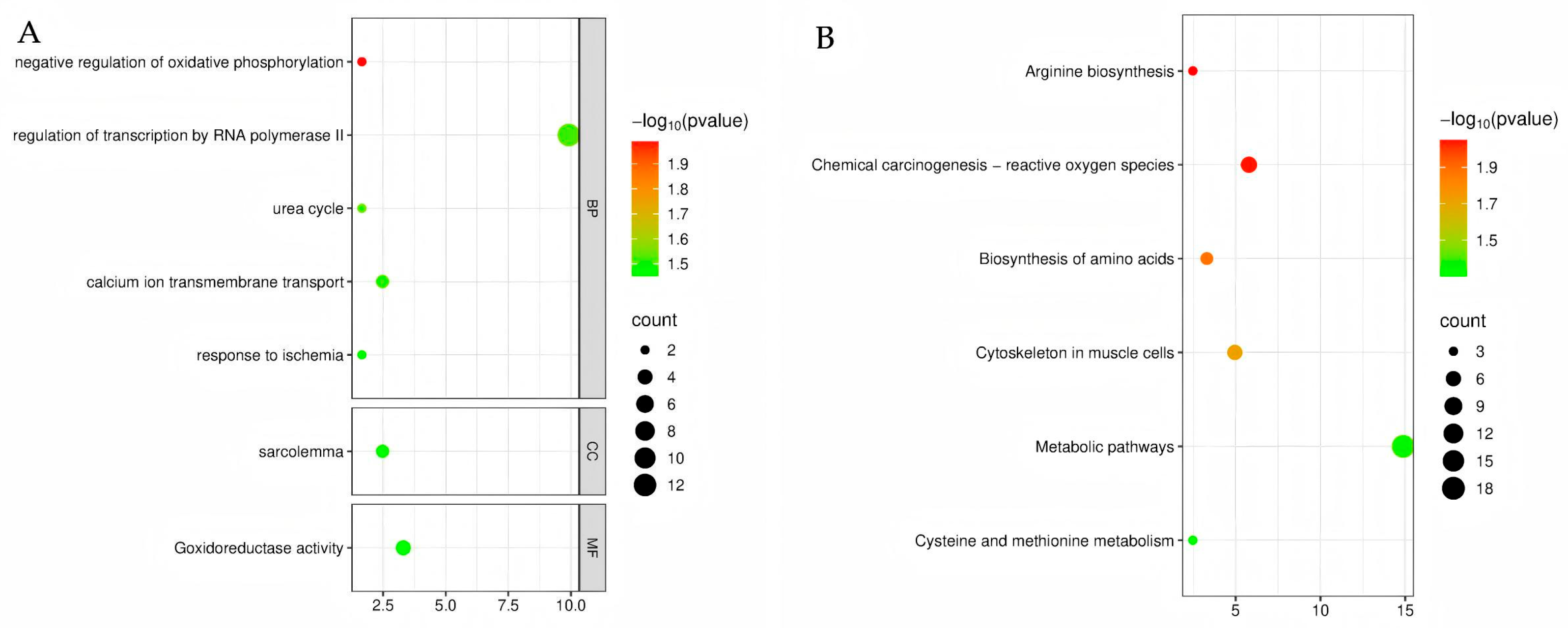
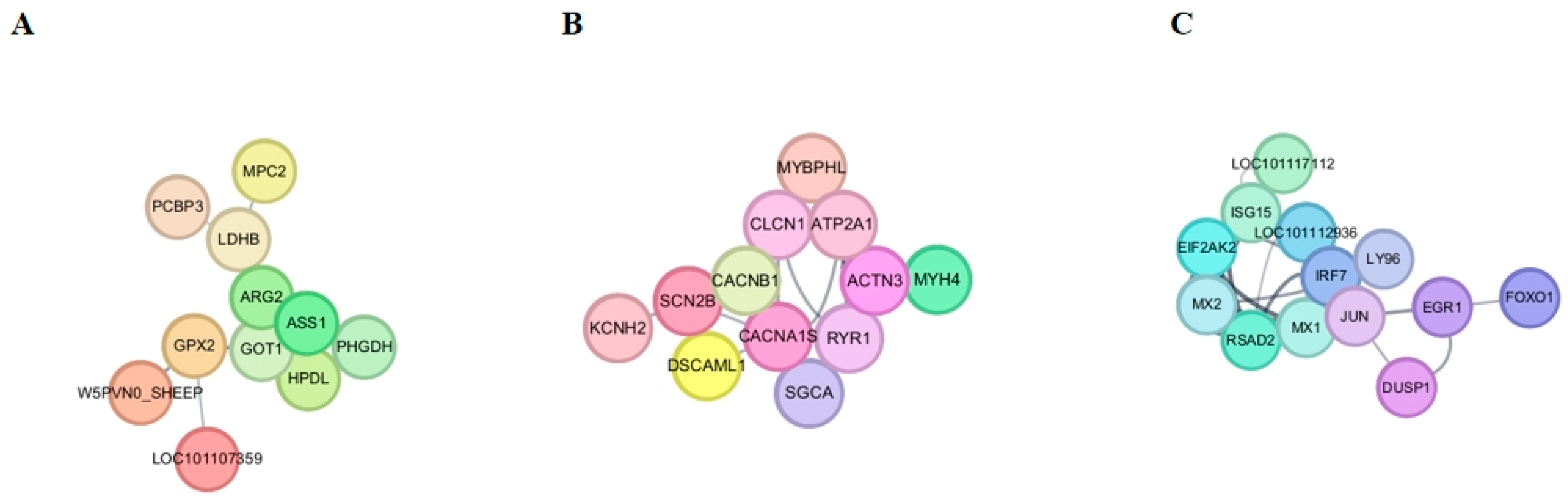

| Age (Days) | Group | Body Weight (kg) | Body Length (cm) | Withers Height (cm) | Chest Girth (cm) | Cannon Circumference (cm) | Hip Width (cm) |
|---|---|---|---|---|---|---|---|
| 30 | MT | 11.09 ± 3.16 a | 43.23 ± 3.82 a | 46.47 ± 3.60 a | 51.14 ± 5.64 a | 6.36 ± 0.60 a | 12.82 ± 1.42 a |
| WT | 8.22 ± 1.48 b | 37.79 ± 2.45 b | 41.61 ± 2.45 b | 42.44 ± 2.03 b | 6.03 ± 0.38 b | 10.62 ± 0.37 b | |
| 60 | MT | 18.32 ± 3.92 a | 52.23 ± 4.46 a | 53.17 ± 3.22 a | 60.10 ± 5.40 a | 6.63 ± 0.65 a | 13.30 ± 1.64 a |
| WT | 16.07 ± 2.91 b | 50.56 ± 3.64 b | 50.82 ± 3.30 b | 58.88 ± 4.52 a | 6.43 ± 0.55 a | 12.10 ± 1.16 b | |
| 90 | MT | 23.94 ± 4.52 a | 59.38 ± 3.69 a | 57.82 ± 3.70 a | 67.82 ± 5.51 a | 7.04 ± 0.56 a | 17.31 ± 1.46 a |
| WT | 20.54 ± 2.92 b | 56.88 ± 3.41 b | 54.85 ± 2.24 b | 65.28 ± 4.05 b | 6.93 ± 0.52 a | 15.17 ± 1.06 b | |
| 120 | MT | 28.52 ± 5.15 a | 61.39 ± 4.62 a | 58.81 ± 2.69 a | 72.55 ± 5.18 a | 7.05 ± 0.57 a | 19.11 ± 1.58 a |
| WT | 24.11 ± 3.49 b | 58.56 ± 3.71 b | 56.38 ± 2.35 b | 69.27 ± 4.60 b | 6.78 ± 0.57 b | 16.58 ± 1.21 b | |
| 160 | MT | 35.18 ± 5.85 a | 66.22 ± 4.54 a | 62.71 ± 3.14 a | 78.46 ± 5.76 a | 7.55 ± 0.73 a | 21.74 ± 2.26 a |
| WT | 29.20 ± 3.71 b | 60.24 ± 3.20 b | 59.24 ± 3.36 b | 74.62 ± 4.00 b | 7.39 ± 0.59 a | 18.46 ± 1.28 b | |
| 190 | MT | 38.39 ± 6.56 a | 67.23 ± 4.35 a | 64.00 ± 2.77 a | 82.30 ± 5.40 a | 7.70 ± 0.63 a | 19.70 ± 1.63 a |
| WT | 31.93 ± 4.88 b | 64.91 ± 4.11 b | 61.58 ± 2.81 b | 77.43 ± 4.35 b | 7.52 ± 0.58 a | 17.71 ± 1.28 b | |
| 365 | MT | 61.07 ± 10.59 a | 80.85 ± 5.86 a | 71.37 ± 4.48 a | 113.16 ± 8.80 a | 11.20 ± 1.35 a | 32.40 ± 2.93 a |
| WT | 50.20 ± 7.97 b | 75.60 ± 5.29 b | 68.23 ± 4.06 b | 104.70 ± 7.36 b | 11.08 ± 1.14 a | 27.50 ± 2.41 b |
| Indicator | Unit | MT Group | WT Group | p-Value |
|---|---|---|---|---|
| White Blood Cell Count | 109/L | 7.35 ± 1.08 | 5.70 ± 1.18 | 0.752 |
| Absolute Neutrophil Count | 109/L | 1.42 ± 0.35 | 1.32 ± 0.38 | 0.374 |
| Absolute Lymphocyte Count | 109/L | 5.37 ± 0.94 | 4.02 ± 0.94 | 0.153 |
| Red Blood Cell Count | 1012/L | 8.98 ± 0.75 | 8.30 ± 1.97 | 0.607 |
| Hemoglobin Concentration | g/L | 84.11 ± 10.49 | 85.71 ± 20.62 | 0.910 |
| Hematocrit | % | 27.83 ± 4.22 | 27.35 ± 7.41 | 0.926 |
| Mean Corpuscular Volume | fL | 30.91 ± 2.50 | 32.74 ± 1.33 | 0.325 |
| Mean Corpuscular Hemoglobin Concentration | g/L | 303.09 ± 9.70 | 315.61 ± 12.45 | 0.242 |
| Platelet Count | 109/L | 168.60 ± 48.88 | 184.20 ± 75.61 | 0.779 |
| Mean Platelet Volume | fL | 6.48 ± 2.09 | 7.18 ± 4.26 | 0.813 |
| Indicator | Unit | MT Group | WT Group | p-Value |
|---|---|---|---|---|
| Albumin | g/L | 36.56 ± 1.42 | 33.83 ± 2.28 | 0.153 |
| Total Protein | g/L | 72.40 ± 4.47 | 73.13 ± 2.64 | 0.819 |
| Albumin/Globulin Ratio | 1.02 ± 0.07 | 0.87 ± 0.18 | 0.240 | |
| Aspartate Aminotransferase | U/L | 178.00 ± 38.31 | 139 ± 23.07 | 0.205 |
| Alanine Aminotransferase | U/L | 27.00 ± 3.46 | 25.33 ± 2.08 | 0.515 |
| Lactate Dehydrogenase | U/L | 595.00 ± 27.62 | 533.67 ± 62.05 | 0.193 |
| Creatine Kinase | U/L | 92.33 ± 11.24 | 73.33 ± 10.50 | 0.099 |
| Creatinine | umol/L | 118.07 ± 10.77 | 160.60 ± 46.97 | 0.201 |
| Urea | mmol/L | 5.78 ± 0.62 | 6.17 ± 0.51 | 0.455 |
| Glucose | mmol/L | 4.66 ± 0.47 | 4.97 ± 0.62 | 0.530 |
| Total Cholesterol | mmol/L | 1.56 ± 0.11 | 1.96 ± 0.36 | 0.134 |
| Triglycerides | mmol/L | 0.52 ± 0.23 | 0.58 ± 0.01 | 0.744 |
| Calcium | mmol/L | 2.40 ± 0.18 | 2.28 ± 0.08 | 0.378 |
| Group | Sample | Total Reads | Total Mapped Reads | Clean Bases (G) | Valid Bases (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|---|
| MT | 1 | 46,957,018 | 46,440,752 | 6.87 | 97.54 | 96.90 | 56.68 |
| 2 | 46,931,116 | 46,325,269 | 6.90 | 97.99 | 96.72 | 56.62 | |
| 3 | 47,106,932 | 46,083,379 | 6.93 | 97.98 | 96.69 | 56.05 | |
| WT | 1 | 48,464,622 | 48,010,755 | 7.06 | 96.99 | 96.97 | 55.90 |
| 2 | 47,057,204 | 46,444,644 | 6.90 | 97.72 | 96.79 | 55.83 | |
| 3 | 46,073,008 | 45,485,260 | 6.78 | 97.98 | 96.69 | 55.99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Li, P.; Wang, X.; Kong, M.; Wu, W.; Zheng, W. Effects of MSTN Gene Knockout on Growth Performance and Muscle Transcriptome in Chinese Merino Sheep (Xinjiang Type). Animals 2025, 15, 3387. https://doi.org/10.3390/ani15233387
Zhang L, Li P, Wang X, Kong M, Wu W, Zheng W. Effects of MSTN Gene Knockout on Growth Performance and Muscle Transcriptome in Chinese Merino Sheep (Xinjiang Type). Animals. 2025; 15(23):3387. https://doi.org/10.3390/ani15233387
Chicago/Turabian StyleZhang, Li, Pengfei Li, Xu Wang, Menghua Kong, Weiwei Wu, and Wenxin Zheng. 2025. "Effects of MSTN Gene Knockout on Growth Performance and Muscle Transcriptome in Chinese Merino Sheep (Xinjiang Type)" Animals 15, no. 23: 3387. https://doi.org/10.3390/ani15233387
APA StyleZhang, L., Li, P., Wang, X., Kong, M., Wu, W., & Zheng, W. (2025). Effects of MSTN Gene Knockout on Growth Performance and Muscle Transcriptome in Chinese Merino Sheep (Xinjiang Type). Animals, 15(23), 3387. https://doi.org/10.3390/ani15233387






