Intestinal Microecological Mechanisms of Aflatoxin B1 Degradation by Black Soldier Fly Larvae (Hermetia illucens): A Review
Simple Summary
Abstract
1. Introduction
2. Research Progress on AFB1 Detoxification Technology
3. Advantages of BSFL for the Degradation of AFB1 Contaminated Waste
4. Composition, Source, and Colonization of BSFL Intestinal Microbiota
5. Synergistic Degradation of AFB1 by Endogenous Enzymes and Intestinal Microbiota of BSFL
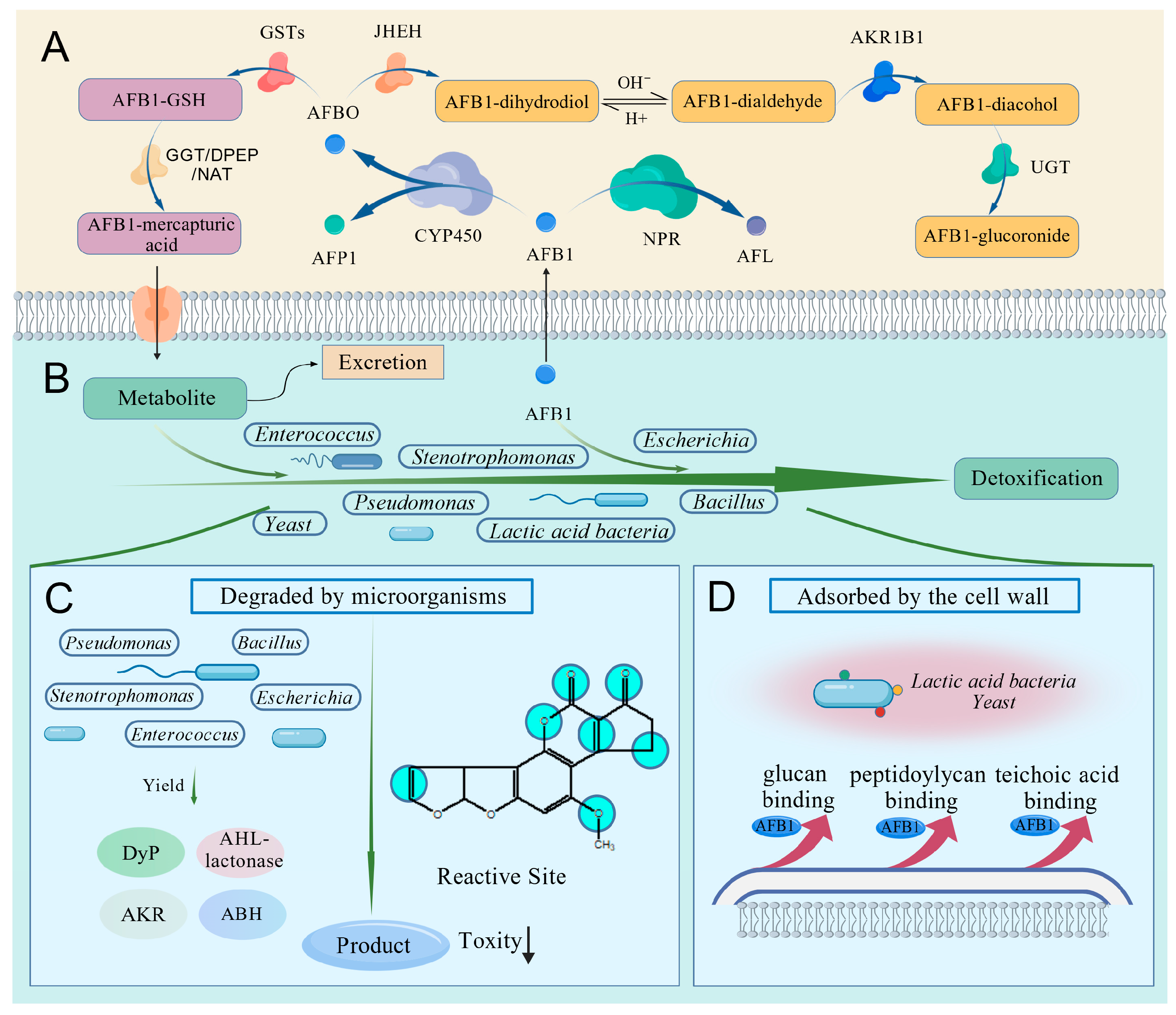
6. Possible Pathways of AFB1 Degradation by the Intestinal Microbiota in BSFL
7. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AFB1 | Aflatoxin B1 |
| BSFL | black soldier fly larvae |
| AF | Aflatoxin |
| AFB2 | Aflatoxin B2 |
| AFG1 | Aflatoxin G1 |
| AFG2 | Aflatoxin G2 |
| AFM1 | Aflatoxin M1 |
| AFM2 | Aflatoxin M2 |
| CYP450 | cytochrome P450 monooxygenase |
| AFBO | AFB1-8,9-epoxide |
| LCs | laccases |
| PODs | peroxidases |
| FDRs | F420H2-dependent reductases |
| MnP | manganese peroxidase |
| DyP | dye-decolorizing peroxidase |
| DON | deoxynivalenol |
| OTA | ochratoxin A |
| ZEN | zearalenone |
| AFL | aflatoxicol |
| AMG | anterior midgut |
| MMG | middle midgut |
| PMG | posterior midgut |
| AFP1 | aflatoxin P1 |
| NPR | NADPH-dependent reductase |
| JHEH | juvenile hormone epoxide hydrolase 1 |
| AKR1B1 | aldo-keto reductase family 1 member B1 |
| GSH | glutathione |
| GST | glutathione-S-transferase |
| AFB1-GSH | AFB1-glutathione conjugate |
| GGT | γ-glutamyl transferase |
| DPEP | dipeptidase |
| NAT | N-acetyltransferase |
| UGT-2 | UDP-glucosyltransferase 2 |
| MRP2 | multidrug resistance protein 2 |
| AHL | N-acyl homoserine lactone-degrading enzyme |
| BacC | bacilysin biosynthesis oxidoreductase |
| Nrf2 | nuclear factor erythroid 2-related factor 2 |
| BsDyP | Bacillus subtilis dye-decolorizing peroxidase |
| AFQ1 | Aflatoxin Q1 |
| AFD1 | Aflatoxin D1 |
| TCA | the citric acid |
References
- Meneely, J.P.; Kolawole, O.; Haughey, S.A.; Miller, S.J.; Krska, R.; Elliott, C.T. The challenge of global aflatoxins legislation with a focus on peanuts and peanut products: A systematic review. Expos. Health 2023, 15, 467–487. [Google Scholar] [CrossRef]
- Hameed, R.M.; Mukheef, M.A.; Khader, H.H.; Fatima, G.; Anwar, S. Aflatoxin B1: A review on biochemical properties, exposure, mechanisms of action and chronic diseases caused by aflatoxins. Era J. Med. Res. 2023, 10, 61–73. [Google Scholar] [CrossRef]
- Benkerroum, N. Chronic and acute toxicities of aflatoxins: Mechanisms of action. Int. J. Environ. Res. Public Health 2020, 17, 423. [Google Scholar] [CrossRef]
- Mutegi, C.K.; Cotty, P.J.; Bandyopadhyay, R. Prevalence and mitigation of aflatoxins in Kenya (1960-to date). World Mycotoxin J. 2018, 11, 341–357. [Google Scholar] [CrossRef]
- Krishnamachari, K.A.; Bhat, R.V.; Nagarajan, V.; Tilak, T.B. Hepatitis due to aflatoxicosis: An outbreak in western India. Lancet 1975, 305, 1061–1063. [Google Scholar] [CrossRef] [PubMed]
- Kinyenje, E.; Kishimba, R.; Mohamed, M.; Mwafulango, A.; Eliakimu, E.; Kwesigabo, G. Aflatoxicosis outbreak and its associated factors in Kiteto, Chemba and Kondoa Districts, Tanzania. PLoS Glob. Public Health 2023, 3, e0002191. [Google Scholar] [CrossRef]
- Lewis, L.; Onsongo, M.; Njapau, H.; Schurz-Rogers, H.; Luber, G.; Kieszak, S.; Nyamongo, J.; Backer, L.; Dahiye Abdikher, M.; Misore, A.; et al. Aflatoxin Contamination of Commercial Maize Products during an Outbreak of Acute Aflatoxicosis in Eastern and Central Kenya. Environ. Health Perspect. 2005, 113, 1763–1767. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhao, L.; Ma, Q.; Ji, C. Novel strategies for degradation of aflatoxins in food and feed: A review. Food Res. Int. 2021, 140, 109878. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.J.; Hsieh, D.P. Mutagenicity of aflatoxins related to their metabolism and carcinogenic potential. Proc. Natl. Acad. Sci. USA 1976, 73, 2241–2244. [Google Scholar] [CrossRef]
- Martínez, J.; Hernández-Rodríguez, M.; Méndez-Albores, A.; Téllez-Isaías, G.; Mera Jiménez, E.; Nicolás-Vázquez, M.I.; Miranda Ruvalcaba, R. Computational studies of aflatoxin B1 (AFB1): A review. Toxins 2023, 15, 135. [Google Scholar] [CrossRef]
- Lalah, J.O.; Omwoma, S.; Orony, D.A.O. Aflatoxin B1: Chemistry, Environmental and Diet Sources and Potential Exposure in Human in Kenya. In Aflatoxin B1 Occurrence, Detection and Toxicological Effects; Long, X., Ed.; IntechOpen: Rijeka, Croatia, 2019; pp. 3–35. [Google Scholar]
- Abrehame, S.; Manoj, V.R.; Hailu, M.; Chen, Y.; Lin, Y.; Chen, Y. Aflatoxins: Source, detection, clinical features and prevention. Processes 2023, 11, 204. [Google Scholar] [CrossRef]
- Čolović, R.; Puvača, N.; Cheli, F.; Avantaggiato, G.; Greco, D.; Đuragić, O.; Kos, J.; Pinotti, L. Decontamination of Mycotoxin-Contaminated Feedstuffs and Compound Feed. Toxins 2019, 11, 617. [Google Scholar] [CrossRef]
- Yazdanpanah, H.; Mohammadi, T.; Abouhossain, G.; Cheraghali, A.M. Effect of roasting on degradation of aflatoxins in contaminated pistachio nuts. Food Chem. Toxicol. 2005, 43, 1135–1139. [Google Scholar] [CrossRef]
- Liu, M.; Zhao, L.; Gong, G.; Zhang, L.; Shi, L.; Dai, J.; Han, Y.; Wu, Y.; Khalil, M.M.; Sun, L. Invited review: Remediation strategies for mycotoxin control in feed. J. Anim. Sci. Biotechnol. 2022, 13, 19. [Google Scholar] [CrossRef]
- Elliott, C.T.; Connolly, L.; Kolawole, O. Potential adverse effects on animal health and performance caused by the addition of mineral adsorbents to feeds to reduce mycotoxin exposure. Mycotoxin Res. 2020, 36, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Vijayanandraj, S.; Brinda, R.; Kannan, K.; Adhithya, R.; Vinothini, S.; Senthil, K.; Chinta, R.R.; Paranidharan, V.; Velazhahan, R. Detoxification of aflatoxin B1 by an aqueous extract from leaves of Adhatoda vasica Nees. Microbiol. Res. 2014, 169, 294–300. [Google Scholar] [CrossRef]
- Brinda, R.; Vijayanandraj, S.; Uma, D.; Malathi, D.; Paranidharan, V.; Velazhahan, R. Role of Adhatoda vasica (L.) Nees leaf extract in the prevention of aflatoxin-induced toxicity in Wistar rats. J. Sci. Food Agric. 2013, 93, 2743–2748. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, A.C.; Barroso, J.G.; Pedro, L.G.; Scheffer, J.J.C. Factors affecting secondary metabolite production in plants: Volatile components and essential oils. Flavour Fragr. J. 2008, 23, 213–226. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, L.; Zhang, Y.; Ran, R.; Meng, X.; Liu, S. Research advances in the degradation of aflatoxin by lactic acid bacteria. J. Venom. Anim. Toxins Incl. Trop. Dis. 2023, 29, e20230029. [Google Scholar] [CrossRef]
- Bovo, F.; Franco, L.T.; Rosim, R.E.; Barbalho, R.; Oliveira, C.A.F.d. In vitro ability of beer fermentation residue and yeast-based products to bind aflatoxin B1. Braz. J. Microbiol. 2015, 46, 577–581. [Google Scholar] [CrossRef]
- Arun, C.S.; Mahalingam, P.U.; Parengal, H.; Thomas, J. Potential of Animal Excreta as a Source of Probiotic Lactic Acid Bacteria for Aflatoxin B1 Detoxification by the Surface Binding Mechanism. J. Pure Appl. Microbiol. 2023, 17, 2386–2401. [Google Scholar] [CrossRef]
- Poloni, V.; Dogi, C.; Pereyra, C.M.; Fernández Juri, M.G.; Köhler, P.; Rosa, C.A.; Dalcero, A.M.; Cavaglieri, L.R. Potentiation of the effect of a commercial animal feed additive mixed with different probiotic yeast strains on the adsorption of aflatoxin B1. Food Addit. Contam. Part A 2015, 32, 970–976. [Google Scholar] [CrossRef]
- González Pereyra, M.L.; Martínez, M.P.; Cavaglieri, L.R. Presence of aiiA homologue genes encoding for N-Acyl homoserine lactone-degrading enzyme in aflatoxin B1-decontaminating Bacillus strains with potential use as feed additives. Food Chem. Toxicol. 2019, 124, 316–323. [Google Scholar] [CrossRef]
- Petchkongkaew, A.; Taillandier, P.; Gasaluck, P.; Lebrihi, A. Isolation of Bacillus spp. from Thai fermented soybean (Thua-nao): Screening for aflatoxin B1 and ochratoxin A detoxification. J. Appl. Microbiol. 2008, 104, 1495–1502. [Google Scholar] [CrossRef]
- Wang, L.; Huang, W.; Sha, Y.; Yin, H.; Liang, Y.; Wang, X.; Shen, Y.; Wu, X.; Wu, D.; Wang, J. Co-Cultivation of Two Bacillus Strains for Improved Cell Growth and Enzyme Production to Enhance the Degradation of Aflatoxin B1. Toxins 2021, 13, 435. [Google Scholar] [CrossRef]
- Verheecke, C.; Liboz, T.; Mathieu, F. Microbial degradation of aflatoxin B1: Current status and future advances. Int. J. Food Microbiol. 2016, 237, 1–9. [Google Scholar] [CrossRef]
- El-Deeb, B.; Altalhi, A.; Khiralla, G.; Hassan, S.; Gherbawy, Y. Isolation and Characterization of Endophytic Bacilli bacterium from Maize Grains Able to Detoxify Aflatoxin B1. Food Biotechnol. 2013, 27, 199–212. [Google Scholar] [CrossRef]
- Rao, K.R.; Vipin, A.; Hariprasad, P.; Appaiah, K.A.; Venkateswaran, G. Biological detoxification of Aflatoxin B1 by Bacillus licheniformis CFR1. Food Control 2017, 71, 234–241. [Google Scholar] [CrossRef]
- Farzaneh, M.; Shi, Z.; Ghassempour, A.; Sedaghat, N.; Ahmadzadeh, M.; Mirabolfathy, M.; Javan-Nikkhah, M. Aflatoxin B1 degradation by Bacillus subtilis UTBSP1 isolated from pistachio nuts of Iran. Food Control 2012, 23, 100–106. [Google Scholar] [CrossRef]
- Guan, S.; Ji, C.; Zhou, T.; Li, J.; Ma, Q.; Niu, T. Aflatoxin B1 Degradation by Stenotrophomonas maltophilia and Other Microbes Selected Using Coumarin Medium. Int. J. Mol. Sci. 2008, 9, 1489–1503. [Google Scholar] [CrossRef]
- Tang, X.; Cai, Y.; Yu, X.; Zhou, W. Detoxification of aflatoxin B1 by Bacillus aryabhattai through conversion of double bond in terminal furan. J. Appl. Microbiol. 2023, 134, lxad192. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Wu, W.; Zhang, D.; Cao, L.; Cheng, J. AFB1 Microbial Degradation by Bacillus subtilis WJ6 and Its Degradation Mechanism Exploration Based on the Comparative Transcriptomics Approach. Metabolites 2023, 13, 785. [Google Scholar] [CrossRef]
- Guo, J.; Zhang, H.; Zhao, Y.; Hao, X.; Liu, Y.; Li, S.; Wu, R. Identification of a Novel Aflatoxin B1-Degrading Strain, Bacillus halotolerans DDC-4, and Its Response Mechanisms to Aflatoxin B1. Toxins 2024, 16, 256. [Google Scholar] [CrossRef]
- Huang, W.; Chang, J.; Wang, P.; Liu, C.; Yin, Q.; Zhu, Q.; Lu, F.; Gao, T. Effect of the combined compound probiotics with mycotoxin-degradation enzyme on detoxifying aflatoxin B1 and zearalenone. J. Toxicol. Sci. 2018, 43, 377–385. [Google Scholar] [CrossRef]
- Li, T.; Chang, X.; Qiao, Z.; Ren, G.; Zhou, N.; Chen, J.; Jiang, D.; Liu, C. Characterization and genomic analysis of Bacillus megaterium with the ability to degrade aflatoxin B1. Front. Microbiol. 2024, 15, 1407270. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, X.; Dong, L.; He, S. Screening and identification of an aflatoxin B1-degrading strain from the Qinghai-Tibet Plateau and biodegradation products analysis. Front. Microbiol. 2024, 15, 1367297. [Google Scholar] [CrossRef] [PubMed]
- Al-Saadi, H.A.; Al-Sadi, A.M.; Al-Wahaibi, A.; Al-Raeesi, A.; Al-Kindi, M.; Soundra Pandian, S.B.; Al-Harrasi, M.M.A.; Al-Mahmooli, I.H.; Velazhahan, R. Rice Weevil (Sitophilus oryzae L.) Gut Bacteria Inhibit Growth of Aspergillus flavus and Degrade Aflatoxin B1. J. Fungi 2024, 10, 377. [Google Scholar] [CrossRef] [PubMed]
- Adebo, O.A.; Njobeh, P.B.; Sidu, S.; Tlou, M.G.; Mavumengwana, V. Aflatoxin B1 degradation by liquid cultures and lysates of three bacterial strains. Int. J. Food Microbiol. 2016, 233, 11–19. [Google Scholar] [CrossRef]
- Maneeboon, T.; Roopkham, C.; Mahakarnchanakul, W.; Chuaysrinule, C. Exploration of Pseudomonas knackmussii AD02 for the biological mitigation of post-harvest aflatoxin contamination: Characterization and degradation mechanism. J. Stored Prod. Res. 2024, 109, 102470. [Google Scholar] [CrossRef]
- Samuel, M.S.; Sivaramakrishna, A.; Mehta, A. Degradation and detoxification of aflatoxin B1 by Pseudomonas putida. Int. Biodeterior. Biodegrad. 2014, 86, 202–209. [Google Scholar] [CrossRef]
- Sangare, L.; Zhao, Y.; Folly, Y.M.E.; Chang, J.; Li, J.; Selvaraj, J.N.; Xing, F.; Zhou, L.; Wang, Y.; Liu, Y. Aflatoxin B1 degradation by a Pseudomonas strain. Toxins 2014, 6, 3028–3040. [Google Scholar] [CrossRef]
- Krifaton, C.; Kriszt, B.; Szoboszlay, S.; Cserháti, M.; Szűcs, Á.; Kukolya, J. Analysis of aflatoxin-B1-degrading microbes by use of a combined toxicity-profiling method. Mutat. Res./Genet. Toxicol. Environ. Mutag. 2011, 726, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Li, J.; He, Y.; Guan, S.; Wang, N.; Ji, C.; Niu, T. AFB1 Bio-Degradation by a New Strain-Stenotrophomonas sp. Agric. Sci. China 2008, 7, 1433–1437. [Google Scholar] [CrossRef]
- Cai, M.; Qian, Y.; Chen, N.; Ling, T.; Wang, J.; Jiang, H.; Wang, X.; Qi, K.; Zhou, Y. Detoxification of aflatoxin B1 by Stenotrophomonas sp. CW117 and characterization the thermophilic degradation process. Environ. Pollut. 2020, 261, 114178. [Google Scholar] [CrossRef]
- Suo, J.; Liang, T.; Zhang, H.; Liu, K.; Li, X.; Xu, K.; Guo, J.; Luo, Q.; Yang, S. Characteristics of aflatoxin B1 degradation by Stenotrophomonas acidaminiphila and it’s combination with black soldier fly larvae. Life 2023, 13, 234. [Google Scholar] [CrossRef]
- Eshelli, M.; Harvey, L.; Edrada-Ebel, R.; McNeil, B. Metabolomics of the Bio-Degradation Process of Aflatoxin B1 by Actinomycetes at an Initial pH of 6.0. Toxins 2015, 7, 439–456. [Google Scholar] [CrossRef]
- Alberts, J.; Engelbrecht, Y.; Steyn, P.; Holzapfel, W.; Van Zyl, W. Biological degradation of aflatoxin B1 by Rhodococcus erythropolis cultures. Int. J. Food Microbiol. 2006, 109, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Cserháti, M.; Kriszt, B.; Krifaton, C.; Szoboszlay, S.; Háhn, J.; Tóth, S.; Nagy, I.; Kukolya, J. Mycotoxin-degradation profile of Rhodococcus strains. Int. J. Food Microbiol. 2013, 166, 176–185. [Google Scholar] [CrossRef]
- Deng, D.; Tang, J.; Liu, Z.; Tian, Z.; Song, M.; Cui, Y.; Rong, T.; Lu, H.; Yu, M.; Li, J.; et al. Functional Characterization and Whole-Genome Analysis of an Aflatoxin-Degrading Rhodococcus pyridinivorans Strain. Biology 2022, 11, 774. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Tang, Y.; Si, W.; Yin, J.; Xu, Y.; Yang, J. Rhodococcus turbidus PD630 enables efficient biodegradation of aflatoxin B1. LWT-Food Sci. Technol. 2023, 186, 115225. [Google Scholar] [CrossRef]
- Risa, A.; Krifaton, C.; Kukolya, J.; Kriszt, B.; Cserháti, M.; Táncsics, A. Aflatoxin B1 and Zearalenone-Detoxifying Profile of Rhodococcus Type Strains. Curr. Microbiol. 2018, 75, 907–917. [Google Scholar] [CrossRef]
- Teniola, O.D.; Addo, P.A.; Brost, I.M.; Färber, P.; Jany, K.D.; Alberts, J.F.; van Zyl, W.H.; Steyn, P.S.; Holzapfel, W.H. Degradation of aflatoxin B1 by cell-free extracts of Rhodococcus erythropolis and Mycobacterium fluoranthenivorans sp. nov. DSM44556T. Int. J. Food Microbiol. 2005, 105, 111–117. [Google Scholar] [CrossRef]
- Khanafari, A.; Soudi, H.; Mirabou, A.M. Biocontrol of Aspergillus flavus and aflatoxin B1 production in corn. J. Environ. Health Sci. 2007, 4, 163–168. [Google Scholar]
- Zhu, Y.; Xu, Y.; Yang, Q. Antifungal properties and AFB1 detoxification activity of a new strain of Lactobacillus plantarum. J. Hazard. Mater. 2021, 414, 125569. [Google Scholar] [CrossRef]
- Lemmetty, J.; Lee, Y.; Laitila, T.; Bredehorst, S.; Coda, R.; Katina, K.; Maina, N.H. Sequestration of aflatoxin B1 by lactic acid bacteria: Role of binding and biotransformation. Food Res. Int. 2025, 199, 115351. [Google Scholar] [CrossRef]
- Feng, J.; Cao, L.; Du, X.; Zhang, Y.; Cong, Y.; He, J.; Zhang, W. Biological Detoxification of Aflatoxin B1 by Enterococcus faecium HB2-2. Foods 2024, 13, 1887. [Google Scholar] [CrossRef] [PubMed]
- Hormisch, D.; Brost, I.; Kohring, G.W.; Giffhorn, F.; Kroppenstedt, R.M.; Stackebradt, E.; Färber, P.; Holzapfel, W.H. Mycobacterium fluoranthenivorans sp. nov., a Fluoranthene and Aflatoxin B1 Degrading Bacterium from Contaminated Soil of a Former Coal Gas Plant. Syst. Appl. Microbiol. 2004, 27, 653–660. [Google Scholar] [CrossRef]
- Campos-Avelar, I.; Colas de la Noue, A.; Durand, N.; Cazals, G.; Martinez, V.; Strub, C.; Fontana, A.; Schorr-Galindo, S. Aspergillus flavus Growth Inhibition and Aflatoxin B1 Decontamination by Streptomyces Isolates and Their Metabolites. Toxins 2021, 13, 340. [Google Scholar] [CrossRef] [PubMed]
- Harkai, P.; Szabó, I.; Cserháti, M.; Krifaton, C.; Risa, A.; Radó, J.; Balázs, A.; Berta, K.; Kriszt, B. Biodegradation of aflatoxin-B1 and zearalenone by Streptomyces sp. collection. Int. Biodeterior. Biodegrad. 2016, 108, 48–56. [Google Scholar] [CrossRef]
- Adebo, O.A.; Njobeh, P.B.; Mavumengwana, V. Degradation and detoxification of AFB1 by Staphylocococcus warneri, Sporosarcina sp. and Lysinibacillus fusiformis. Food Control 2016, 68, 92–96. [Google Scholar] [CrossRef]
- D’souza, D.H.; Brackett, R.E. The role of trace metal ions in aflatoxin B1 degradation by Flavobacterium aurantiacum. J. Food Prot. 1998, 61, 1666–1669. [Google Scholar] [CrossRef]
- Zhang, R.; Xu, C.; Xie, Y.; Chen, A.; Lu, P.; Wu, M.; Han, G.; Hu, S. Biotransformation of aflatoxin B1 by a novel strain Brevundimonas sp. LF-1. Int. Biodeterior. Biodegrad. 2024, 191, 105810. [Google Scholar] [CrossRef]
- Wang, L.; Wu, J.; Liu, Z.; Shi, Y.; Liu, J.; Xu, X.; Hao, S.; Mu, P.; Deng, F.; Deng, Y. Aflatoxin B1 Degradation and Detoxification by Escherichia coli CG1061 Isolated from Chicken Cecum. Front. Pharmacol. 2019, 9, 1548. [Google Scholar] [CrossRef]
- Mwakinyali, S.E.; Ming, Z.; Xie, H.; Zhang, Q.; Li, P. Investigation and characterization of Myroides odoratimimus strain 3J2MO aflatoxin B1 degradation. J. Agric. Food. Chem. 2019, 67, 4595–4602. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Kong, Q.; Chi, C.; Shan, S.; Guan, B. Biotransformation of aflatoxin B1 and aflatoxin G1 in peanut meal by anaerobic solid fermentation of Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus. Int. J. Food Microbiol. 2015, 211, 1–5. [Google Scholar] [CrossRef]
- Qiu, T.; Wang, H.; Yang, Y.; Yu, J.; Ji, J.; Sun, J.; Zhang, S.; Sun, X. Exploration of biodegradation mechanism by AFB1-degrading strain Aspergillus niger FS10 and its metabolic feedback. Food Control 2021, 121, 107609. [Google Scholar] [CrossRef]
- Verheecke, C.; Liboz, T.; Darriet, M.; Sabaou, N.; Mathieu, F. In vitro interaction of actinomycetes isolates with Aspergillus flavus: Impact on aflatoxins B1 and B2 production. Lett. Appl. Microbiol. 2014, 58, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Moebus, V.F.; Pinto, L.d.A.; Köptcke, F.B.N.; Keller, K.M.; Aronovich, M.; Keller, L.A.M. In Vitro Mycotoxin Decontamination by Saccharomyces cerevisiae Strains Isolated from Bovine Forage. Fermentation 2023, 9, 585. [Google Scholar] [CrossRef]
- Yue, X.; Ren, X.; Fu, J.; Wei, N.; Altomare, C.; Haidukowski, M.; Logrieco, A.F.; Zhang, Q.; Li, P. Characterization and mechanism of aflatoxin degradation by a novel strain of Trichoderma reesei CGMCC3.5218. Front. Microbiol. 2022, 13, 1003039. [Google Scholar] [CrossRef]
- Guo, C.; Fan, L.; Yang, Q.; Ning, M.; Zhang, B.; Ren, X. Characterization and mechanism of simultaneous degradation of aflatoxin B1 and zearalenone by an edible fungus of Agrocybe cylindracea GC-Ac2. Front. Microbiol. 2024, 15, 1292824. [Google Scholar] [CrossRef]
- Jackson, L.W.; Pryor, B.M. Degradation of aflatoxin B1 from naturally contaminated maize using the edible fungus Pleurotus ostreatus. AMB Express 2017, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Zaccaria, M.; Dawson, W.; Russel Kish, D.; Reverberi, M.; Bonaccorsi di Patti, M.C.; Domin, M.; Cristiglio, V.; Chan, B.; Dellafiora, L.; Gabel, F. Experimental-theoretical study of laccase as a detoxifier of aflatoxins. Sci. Rep. 2023, 13, 860. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ogata, M.; Hirai, H.; Kawagishi, H. Detoxification of aflatoxin B1 by manganese peroxidase from the white-rot fungus Phanerochaete sordida YK-624. FEMS Microbiol. Lett. 2011, 314, 164–169. [Google Scholar] [CrossRef]
- Wang, X.; Qin, X.; Hao, Z.; Luo, H.; Yao, B.; Su, X. Degradation of Four Major Mycotoxins by Eight Manganese Peroxidases in Presence of a Dicarboxylic Acid. Toxins 2019, 11, 566. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.C.; Jackson, C.J.; Tattersall, D.B.; French, N.; Peat, T.S.; Newman, J.; Briggs, L.J.; Lapalikar, G.V.; Campbell, P.M.; Scott, C.; et al. Identification and characterization of two families of F420H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation. Mol. Microbiol. 2010, 78, 561–575. [Google Scholar] [CrossRef]
- Subagia, R.; Schweiger, W.; Kunz-Vekiru, E.; Wolfsberger, D.; Schatzmayr, G.; Ribitsch, D.; Guebitz, G.M. Detoxification of aflatoxin B1 by a Bacillus subtilis spore coat protein through formation of the main metabolites AFQ1 and epi-AFQ1. Front. Microbiol. 2024, 15, 1406707. [Google Scholar] [CrossRef]
- Gao, B.; An, W.; Wu, J.; Wang, X.; Han, B.; Tao, H.; Liu, J.; Wang, Z.; Wang, J. Simultaneous Degradation of AFB1 and ZEN by CotA Laccase from Bacillus subtilis ZJ-2019-1 in the Mediator-Assisted or Immobilization System. Toxins 2024, 16, 445. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, L.; Huang, Z.; Guo, Y.; Tang, Y.; Wang, Y.; Ma, Q.; Zhao, L. Combined Strategies for Improving Aflatoxin B1 Degradation Ability and Yield of a Bacillus licheniformis CotA-Laccase. Int. J. Mol. Sci. 2024, 25, 6455. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, Y.; Liu, L.; Tang, Y.; Wang, Y.; Ma, Q.; Zhao, L. Improvement of aflatoxin B1 degradation ability by Bacillus licheniformis CotA-laccase Q441A mutant. Heliyon 2023, 9, e22388. [Google Scholar] [CrossRef]
- Xiong, D.; Wen, J.; Lu, G.; Li, T.; Long, M. Isolation, purification, and characterization of a laccase-degrading aflatoxin B1 from Bacillus amyloliquefaciens B10. Toxins 2022, 14, 250. [Google Scholar] [CrossRef]
- Hao, W.; Gu, X.; Yu, X.; Zhao, Y.; Li, C.; Jia, M.; Du, X. Laccase Lac-W detoxifies aflatoxin B1 and degrades five other major mycotoxins in the absence of redox mediators. Environ. Pollut. 2023, 338, 122581. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, R.; Ng, T.B.; Lai, Y.; Yang, J.; Ye, X. A new laccase of Lac 2 from the white rot fungus Cerrena unicolor 6884 and Lac 2-mediated degradation of aflatoxin B1. Toxins 2020, 12, 476. [Google Scholar] [CrossRef]
- Liu, Y.; Mao, H.; Hu, C.; Tron, T.; Lin, J.; Wang, J.; Sun, B. Molecular docking studies and in vitro degradation of four aflatoxins (AFB1, AFB2, AFG1, and AFG2) by a recombinant laccase from Saccharomyces cerevisiae. J. Food Sci. 2020, 85, 1353–1360. [Google Scholar] [CrossRef]
- Loi, M.; Fanelli, F.; Cimmarusti, M.T.; Mirabelli, V.; Haidukowski, M.; Logrieco, A.F.; Caliandro, R.; Mule, G. In vitro single and combined mycotoxins degradation by Ery4 laccase from Pleurotus eryngii and redox mediators. Food Control 2018, 90, 401–406. [Google Scholar] [CrossRef]
- Qin, X.; Xin, Y.; Zou, J.; Su, X.; Wang, X.; Wang, Y.; Zhang, J.; Tu, T.; Yao, B.; Luo, H.; et al. Efficient Degradation of Aflatoxin B1 and Zearalenone by Laccase-like Multicopper Oxidase from Streptomyces thermocarboxydus in the Presence of Mediators. Toxins 2021, 13, 754. [Google Scholar] [CrossRef]
- Dragičević, T.; Hren, M.; Gmajnić, M.; Pelko, S.; Kungulovski, D.; Kungulovski, I.; Čvek, D.; Frece, J.; Markov, K.; Delaš, F. Biodegradation of Olive Mill Wastewater by Trichosporon cutaneum and Geotrichum candidum. Arh. Hig. Rada. Toksikol. 2010, 61, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lin, P.; Lu, Y.; Jia, R. Degradation and detoxification of aflatoxin B1 by two peroxidase enzymes from Irpex lacteus F17. Bioprocess Biosyst. Eng. 2025, 48, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Loi, M.; Renaud, J.B.; Rosini, E.; Pollegioni, L.; Vignali, E.; Haidukowski, M.; Sumarah, M.W.; Logrieco, A.F.; Mulè, G. Enzymatic transformation of aflatoxin B1 by Rh_DypB peroxidase and characterization of the reaction products. Chemosphere 2020, 250, 126296. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Su, X.; Tu, T.; Zhang, J.; Wang, X.; Wang, Y.; Wang, Y.; Bai, Y.; Yao, B.; Luo, H.; et al. Enzymatic Degradation of Multiple Major Mycotoxins by Dye-Decolorizing Peroxidase from Bacillus subtilis. Toxins 2021, 13, 429. [Google Scholar] [CrossRef]
- Xu, T.; Xie, C.; Yao, D.; Zhou, C.; Liu, J. Crystal structures of Aflatoxin-oxidase from Armillariella tabescens reveal a dual activity enzyme. Biochem. Biophys. Res. Commun. 2017, 494, 621–625. [Google Scholar] [CrossRef]
- Xu, L.; Eisa Ahmed, M.F.; Sangare, L.; Zhao, Y.; Selvaraj, J.N.; Xing, F.; Wang, Y.; Yang, H.; Liu, Y. Novel Aflatoxin-Degrading Enzyme from Bacillus shackletonii L7. Toxins 2017, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Guan, S.; Gao, X.; Ma, Q.; Lei, Y.; Bai, X.; Ji, C. Preparation, purification and characteristics of an aflatoxin degradation enzyme from Myxococcus fulvus ANSM068. J. Appl. Microbiol. 2011, 110, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Weng, L.; Dong, Y.; Zhang, L. Specificity and Enzyme Kinetics of the Quorum-quenching N-Acyl Homoserine Lactone Lactonase (AHL-lactonase). J. Biol. Chem. 2004, 279, 13645–13651. [Google Scholar] [CrossRef]
- Singh, J.; Mehta, A. The main Aflatoxin B1 degrading enzyme in Pseudomonas putida is thermostable lipase. Heliyon 2022, 8, e10809. [Google Scholar] [CrossRef]
- Bui, S.; Gil-Guerrero, S.; Van der Linden, P.; Carpentier, P.; Ceccarelli, M.; Jambrina, P.G.; Steiner, R.A. Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the α/β-hydrolase fold. Chem. Sci. 2023, 14, 10547–10560. [Google Scholar] [CrossRef]
- Afsharmanesh, H.; Perez-Garcia, A.; Zeriouh, H.; Ahmadzadeh, M.; Romero, D. Aflatoxin degradation by Bacillus subtilis UTB1 is based on production of an oxidoreductase involved in bacilysin biosynthesis. Food Control 2018, 94, 48–55. [Google Scholar] [CrossRef]
- Niermans, K.; Meyer, A.M.; Hoek-van den Hil, E.F.; van Loon, J.J.A.; van der Fels-Klerx, H.J. A systematic literature review on the effects of mycotoxin exposure on insects and on mycotoxin accumulation and biotransformation. Mycotoxin Res. 2021, 37, 279–295. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Campbell, B.C. In vitro metabolism of aflatoxin B1 by larvae of navel orangeworm, Amyelois transitella (Walker) (Insecta, Lepidoptera, Pyralidae) and codling moth, Cydia pomonella (L.) (Insecta, Lepidoptera, Tortricidae). Arch. Insect Biochem. Physiol. 2000, 45, 166–174. [Google Scholar] [CrossRef]
- Niu, G.; Wen, Z.; Rupasinghe, S.G.; Zeng, R.; Berenbaum, M.R.; Schuler, M.A. Aflatoxin B1 detoxification by CYP321A1 in Helicoverpa zea. Arch. Insect Biochem. Physiol. 2008, 69, 32–45. [Google Scholar] [CrossRef]
- Bosch, G.; Van Der Fels-Klerx, H.; de Rijk, T.C.; Oonincx, D.G. Aflatoxin B1 tolerance and accumulation in black soldier fly larvae (Hermetia illucens) and yellow mealworms (Tenebrio molitor). Toxins 2017, 9, 185. [Google Scholar] [CrossRef]
- Zhao, D.; Xie, H.; Gao, L.; Zhang, J.; Li, Y.; Mao, G.; Zhang, H.; Wang, F.; Lam, S.; Song, A. Detoxication and bioconversion of aflatoxin B1 by yellow mealworms (Tenebrio molitor): A sustainable approach for valuable larval protein production from contaminated grain. Ecotoxicol. Environ. Saf. 2022, 242, 113935. [Google Scholar] [CrossRef]
- Meijer, N.; Nijssen, R.; Bosch, M.; Boers, E.; Van der Fels-Klerx, H. Aflatoxin B1 metabolism of reared Alphitobius diaperinus in different life-stages. Insects 2022, 13, 357. [Google Scholar] [CrossRef]
- Shah, P.N.; Niermans, K.; Hoek-van den Hil, E.F.; Dicke, M.; van Loon, J.J.A. Effects of aflatoxin B1 on metabolism- and immunity-related gene expression in Hermetia illucens L. (Diptera: Stratiomyidae). Pestic. Biochem. Physiol. 2024, 202, 105944. [Google Scholar] [CrossRef] [PubMed]
- Bruno, D.; Bonelli, M.; De Filippis, F.; Di Lelio, I.; Tettamanti, G.; Casartelli, M.; Ercolini, D.; Caccia, S. The intestinal microbiota of Hermetia illucens larvae is affected by diet and shows a diverse composition in the different midgut regions. Appl. Environ. Microbiol. 2019, 85, e01864-18. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Lei, A.; Wang, M.; Sun, M.; Yang, S.; Liu, X.; Liu, L.; Chen, H. Novel tetracycline-degrading enzymes from the gut microbiota of black soldier fly: Discovery, performance, degradation pathways, mechanisms, and application potential. J. Hazard. Mater. 2025, 488, 137286. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Bonelli, M.; Bruno, D.; Sequino, G.; Montali, A.; Reguzzoni, M.; Pasolli, E.; Savy, D.; Cangemi, S.; Cozzolino, V. Plastics shape the black soldier fly larvae gut microbiome and select for biodegrading functions. Microbiome 2023, 11, 205. [Google Scholar] [CrossRef]
- Bonelli, M.; Bruno, D.; Caccia, S.; Sgambetterra, G.; Cappellozza, S.; Jucker, C.; Tettamanti, G.; Casartelli, M. Structural and Functional Characterization of Hermetia illucens Larval Midgut. Front. Physiol. 2019, 10, 204. [Google Scholar] [CrossRef]
- Zheng, S.; Li, R.; Huang, Y.; Yang, M.; Chen, W.; Mo, S.; Qi, R.; Wang, W.; Wan, D.; Yin, Y. Gut microbiome of black soldier fly larvae for efficient use and purification of organic waste: An environmentally friendly development concept. Innov. Life 2025, 3, 100134. [Google Scholar] [CrossRef]
- Amrul, N.F.; Kabir Ahmad, I.; Ahmad Basri, N.E.; Suja, F.; Abdul Jalil, N.A.; Azman, N.A. A review of organic waste treatment using black soldier fly (Hermetia illucens). Sustainability 2022, 14, 4565. [Google Scholar] [CrossRef]
- Boakye-Yiadom, K.A.; Ilari, A.; Duca, D. Greenhouse gas emissions and life cycle assessment on the black soldier fly (Hermetia illucens L.). Sustainability 2022, 14, 10456. [Google Scholar] [CrossRef]
- Purschke, B.; Scheibelberger, R.; Axmann, S.; Adler, A.; Jäger, H. Impact of substrate contamination with mycotoxins, heavy metals and pesticides on the growth performance and composition of black soldier fly larvae (Hermetia illucens) for use in the feed and food value chain. Food Addit. Contam. Part A 2017, 34, 1410–1420. [Google Scholar] [CrossRef] [PubMed]
- Meijer, N.; Stoopen, G.; van der Fels-Klerx, H.J.; van Loon, J.J.A.; Carney, J.; Bosch, G. Aflatoxin B1 Conversion by Black Soldier Fly (Hermetia illucens) Larval Enzyme Extracts. Toxins 2019, 11, 532. [Google Scholar] [CrossRef]
- Camenzuli, L.; Van Dam, R.; De Rijk, T.; Andriessen, R.; Van Schelt, J.; Van der Fels-Klerx, H.J. Tolerance and Excretion of the Mycotoxins Aflatoxin B1, Zearalenone, Deoxynivalenol, and Ochratoxin A by Alphitobius diaperinus and Hermetia illucens from Contaminated Substrates. Toxins 2018, 10, 91. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The gut microbiota of insects-diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Varotto Boccazzi, I.; Ottoboni, M.; Martin, E.; Comandatore, F.; Vallone, L.; Spranghers, T.; Eeckhout, M.; Mereghetti, V.; Pinotti, L.; Epis, S. A survey of the mycobiota associated with larvae of the black soldier fly (Hermetia illucens) reared for feed production. PLoS ONE 2017, 12, e0182533. [Google Scholar] [CrossRef]
- Klüber, P.; Müller, S.; Schmidt, J.; Zorn, H.; Rühl, M. Isolation of bacterial and fungal microbiota associated with Hermetia illucens larvae reveals novel insights into entomopathogenicity. Microorganisms 2022, 10, 319. [Google Scholar] [CrossRef] [PubMed]
- Tegtmeier, D.; Hurka, S.; Klüber, P.; Brinkrolf, K.; Heise, P.; Vilcinskas, A. Cottonseed Press Cake as a Potential Diet for Industrially Farmed Black Soldier Fly Larvae Triggers Adaptations of Their Bacterial and Fungal Gut Microbiota. Front. Microbiol. 2021, 12, 634503. [Google Scholar] [CrossRef]
- Tanga, C.M.; Waweru, J.W.; Tola, Y.H.; Onyoni, A.A.; Khamis, F.M.; Ekesi, S.; Paredes, J.C. Organic Waste Substrates Induce Important Shifts in Gut Microbiota of Black Soldier Fly (Hermetia illucens L.): Coexistence of Conserved, Variable, and Potential Pathogenic Microbes. Front. Microbiol. 2021, 12, 635881. [Google Scholar] [CrossRef]
- Auger, L.; Deschamps, M.H.; Vandenberg, G.; Derome, N. Microbiota is structured by gut regions, life stage, and diet in the Black Soldier Fly (Hermetia illucens). Front. Microbiol. 2023, 14, 1221728. [Google Scholar] [CrossRef]
- Jeon, H.; Park, S.; Choi, J.; Jeong, G.; Lee, S.; Choi, Y.; Lee, S. The intestinal bacterial community in the food waste-reducing larvae of Hermetia illucens. Curr. Microbiol. 2011, 62, 1390–1399. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Crippen, T.L.; Singh, B.; Tarone, A.M.; Dowd, S.; Yu, Z.; Wood, T.K.; Tomberlin, J.K. A survey of bacterial diversity from successive life stages of black soldier fly (Diptera: Stratiomyidae) by using 16S rDNA pyrosequencing. J. Med. Entomol. 2013, 50, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Crippen, T.L.; Holmes, L.; Singh, B.; Pimsler, M.L.; Benbow, M.E.; Tarone, A.M.; Dowd, S.; Yu, Z.; Vanlaerhoven, S.L. Bacteria mediate oviposition by the black soldier fly, Hermetia illucens (L.), (Diptera: Stratiomyidae). Sci. Rep. 2013, 3, 2563. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Ma, S.; Hu, R.; Tomberlin, J.K.; Thomashow, L.S.; Zheng, L.; Li, W.; Yu, Z.; Zhang, J. Rapidly mitigating antibiotic resistant risks in chicken manure by Hermetia illucens bioconversion with intestinal microflora. Environ. Microbiol. 2018, 20, 4051–4062. [Google Scholar] [CrossRef]
- Cai, M.; Ma, S.; Hu, R.; Tomberlin, J.K.; Yu, C.; Huang, Y.; Zhan, S.; Li, W.; Zheng, L.; Yu, Z. Systematic characterization and proposed pathway of tetracycline degradation in solid waste treatment by Hermetia illucens with intestinal microbiota. Environ. Pollut. 2018, 242, 634–642. [Google Scholar] [CrossRef]
- Jiang, C.; Jin, W.; Tao, X.; Zhang, Q.; Zhu, J.; Feng, S.; Xu, X.; Li, H.; Wang, Z.; Zhang, Z. Black soldier fly larvae (Hermetia illucens) strengthen the metabolic function of food waste biodegradation by gut microbiome. Microb. Biotechnol. 2019, 12, 528–543. [Google Scholar] [CrossRef]
- Wynants, E.; Frooninckx, L.; Crauwels, S.; Verreth, C.; De Smet, J.; Sandrock, C.; Wohlfahrt, J.; Van Schelt, J.; Depraetere, S.; Lievens, B. Assessing the microbiota of black soldier fly larvae (Hermetia illucens) reared on organic waste streams on four different locations at laboratory and large scale. Microb. Ecol. 2019, 77, 913–930. [Google Scholar] [CrossRef]
- Zhan, S.; Fang, G.; Cai, M.; Kou, Z.; Xu, J.; Cao, Y.; Bai, L.; Zhang, Y.; Jiang, Y.; Luo, X. Genomic landscape and genetic manipulation of the black soldier fly Hermetia illucens, a natural waste recycler. Cell Res. 2020, 30, 50–60. [Google Scholar] [CrossRef]
- Liu, C.; Yao, H.; Chapman, S.J.; Su, J.; Wang, C. Changes in gut bacterial communities and the incidence of antibiotic resistance genes during degradation of antibiotics by black soldier fly larvae. Environ. Int. 2020, 142, 105834. [Google Scholar] [CrossRef] [PubMed]
- Ao, Y.; Yang, C.; Wang, S.; Hu, Q.; Yi, L.; Zhang, J.; Yu, Z.; Cai, M.; Yu, C. Characteristics and nutrient function of intestinal bacterial communities in black soldier fly (Hermetia illucens L.) larvae in livestock manure conversion. Microb. Biotechnol. 2021, 14, 886–896. [Google Scholar] [CrossRef]
- Cifuentes, Y.; Glaeser, S.P.; Mvie, J.; Bartz, J.O.; Müller, A.; Gutzeit, H.O.; Vilcinskas, A.; Kämpfer, P. The gut and feed residue microbiota changing during the rearing of Hermetia illucens larvae. Antonie Van Leeuwenhoek 2020, 113, 1323–1344. [Google Scholar] [CrossRef]
- Callegari, M.; Jucker, C.; Fusi, M.; Leonardi, M.G.; Daffonchio, D.; Borin, S.; Savoldelli, S.; Crotti, E. Hydrolytic Profile of the Culturable Gut Bacterial Community Associated with Hermetia illucens. Front. Microbiol. 2020, 11, 1965. [Google Scholar] [CrossRef]
- Klammsteiner, T.; Walter, A.; Bogataj, T.; Heussler, C.D.; Stres, B.; Steiner, F.M.; Schlick-Steiner, B.C.; Arthofer, W.; Insam, H. The Core Gut Microbiome of Black Soldier Fly (Hermetia illucens) Larvae Raised on Low-Bioburden Diets. Front. Microbiol. 2020, 11, 993. [Google Scholar] [CrossRef]
- Raimondi, S.; Spampinato, G.; Macavei, L.I.; Lugli, L.; Candeliere, F.; Rossi, M.; Maistrello, L.; Amaretti, A. Effect of rearing temperature on growth and microbiota composition of Hermetia illucens. Microorganisms 2020, 8, 902. [Google Scholar] [CrossRef] [PubMed]
- Shelomi, M.; Wu, M.; Chen, S.; Huang, J.; Burke, C.G. Microbes associated with black soldier fly (Diptera: Stratiomiidae) degradation of food waste. Environ. Entomol. 2020, 49, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Wang, X.; Xu, X.; Cai, R.; Xie, S. Effects of heavy metals on the bioaccumulation, excretion and gut microbiome of black soldier fly larvae (Hermetia illucens). Ecotoxicol. Environ. Saf. 2020, 192, 110323. [Google Scholar] [CrossRef]
- Galassi, G.; Jucker, C.; Parma, P.; Lupi, D.; Crovetto, G.M.; Savoldelli, S.; Colombini, S. Impact of agro-industrial byproducts on bioconversion, chemical composition, in vitro digestibility, and microbiota of the black soldier fly (Diptera: Stratiomyidae) larvae. J. Insect Sci. 2021, 21, 8. [Google Scholar] [CrossRef]
- Tegtmeier, D.; Hurka, S.; Mihajlovic, S.; Bodenschatz, M.; Schlimbach, S.; Vilcinskas, A. Culture-independent and culture-dependent characterization of the black soldier fly gut microbiome reveals a large proportion of culturable bacteria with potential for industrial applications. Microorganisms 2021, 9, 1642. [Google Scholar] [CrossRef]
- Li, X.; Zhou, S.; Zhang, J.; Zhou, Z.; Xiong, Q. Directional changes in the intestinal bacterial community in black soldier fly (Hermetia illucens) larvae. Animals 2021, 11, 3475. [Google Scholar] [CrossRef] [PubMed]
- Gorrens, E.; Van Moll, L.; Frooninckx, L.; De Smet, J.; Van Campenhout, L. Isolation and Identification of Dominant Bacteria from Black Soldier Fly Larvae (Hermetia illucens) Envisaging Practical Applications. Front. Microbiol. 2021, 12, 665546. [Google Scholar] [CrossRef]
- Klammsteiner, T.; Walter, A.; Bogataj, T.; Heussler, C.D.; Stres, B.; Steiner, F.M.; Schlick-Steiner, B.C.; Insam, H. Impact of Processed Food (Canteen and Oil Wastes) on the Development of Black Soldier Fly (Hermetia illucens) Larvae and Their Gut Microbiome Functions. Front. Microbiol. 2021, 12, 619112. [Google Scholar] [CrossRef]
- Osimani, A.; Ferrocino, I.; Corvaglia, M.R.; Roncolini, A.; Milanović, V.; Garofalo, C.; Aquilanti, L.; Riolo, P.; Ruschioni, S.; Jamshidi, E. Microbial dynamics in rearing trials of Hermetia illucens larvae fed coffee silverskin and microalgae. Food Res. Int. 2021, 140, 110028. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, J.; Jiang, L.; Yu, X.; Zhu, H.; Zhang, J.; Feng, Z.; Zhang, X.; Chen, G.; Zhang, Z. Black soldier fly (Hermetia illucens) larvae significantly change the microbial community in chicken manure. Curr. Microbiol. 2021, 78, 303–315. [Google Scholar] [CrossRef]
- Shumo, M.; Khamis, F.M.; Ombura, F.L.; Tanga, C.M.; Fiaboe, K.K.; Subramanian, S.; Ekesi, S.; Schlüter, O.K.; van Huis, A.; Borgemeister, C. A molecular survey of bacterial species in the guts of black soldier fly larvae (Hermetia illucens) reared on two urban organic waste streams in Kenya. Front. Microbiol. 2021, 12, 687103. [Google Scholar] [CrossRef]
- Cifuentes, Y.; Vilcinskas, A.; Kämpfer, P.; Glaeser, S.P. Isolation of Hermetia illucens larvae core gut microbiota by two different cultivation strategies. Antonie Van Leeuwenhoek 2022, 115, 821–837. [Google Scholar] [CrossRef]
- Pei, Y.; Zhao, S.; Chen, X.; Zhang, J.; Ni, H.; Sun, M.; Lin, H.; Liu, X.; Chen, H.; Yang, S. Bacillus velezensis EEAM 10B Strengthens Nutrient Metabolic Process in Black Soldier Fly Larvae (Hermetia illucens) via Changing Gut Microbiome and Metabolic Pathways. Front. Nutr. 2022, 9, 880488. [Google Scholar] [CrossRef]
- Yang, F.; Tomberlin, J.K.; Jordan, H.R. Starvation Alters Gut Microbiome in Black Soldier Fly (Diptera: Stratiomyidae) Larvae. Front. Microbiol. 2021, 12, 601253. [Google Scholar] [CrossRef]
- Greenwood, M.P.; Hull, K.L.; Brink-Hull, M.; Lloyd, M.; Rhode, C. Feed and host genetics drive microbiome diversity with resultant consequences for production traits in mass-reared black soldier fly (Hermetia illucens) larvae. Insects 2021, 12, 1082. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Ma, Y.; Tang, B.; Zeng, R.; Zhou, Q. Intestinal microbiota and functional characteristics of black soldier fly larvae (Hermetia illucens). Ann. Microbiol. 2021, 71, 13. [Google Scholar] [CrossRef]
- Soomro, A.; Cai, M.; Laghari, Z.; Zheng, L.; Ur Rehman, K.; Xiao, X.; Hu, S.; Yu, Z.; Zhang, J. Impact of heat treatment on microbiota of black soldier fly larvae reared on soybean curd residues. J. Insects Food Feed 2021, 7, 329–344. [Google Scholar] [CrossRef]
- Gorrens, E.; De Smet, J.; Vandeweyer, D.; Bossaert, S.; Crauwels, S.; Lievens, B.; Van Campenhout, L. The bacterial communities of black soldier fly larvae (Hermetia illucens) during consecutive, industrial rearing cycles. J. Insects Food Feed 2022, 8, 1061–1076. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, X.; Elhag, O.; Cai, M.; Zheng, L.; Huang, F.; Jordan, H.R.; Tomberlin, J.K.; Sze, S.H.; Yu, Z. Hermetia illucens L. larvae–associated intestinal microbes reduce the transmission risk of zoonotic pathogens in pig manure. Microb. Biotechnol. 2022, 15, 2631–2644. [Google Scholar] [CrossRef]
- Vitenberg, T.; Opatovsky, I. Assessing fungal diversity and abundance in the black soldier fly and its environment. J. Insect Sci. 2022, 22, 3. [Google Scholar] [CrossRef]
- Querejeta, M.; Hervé, V.; Perdereau, E.; Marchal, L.; Herniou, E.A.; Boyer, S.; Giron, D. Changes in Bacterial Community Structure Across the Different Life Stages of Black Soldier Fly (Hermetia illucens). Microb. Ecol. 2023, 86, 1254–1267. [Google Scholar] [CrossRef]
- Wang, X.; Tian, X.; Liu, Z.; Liu, Z.; Shang, S.; Li, H.; Qu, J.; Chen, P. Rearing of black soldier fly larvae with corn straw and the assistance of gut microorganisms in digesting corn straw. Insects 2024, 15, 734. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, J.; Zhu, F.; Fan, M.; Zheng, J.; Cai, M.; Zheng, L.; Huang, F.; Yu, Z.; Zhang, J. Enhanced protein degradation by black soldier fly larvae (Hermetia illucens L.) and its gut microbes. Front. Microbiol. 2023, 13, 1095025. [Google Scholar] [CrossRef] [PubMed]
- Ruan, L.; Ye, K.; Wang, Z.; Xiong, A.; Qiao, R.; Zhang, J.; Huang, Z.; Cai, M.; Yu, C. Characteristics of gut bacterial microbiota of black soldier fly (Diptera: Stratiomyidae) larvae effected by typical antibiotics. Ecotoxicol. Environ. Saf. 2024, 270, 115861. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Liu, C.; Li, Y.; Qin, Y.; Wang, C.; Wang, T. The underlying mechanisms of oxytetracycline degradation mediated by gut microbial proteins and metabolites in Hermetia illucens. Sci. Total Environ. 2024, 946, 174224. [Google Scholar] [CrossRef]
- Li, X.; Mei, C.; Luo, X.; Wulamu, D.; Zhan, S.; Huang, Y.; Yang, H. Dynamics of the intestinal bacterial community in black soldier fly larval guts and its influence on insect growth and development. Insect Sci. 2023, 30, 947–963. [Google Scholar] [CrossRef]
- Ma, C.; Huang, Z.; Feng, X.; Memon, F.U.; Cui, Y.; Duan, X.; Zhu, J.; Tettamanti, G.; Hu, W.; Tian, L. Selective breeding of cold-tolerant black soldier fly (Hermetia illucens) larvae: Gut microbial shifts and transcriptional patterns. Waste Manag. 2024, 177, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Vandeweyer, D.; Bruno, D.; Bonelli, M.; IJdema, F.; Lievens, B.; Crauwels, S.; Casartelli, M.; Tettamanti, G.; De Smet, J. Bacterial biota composition in gut regions of black soldier fly larvae reared on industrial residual streams: Revealing community dynamics along its intestinal tract. Front. Microbiol. 2023, 14, 1276187. [Google Scholar] [CrossRef]
- Xia, J.; Ge, C.; Yao, H. Antimicrobial peptides from black soldier fly (Hermetia illucens) as potential antimicrobial factors representing an alternative to antibiotics in livestock farming. Animals 2021, 11, 1937. [Google Scholar] [CrossRef]
- Almazroo, O.A.; Miah, M.K.; Venkataramanan, R. Drug Metabolism in the Liver. Clin. Liver Dis. 2017, 21, 1–20. [Google Scholar] [CrossRef]
- Benkerroum, N. Retrospective and prospective look at aflatoxin research and development from a practical standpoint. Int. J. Environ. Res. Public Health 2019, 16, 3633. [Google Scholar] [CrossRef]
- Jiang, S.; Li, H.; Zhang, L.; Mu, W.; Zhang, Y.; Chen, T.; Wu, J.; Tang, H.; Zheng, S.; Liu, Y.; et al. Generic Diagramming Platform (GDP): A comprehensive database of high-quality biomedical graphics. Nucleic Acids Res. 2025, 53, D1670–D1676. [Google Scholar] [CrossRef] [PubMed]
- Farzaneh, M.; Shi, Z.Q.; Ahmadzadeh, M.; Hu, L.B.; Ghassempour, A. Inhibition of the Aspergillus flavus growth and aflatoxin B1 contamination on pistachio nut by fengycin and surfactin-producing Bacillus subtilis UTBSP1. Plant Pathol. J. 2016, 32, 209–215. [Google Scholar] [CrossRef]
- Mukherjee, A.K.; Bordoloi, N.K. Biodegradation of benzene, toluene, and xylene (BTX) in liquid culture and in soil by Bacillus subtilis and Pseudomonas aeruginosa strains and a formulated bacterial consortium. Environ. Sci. Pollut. Res. 2012, 19, 3380–3388. [Google Scholar] [CrossRef]
- Gai, Z.; Zhang, Z.; Wang, X.; Tao, F.; Tang, H.; Xu, P. Genome Sequence of Pseudomonas aeruginosa DQ8, an Efficient Degrader of n-Alkanes and Polycyclic Aromatic Hydrocarbons. J. Bacteriol. 2012, 194, 6304–6305. [Google Scholar] [CrossRef] [PubMed]
- Sugimori, D.; Utsue, T. A study of the efficiency of edible oils degraded in alkaline conditions by Pseudomonas aeruginosa SS-219 and Acinetobacter sp. SS-192 bacteria isolated from Japanese soil. World J. Microbiol. Biotechnol. 2012, 28, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Beranová, J.; Mansilla, M.C.; Mendoza, D.d.; Elhottová, D.; Konopásek, I. Differences in Cold Adaptation of Bacillus subtilis under Anaerobic and Aerobic Conditions. J. Bacteriol. 2010, 192, 4164–4171. [Google Scholar] [CrossRef]
- Wu, M.; Guina, T.; Brittnacher, M.; Nguyen, H.; Eng, J.; Miller, S.I. The Pseudomonas aeruginosa Proteome during Anaerobic Growth. J. Bacteriol. 2005, 187, 8185–8190. [Google Scholar] [CrossRef]
- Tseng, C.P.; Albrecht, J.; Gunsalus, R.P. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J. Bacteriol. 1996, 178, 1094–1098. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Goel, S.S.; Siebner, H.; Ronen, Z.; Ramanathan, G. Transformation of 2, 4, 6-trinitrotoluene by Stenotrophomonas strain SG1 under aerobic and anaerobic conditions. Chemosphere 2023, 311, 137085. [Google Scholar] [CrossRef]
- Al-Fatlawi, A.H.; Raheem, S.A. Inactivation of Enterococcus faecalis in drinking water using silver nanoparticles embedded paper. Indian J. Forensic Med. Toxicol. 2020, 14, 1117–1121. [Google Scholar] [CrossRef]
- Song, J.; Sun, D.; Zhao, L.; Jiang, H.; Zhu, C. High Power Generation by a Strain of Facultative Anaerobe in Double-Chamber Microbial Fuel Cell. Adv. Mater. Res. 2012, 347, 2616–2621. [Google Scholar] [CrossRef]
- Singh, S.; Singh, A.K.; Singh, S.K.; Yadav, V.B.; Kumar, A.; Nath, G. Current update on the antibiotic resistance profile and the emergence of colistin resistance in Enterobacter isolates from hospital-acquired infections. Microbe 2025, 8, 100432. [Google Scholar] [CrossRef]
- Kong, D.; Park, J.; Lee, C.; Khandelwal, H.; Kim, M.; Sakuntala, M.; Kim, T.; Jeon, B.; Kim, J.; Kim, C. Reprint of “A newly isolated Klebsiella variicola JYP01 strain with iron-interaction capability for energy-efficient production of 1,3-propanediol”. J. Taiwan Inst. Chem. Eng. 2025, 177, 106443. [Google Scholar] [CrossRef]
- Tarrand, J.J.; Han, X.; Kontoyiannis, D.P.; May, G.S. Aspergillus Hyphae in Infected Tissue: Evidence of Physiologic Adaptation and Effect on Culture Recovery. J. Clin. Microbiol. 2005, 43, 382–386. [Google Scholar] [CrossRef]
- Yang, S.; Liu, Q.; Shen, Z.; Wang, H.; He, L. Molecular Epidemiology of Myroides odoratimimus in Nosocomial Catheter-Related Infection at a General Hospital in China. Infect. Drug Resist. 2020, 13, 1981–1993. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Song, Y.; Chen, F.; Zheng, S.; Wang, G. Lysinibacillus manganicus sp. nov., isolated from manganese mining soil. Int. J. Syst. Evol. Microbiol. 2013, 63, 3568–3573. [Google Scholar] [CrossRef]
- Egorova, D.O.; Demakov, V.A.; Plotnikova, E.G. Bioaugmentation of a polychlorobiphenyl contaminated soil with two aerobic bacterial strains. J. Hazard. Mater. 2013, 261, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Chen, J.; Nepovimova, E.; Long, M.; Wu, W.; Kuca, K. Aflatoxin detoxification using microorganisms and enzymes. Toxins 2021, 13, 46. [Google Scholar] [CrossRef] [PubMed]
- Faucet-Marquis, V.; Joannis-Cassan, C.; Hadjeba-Medjdoub, K.; Ballet, N.; Pfohl-Leszkowicz, A. Development of an in vitro method for the prediction of mycotoxin binding on yeast-based products: Case of aflatoxin B 1, zearalenone and ochratoxin A. Appl. Microbiol. Biotechnol. 2014, 98, 7583–7596. [Google Scholar] [CrossRef] [PubMed]
- Repečkienė, J.; Levinskaitė, L.; Paškevičius, A.; Raudonienė, V. Toxin-producing fungi on feed grains and application of yeasts for their detoxification. Pol. J. Vet. Sci. 2013, 16, 391–393. [Google Scholar] [CrossRef]
- Marlida, Y.; Harnentis; Anggraini, L.; Ardani, L.; Huda, N. Yeast Probiotic Isolated from Fish Fermented (Budu) with Promising AFB1 Biodetoxify. Int. J. Vet. Sci. 2025, 14, 310–315. [Google Scholar] [CrossRef]
- Bzducha Wróbel, A.; Bryła, M.; Gientka, I.; Błażejak, S.; Janowicz, M. Candida utilis ATCC 9950 Cell Walls and β(1,3)/(1,6)-Glucan Preparations Produced Using Agro-Waste as a Mycotoxins Trap. Toxins 2019, 11, 192. [Google Scholar] [CrossRef]
- Sidari, R.; Tofalo, R. Dual Role of Yeasts and Filamentous Fungi in Fermented Sausages. Foods 2024, 13, 2547. [Google Scholar] [CrossRef]
- Rodríguez-Rivera, V.; Estrada-García, J.; Sales-Pérez, R.E.; Hernández-Martínez, J.M.; Méndez-Contreras, J.M. Valorization of Agro-industrial Waste to Produce a Probiotic-bio-stimulant Through the Anaerobic Co-fermentation Process with Lactobacillus casei: A Circular Economy Approach in Vulnerable Communities of Mexico. Water Air Soil Pollut. 2025, 236, 925. [Google Scholar] [CrossRef]
- Diez, A.M.; Björkroth, J.; Jaime, I.; Rovira, J. Microbial, sensory and volatile changes during the anaerobic cold storage of morcilla de Burgos previously inoculated with Weissella viridescens and Leuconostoc mesenteroides. Int. J. Food Microbiol. 2009, 131, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, Y.; Zhao, Y.; Wang, Z.; Miao, X.; Huo, W.; Chen, L.; Liu, Q.; Wang, C.; Guo, G. Enhancing Alfalfa Hemicellulose Degradation by Anaerobic Bioprocessing with Engineered Xylanase-Secreting Pediococcus pentosaceus. J. Agric. Food. Chem. 2025, 73, 22563–22576. [Google Scholar] [CrossRef]
- Sumi, A.; Morimura, S.; Shigematsu, T.; Takenouchi, H.; Kida, K. Anaerobic Digestion of Wastewater Including High Concentration of Yeast, Pichia pastoris. Jpn. J. Water Treat. Biol. 2005, 41, 213–218. [Google Scholar] [CrossRef][Green Version]
- Wenda, J.M.; Drzewicka, K.; Mulica, P.; Tetaud, E.; di Rago, J.P.; Golik, P.; Łabędzka-Dmoch, K. Candida albicans PPR proteins are required for the expression of respiratory Complex I subunits. Genetics 2024, 228, iyae124. [Google Scholar] [CrossRef]
- Huang, L.; Duan, C.; Zhao, Y.; Gao, L.; Niu, C.; Xu, J.; Li, S. Reduction of aflatoxin B1 toxicity by Lactobacillus plantarum C88: A potential probiotic strain isolated from Chinese traditional fermented food “tofu”. PLoS ONE 2017, 12, e0170109. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Qin, X.; Tang, Y.; Ma, Q.; Zhang, J.; Zhao, L. CotA laccase, a novel aflatoxin oxidase from Bacillus licheniformis, transforms aflatoxin B1 to aflatoxin Q1 and epi-aflatoxin Q1. Food Chem. 2020, 325, 126877. [Google Scholar] [CrossRef] [PubMed]
- Diaz, G.J.; Murcia, H.W. Biotransformation of Aflatoxin B1 and Its Relationship with the Differential Toxicological Response to Aflatoxin in Commercial Poultry Species; INTECH Open Access Publisher: London, UK, 2011. [Google Scholar]
- Gerdemann, A.; Cramer, B.; Degen, G.H.; Veerkamp, J.; Günther, G.; Albrecht, W.; Behrens, M.; Esselen, M.; Ghallab, A.; Hengstler, J.G. Comparative metabolism of aflatoxin B1 in mouse, rat and human primary hepatocytes using HPLC–MS/MS. Arch. Toxicol. 2023, 97, 3179–3196. [Google Scholar] [CrossRef]
- Nakazato, M.; Morozumi, S.; Saito, K.; Fujinuma, K.; Nishima, T.; Kasai, N. Interconversion of aflatoxin B1 and aflatoxicol by several fungi. Appl. Environ. Microbiol. 1990, 56, 1465–1470. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Wang, L.; Liu, X.; Selvaraj, J.N.; Wang, Y.; Zhao, Y.; Liu, Y. Aflatoxin B1 inhibition in Aspergillus flavus by Aspergillus niger through down-regulating expression of major biosynthetic genes and AFB1 degradation by atoxigenic A. flavus. Int. J. Food Microbiol. 2017, 256, 1–10. [Google Scholar] [CrossRef]
- Suresh, G.; Cabezudo, I.; Pulicharla, R.; Cuprys, A.; Rouissi, T.; Brar, S.K. Biodegradation of aflatoxin B1 with cell-free extracts of Trametes versicolor and Bacillus subtilis. Res. Vet. Sci. 2020, 133, 85–91. [Google Scholar] [CrossRef]
- Kumar, V.; Bahuguna, A.; Lee, J.; Sood, A.; Han, S.; Chun, H.; Kim, M. Degradation mechanism of aflatoxin B1 and aflatoxin G1 by salt tolerant Bacillus albus YUN5 isolated from ‘doenjang’, a traditional Korean food. Food Res. Int. 2023, 165, 112479. [Google Scholar] [CrossRef]
- Van Raamsdonk, L.W.D.; Van der Fels-Klerx, H.J.; De Jong, J. New feed ingredients: The insect opportunity. Food Addit. Contam. Part A 2017, 34, 1384–1397. [Google Scholar] [CrossRef]
- Pinotti, L.; Ottoboni, M. Substrate as insect feed for bio-mass production. J. Insects Food Feed 2021, 7, 585–596. [Google Scholar] [CrossRef]
- Wang, Y.; Shelomi, M. Review of Black Soldier Fly (Hermetia illucens) as Animal Feed and Human Food. Foods 2017, 6, 91. [Google Scholar] [CrossRef] [PubMed]
- Committee, E.S. Risk profile related to production and consumption of insects as food and feed. EFSA J. 2015, 13, 4257. [Google Scholar] [CrossRef]
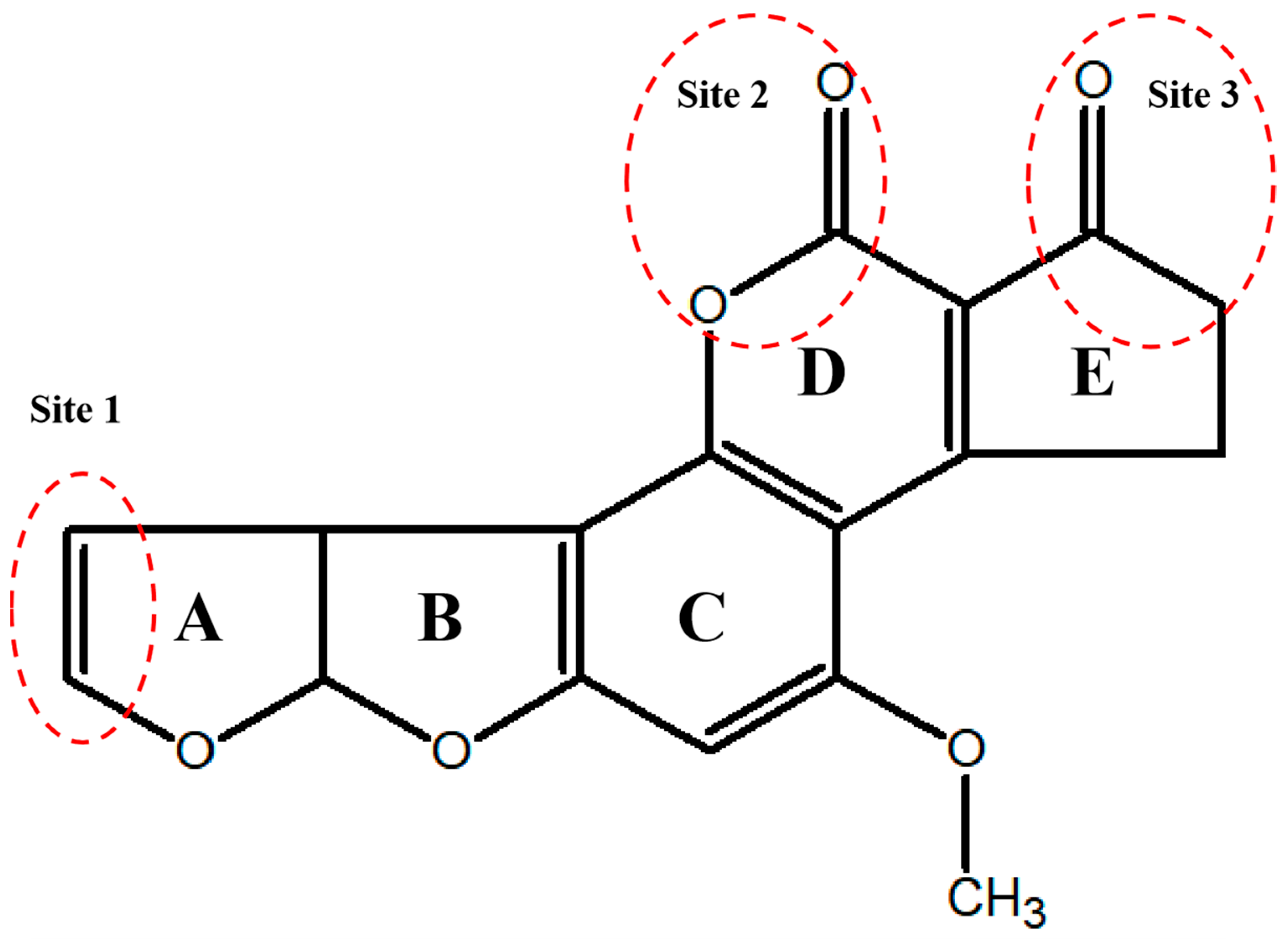

| Class | Source | Name | AFB1 Concentration | Incubation Period | Reduction Efficacy (%) | References |
|---|---|---|---|---|---|---|
| Bacillus | Maize Grains | Bacillus sp. TUBF1 | 10 μg/mL | 48 h/72 h | 81.50/100.00 | [28] |
| Thua-nao | B. licheniformis, B. subtilis | 5 mg/L | 7 d | 74.00/85.00 | [25] | |
| / | B. licheniformis CFR1 | 500 μg/kg | 72 h | 94.70 | [29] | |
| Pistachio nuts | B. subtilis UTBSP1 | 2 μg/kg | 5 d | 95.00 | [30] | |
| Hog deer feces and farm soil | Bacillus sp. | 100 μg/kg | 72 h | 77.80–80.93 | [31] | |
| Plant leaf | B. aryabhattai | 2 μg/mL | 72 h | 82.92 | [32] | |
| Pond mud and soil | Bacillus strains (11) | 500 ng/mL | 48 h | 27.78–79.78 | [24] | |
| Rotten feed | B. subtilis WJ6 | 5 μg/mL | 48 h | 81.57 | [33] | |
| / | B. halotolerans DDC-4 | 1 μg/mL | 72 h | 76.30 | [34] | |
| / | B. subtilis | 40 μg/L | 24 h | 38.38 | [35] | |
| Polygalae | B. megaterium SX1-1 | 0.1 ng/mL | 72 h | 97.45 | [36] | |
| Qinghai–Tibet Plateau | B. amyloliquefaciens YUAD7 | 10 μg/mL | 72 h | 91.70 | [37] | |
| Sitophilus oryzae gut | B. subtilis RWBG1, B. oceanisediminis RWGB2, B. firmus RWGB3 | 1 μg/kg | 48 h | 63.60–84.20 | [38] | |
| Pseudomonas | Gold mine aquifer | P. anguilliseptica VGF1 | 5 μg/kg | 48 h | 51.70 | [39] |
| P. fluorescens | 47.70 | |||||
| Peanut-growing soils | P. knackmussii AD02 | 100 ng/mL | 24 h | 90.00 | [40] | |
| / | P. putida MTCC 1274 and 2445 | 0.2 μg/mL | 24 h | 90.00 | [41] | |
| Sitophilus oryzae gut | P. aeruginosa RWGB4 | 5 μg/kg | 48 h | 48.90 | [38] | |
| Farm soils, maize and rice | P. aeruginosa N17-1 | 100 μg/kg | 72 h | 82.80 | [42] | |
| Hydrocarbon-contaminated sites | Pseudomonas sp. (6) | 2 μg/mL | 72 h | 80.14–97.61 | [43] | |
| Stenotrophomonas | South American tapir feces | Stenotrophomonas maltophilia 35-3 | 100 μg/kg | 72 h | 82.50 | [31] |
| / | Stenotrophomonas sp. NMO-3 | 100 μg/kg | 72 h | 85.70 | [44] | |
| / | Stenotrophomonas sp. CW117 | 45 μg/L | 24 h | 100.00 | [45] | |
| Black soldier fly larval gut | Stenotrophomonas acidaminiphila A2 | 0.1 μg/mL | 48 h | 94.00 | [46] | |
| Rhodococcus | Ostrich feces | Rhodococcus sp. | 100 μg/kg | 72 h | 73.92 | [31] |
| / | R. erythropolis ATTC 4277 | 20 μg/mL | 24 h | 96.00 | [47] | |
| Polycyclic aromatic hydrocarbons contaminated soils | R. erythropolis | 1.75 mg/kg | 72 h | 66.80 | [48] | |
| Hydrocarbon-contaminated sites | Rhodococcus sp. (16) | 2 μg/mL | 72 h | 20.79–99.98 | [43] | |
| Oil-contaminated soil and Natural soil | Rhodococcus sp. (32) | 2 mg/kg | 72 h | 20.00–100.00 | [49] | |
| Soil | R. pyridinivorans 4-4 | 0.1 μg/mL | 24 h | 84.90 | [50] | |
| / | R. turbidus PD630 | 0.4 μg/mL | 72 h | 93.04 | [51] | |
| / | Rhodococcus Strains (42) | 3 μg/mL | 3 d | 17.00–100.00 | [52] | |
| Polycyclic aromatic hydrocarbons contaminated soils | R. erythropolis DSM 14303 | 1.75 mg/kg | 72 h | 94.00–97.00 | [53] | |
| Lactic acid bacteria | / | Lactobacillus plantarum PTCC 1058 | 240 mg/kg | 4–7 d | 77.00 | [54] |
| / | Lactobacillus casein | 40 μg/L | 24 h | 26.06 | [35] | |
| Fermented foods | Lactobacillus plantarum | 150 μg/L | 24 h | 89.50 | [55] | |
| / | Levilactobacillus brevis (2), Lactobacillus helveticus (9), Lactoplantibacillus plantarum (4), Leuconostoc sp. (4), Pediococcus claussenii, Weissella sp. (9) | 1 μg/mL | 1.5 h | Binding: 16.10–40.90 (viable)/29.60–65.70 (non-viable) | [56] | |
| Grassland soil | Enterococcus faecium HB2-2 | 105.1 µg/kg | 96 h | 82.90 | [57] | |
| Other Bacterial Genera | Polycyclic aromatic hydrocarbon-contaminated soils | Mycobacterium fluoranthenivorans sp. nov. DSM44556T | 1.75 mg/kg | 24 h | ~100.00 | [53] |
| Contaminated soil of a former coal gas plant | Mycobacterium fluoranthenivorans FA4T | 2.5 mg/kg | 72 h | Leaving no detectable AFB1 | [58] | |
| Soil | Streptomyces (59) | 2 μg/mL | 5 d | 43.00–94.00 | [59] | |
| / | Str. lividans TK 24, Str. aureofaciens ATCC 10762 | 20 μg/mL | 24 h | 88.00/86.00 | [47] | |
| / | Streptomyces sp. | 1 mg/L | 5 d | 88.40 | [60] | |
| Hydrocarbon-contaminated sites | Streptomyces sp. (2), Arthrobacter protophormiae, Microbacterium sp. (2), Pseudoxanthomonas sp. (2), Chryseobacterium sp. (2) | 2 μg/mL | 72 h | 56.88–80.22 | [43] | |
| Gold mine aquifer | Lysinibacillus fusisormis, Sporosarcina sp., Staphylococcus warneri | 2.5 μg/mL | 48 h | 46.90–61.30 | [61] | |
| / | Staphylococcus sp. VGF2 | 5 μg/kg | 48 h | 56.80 | [39] | |
| / | Flavobacterium aurantiacum NRRL B-184 | 10 μg/mL | 48 h | 81.10 | [62] | |
| Corn-planted soil | Brevundimonas sp. LF-1, Brevundimonas sp. (2), Brachybacterium sp., Klebsiella sp., Enterobacter sp., Cellulosimicrobium sp. | 2 mg/L | 72 h | 86.90 | [63] | |
| Animal feces, farm soil | 100 μg/kg | 72 h | 73.75–78.10 | [31] | ||
| Chicken Cecum | Escherichia coli CG1061 | 2.5 μg/mL | 72 h | 93.70 | [64] | |
| / | Myroides odoratimimus Strain 3J2MO | 100 μg/kg | 48 h | 93.82 | [65] | |
| Mixed strains | / | Bacillus subtilis, Lactobacillus casein, Candida utilis | 40 μg/L | 24 h | 45.49 | [35] |
| / | Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus | 10.5 μg/kg | 3 d | 100.00 | [66] | |
| Fungi | Fermented soybean | Aspergillus niger FS10 | 1 μg/mL | 72 h | 98.65 | [67] |
| / | Aspergillus flavus | 5 mg/kg | 4 d | 0–84.40 | [68] | |
| / | Candida utilis | 40 μg/L | 24 h | 21.08 | [35] | |
| Bovine forage | Saccharomyces cerevisiae (3) | 1261 μg/mL | 48 h | 20.00–55.00 (binding) | [69] | |
| Soil, rotten wood, olive, et al. | Trichoderma sp. (65) | 50 ng/kg | 7 d | 17.80–100.00 | [70] | |
| / | Agrocybe cylindracea GC-Ac2 | 100 ng/mL | 37.9 h | 96.00 | [71] | |
| / | Pleurotus ostreatus | 2500 ng/g | 6 w | >80.00 | [72] |
| Enzyme | Producing Organism | Optimal Conditions | References | |
|---|---|---|---|---|
| Laccase | BsCotA | Bacillus subtilis | pH 7.0, 70 °C, aerobic | [77,78] |
| CotA-Laccase | Bacillus licheniformis | pH 2.5–4.5, 80–90 °C, aerobic | [79,80] | |
| B10 laccase | Bacillus amyloliquefaciens B10 | pH 6.0–8.0, 40 °C, aerobic | [81] | |
| Lac-W | Weizmannia coagulans 36D1 | pH 9.0, 30 °C, aerobic | [82] | |
| Lac 2 | Cerrena unicolor 6884 | pH 7.0, 45–55 °C, aerobic | [83] | |
| C30 laccase | Saccharomyces cerevisiae | pH 5.7, 30 °C, aerobic | [84] | |
| Ery4 laccase | Pleurotus eryngii | aerobic | [85] | |
| StMCO | Streptomyces thermocarboxydus | pH 4.0, aerobic | [86] | |
| Peroxidase | IlMnP1,2,4,5,6, PcMnP1, CsMnP, and NfMnP | Irpex lacteus, Phanerochaete chrysosporium, Ceriporiopsis subvermispora, and Nematoloma frowardii | pH 3.0–4.5, 25–37 °C, they do not rely on O2 but utilize H2O2 as the electron acceptor. | [75,87] |
| MnP | Phanerochaete sordida YK-624 | [74] | ||
| Il-MnP1, Il-DyP4 | Irpex lacteus F17 | [88] | ||
| Rh_DypB | Rhodococcus jostii | [89] | ||
| BsDyP | Bacillus subtilis SCK6 | [90] | ||
| F420H2-dependent reductases | FDR-A and FDR-B | Mycobacterium smegmatis mc2155 | Not rely on O2 but utilize NAD(P)H to provide the reduction equivalent. | [76] |
| Others | Aflatoxin Oxidase | Armillariella tabescens | aerobic | [91] |
| Bacillus aflatoxin-degrading enzyme | Bacillus shackletonii L7 | pH 8.0, 70 °C, aerobic | [92] | |
| Myxobacteria aflatoxin degradation enzyme | Myxococcus fulvus ANSM068 | pH 6.0, 35 °C, aerobic | [93] | |
| N-acyl-homoserine lactonase | Bacillus sp. | pH 4.7, 37 °C, they do not rely on O2. | [24,94] | |
| Thermostable lipase | Pseudomonas putida | 50–70 °C, they do not rely on O2. | [95] | |
| α/β hydrolase | Bacillus halotolerans | Do not rely on O2. | [34,96] | |
| Aldo/Keto reductase | ||||
| Bacilysin bio-synthesis oxidoreductase | Bacillus subtilis | aerobic | [97] | |
| Substrates | Dominant Phyla | Dominant Genera | References |
|---|---|---|---|
| Food waste, calf forage, cooked rice | Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Proteobacteria | Citrobacter, Enterococcus, Klebsiella, Leminorella, Morganella, and Providencia | [121] |
| Gainesville diet | Acidobacteria, Verrucomicrobia, Firmicutes, Actinobacteria, Proteobacteria, and Bacteroidetes | Providencia, Bacteroides, Sphyngobacterium, Dysgonomonas, and Sanguibacter | [122] |
| Gainesville diet | Firmicutes, Actinobacteria, Bacteroidetes, and Proteobacteria | Bacillus, Cellulomonas, Empedobacter, Enterobacter, Gordonia, Kurthia, Microbacterium, and Micrococcus | [123] |
| Chicken feed and vegetable waste | / | Debaryomyces, Rhodotorula, Pichia, Geotrichum, and Trichosporon | [116] |
| Chicken manure | Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria | Providencia, Enterococcus, Morganella, and Dysgonomonas | [124] |
| Wheat bran containing tetracycline | Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, and Fusobacteria | Bacteria: Flavisolibacter, Proteus, Klebsiella, Actinomyces, Globicatella, Providencia, Enterococcus, and Ignatzschineria Fungi: Entyloma, Lysurus, and Trichophyton | [125] |
| Mixture of vegetables and fish meal | Actinobacteria, Firmicutes, Bacteroidetes, and Proteobacteria | Dysgonomonas, Providencia, Blautia, Shingobacterium, Morganella, and Bacillus | [105] |
| Raw food waste | Firmicutes, Bacteroidetes, and Proteobacteria | Bacillus, Lactobacillus, Dysgonomonas, Enterococcus, and Providencia | [126] |
| Fruit/vegetable waste, manure, wheat bran, et al. | Proteobacteria and Firmicutes | Morganella, Bacillus, Enterococcus, Providencia, and Lactobacillus | [127] |
| Food waste and manure | Firmicutes, Bacteroidetes, and Proteobacteria | / | [128] |
| Soya meal supplemented with oxytetracycline | Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria | Enterococcus, Ignatzschineria, Providencia, and Morganella | [129] |
| Swine/chicken manure | Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria | Enterococcus, Providencia, Morganella, Klebsiella, Ignatzschineria, and Clostridium | [130] |
| Commercial chicken feed | Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria | Morganella, Klebsiella, Providencia, Enterobacter, Enterococcus, Bacillus, uncultured Lachnospiraceae, Actinomyces, and Dysgonomonas | [131] |
| Mixture of wheat germ, alfalfa and corn flour | Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes | Providencia, Morganella, Klebsiella, Escherichia, Acinetobacter, Stenotrophomonas, Pseudomonas, and Enterococcus | [132] |
| Chicken feed, grass, vegetables | Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria | Actinomyces, Dysgonomonas, Enterococcus, and unclassified Actinomycetales | [133] |
| Mixture of mill waste, wheat bran, and alfa flour | Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria | Providencia, Klebsiella, Bacillus, Morganella, Alcaligenes, Bordetella, and Kerstersia | [134] |
| Soy pulp and cafeteria waste | Bacteroidetes and Firmicutes | Bacillus, Citrobacter, Dysgonomonas, Porphyromonas and Parabacteroides | [135] |
| Wheat bran exposed to heavy metals (Cu and Cd) | Bacteroidetes, Actinobacteria, Proteobacteria, and Firmicutes | Salana, Parabacteroidetes, and Campylobacter | [136] |
| Hen diet, okara, maize distillers with soluble, brewer’s grains | Bacteroidetes, Actinobacteria, Proteobacteria, and Firmicutes | Providencia, Morganella, and Klebsiella | [137] |
| Chicken feed, cottonseed press cake | Proteobacteria and Firmicutes | Bacteria: Enterobacteriaceae, Pseudomonas, Curtobacterium, Bacillus, Enterococcaceae, and Actinomycetaceae Fungi: Trichosporon, Cladosporium, Diutina, Aspergillus, Xeromyces, and Acaulium | [118] |
| Chicken feed | Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria | Morganella, Enterococcus, Proteus, Providencia, Actinomyces, Lachnospiraceae, Enterobacteriaceae, Klebsiella, Escherichia-Shigella, and Citrobacter | [138] |
| Food waste | Proteobacteria, Firmicutes, and Bacteroidetes | Ignatzschineria, Providencia, Proteus, Klebsiella, and Vagococcus | [139] |
| Chicken feed; fiber-rich ingredients | Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria | Enterococcus, Escherichia, Klebsiella, Providencia, Enterobacter, and Morganella | [140] |
| Chicken feed; food waste; oil waste | Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria | Morganella, Providencia, Dysgonomonas, Lactobacillus, and Enterobacteriaceae | [141] |
| Mixtures of coffee byproducts and microalgae | Firmicutes and Proteobacteria | Morganella, Paenibacillus, Lysinibacillus, Brevundimonas, Enterococcus, Parococcus, Solibacillus, and Paracoccus | [142] |
| Chicken manure | Firmicutes and Actinobacteria | Bacteria: Unclassified_f_peptostreptococcaceae, Enterococcus, and Turicibacter Fungi: Penicillium, Aspergillus, Kernia, and Microascus | [143] |
| Chicken manure; kitchen waste | Proteobacteria, Actinobacteria, and Firmicutes | Providencia, Morganella, Brevibacterium, Staphylococcus, and Bordetella | [144] |
| Chicken feed | Proteobacteria, Firmicutes, and Actinobacteria | Providencia, Proteus, Morganella, Enterococcus, Bacillus, and Enterobacteriaceae | [145] |
| Wheat bran, food waste and peanut shell | Firmicutes | Bacillus, unclassified_f_Caloramatoraceae, Cerasibacillus, and Gracilibacillus | [146] |
| Gainesville diet; starvation | Actinobacteria, Proteobacteria, Firmicutes, Euryarchaeota, and Bacteroidetes | Actinomyces, Campylobacter, Microbacterium, Enterococcus, and Enterobacter | [147] |
| Standard feed, brewer’s spent grain, plant-based sweetener, and vegetable waste | Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria | Morganella, Providencia, Lactobacillus, Enterococcus, and Proteus. | [148] |
| Wheat bran and soybean powder, food waste | Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria | Enterococcus, Acinetobacter, Providencia, Enterobacter, and Myroides | [149] |
| Corn flour and bran | Proteobacteria, Firmicutes, Bacteroidetes, and Actinomycetes | Morganella, Sedimentibacter, Dysgonomonas, Enterococcus, and Providencia | [150] |
| Chicken feed, mixture of vegetable coproducts, pig feed, and wheat bran | Firmicutes and Proteobacteria | Lactiplantibacillus, Weissella, Enterococcus, Morganella, Providencia, Lactobacillus, Corynebacterium, Proteus, Oceanobacillus, Cerasibacillus, Enterobacter, and Bacillus | [151] |
| Pig manure | Actinobacteria, Proteobacteria, and Bacteroidetes | Enterococcus, Providencia, Dysgonomonas, Koukoulia, Pseudomonas, Sphingobacterium, and Clostridiaceae | [152] |
| Palm kernel meal | Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria | Bacteria: Klebsiella, Enterococcus, and Sphingobacterium Fungi: Trichosporon, Candida, Lichtheimia, Fusarium, Pichia, Suhomyces, Diutina, and Kluyveromyces | [117] |
| Household composts | / | Nectriaceae, Meyerozyma, Kodamaeae, Gibberella, Diplodascaceae, Cyberlindnera, and Candida | [153] |
| Gainesville diet | Actinobacteria, Firmicutes, Proteobacteria, and Bacteroidetes | Acetobacter, Pseudomonas, Dysgonomonas, Acinetobacter, Providencia, Myroides, Alcaligenes, and Corynebacterium | [154] |
| Corn Straw | Proteobacteria, Bacteroidetes, and Firmicutes | Acinetobacter, Dysgonomonas, and unclassified Enterobacteriaceae | [155] |
| Wheat bran, wheat middling | Proteobacteria, Firmicutes, and Bacteroidetes | Bacteria: Dysgonomonas, Campylobacter, Enterococcus, Actinomyces, Pseudomonas, Klebsiella, Pediococcus, Lactobacillus, Bacillus, Orbus., and Providencia Fungi: Issatchenkia, Candida, Aspergillus, and Wickerhamomyces | [156] |
| Artificial diet with the addition of antibiotics | Proteobacteria, Firmicutes, and Actinobacteria | Pseudomonas, Actinomyces, Morganella, Providencia, and Klebsiella | [157] |
| Soya meal substrate containing oxytetracycline | / | Enterococcus, Psychrobacter, Providencia, Myroides, Enterobacteriaceae, and Lactobacillales | [158] |
| Mixtures of corn meal, wheat bran, and moisture content | Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria | Klebsiella, Clostridium, Acinetobacter, Pseudomonas, Providencia, Dysgonomonas, Morganella, Acetobacter, Enterococcus, Chryseobacterium, and Actinomyces | [159] |
| Gainesville Diet | / | Morganella, Dysgonomonas, Salmonella, Pseudochrobactrum, and Klebsiella (12 °C); Acinetobacter, Pseudochrobactrum, Enterococcus, Comamonas, and Leucobacter (16 °C) | [160] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Q.; Xia, J.; Ge, C.; Yao, H. Intestinal Microecological Mechanisms of Aflatoxin B1 Degradation by Black Soldier Fly Larvae (Hermetia illucens): A Review. Animals 2025, 15, 3351. https://doi.org/10.3390/ani15223351
Yuan Q, Xia J, Ge C, Yao H. Intestinal Microecological Mechanisms of Aflatoxin B1 Degradation by Black Soldier Fly Larvae (Hermetia illucens): A Review. Animals. 2025; 15(22):3351. https://doi.org/10.3390/ani15223351
Chicago/Turabian StyleYuan, Qiwen, Jing Xia, Chaorong Ge, and Huaiying Yao. 2025. "Intestinal Microecological Mechanisms of Aflatoxin B1 Degradation by Black Soldier Fly Larvae (Hermetia illucens): A Review" Animals 15, no. 22: 3351. https://doi.org/10.3390/ani15223351
APA StyleYuan, Q., Xia, J., Ge, C., & Yao, H. (2025). Intestinal Microecological Mechanisms of Aflatoxin B1 Degradation by Black Soldier Fly Larvae (Hermetia illucens): A Review. Animals, 15(22), 3351. https://doi.org/10.3390/ani15223351







