Compositional Changes and Comparative Analysis of Oral Microbial Community During the Formation of Canine Dental Calculus
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Collection and Eligibility Criteria
2.2. DNA Extraction, PCR Amplification and PacBio Sequencing
2.3. Statistical Analysis
3. Results
3.1. Sequence Analysis
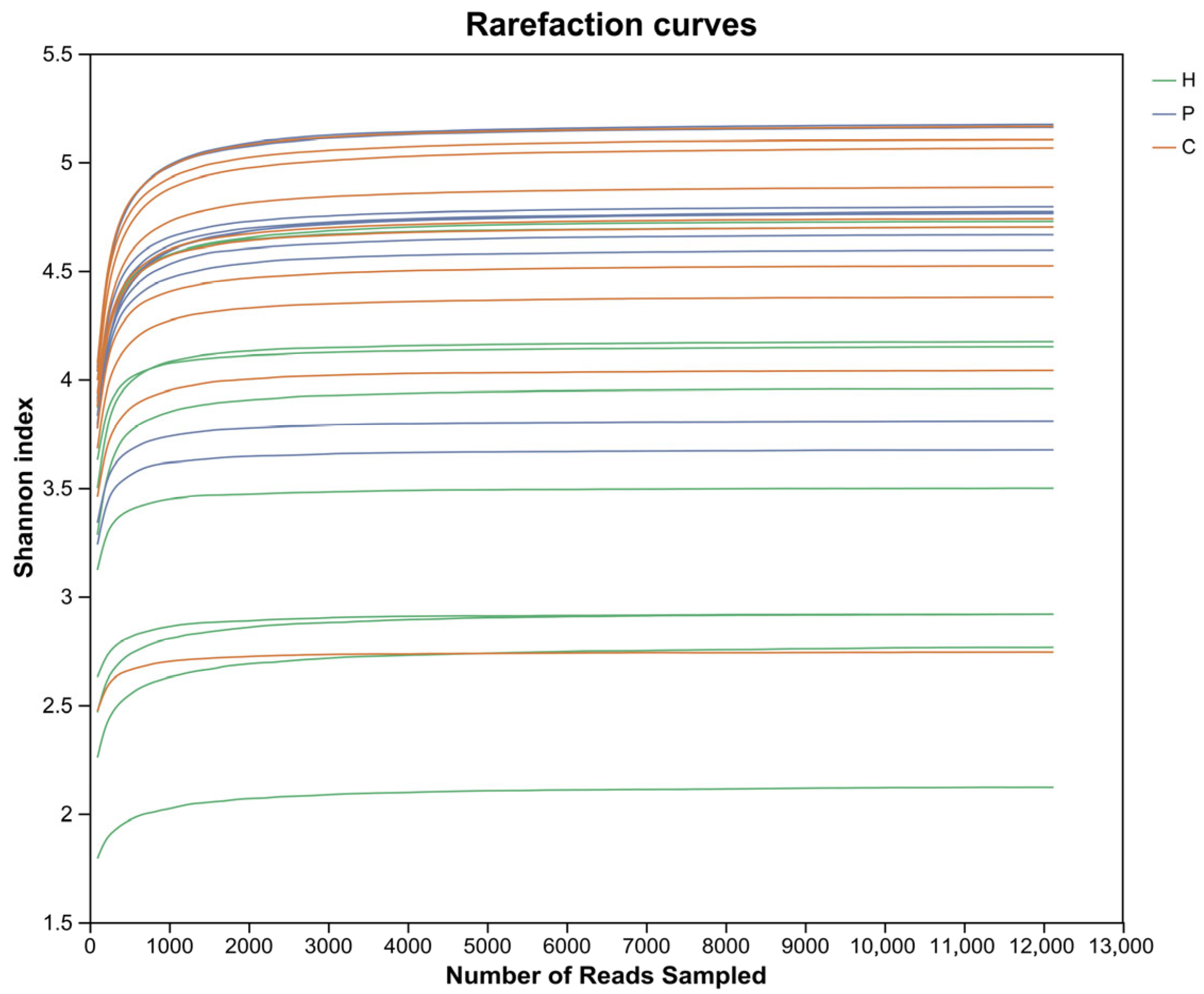
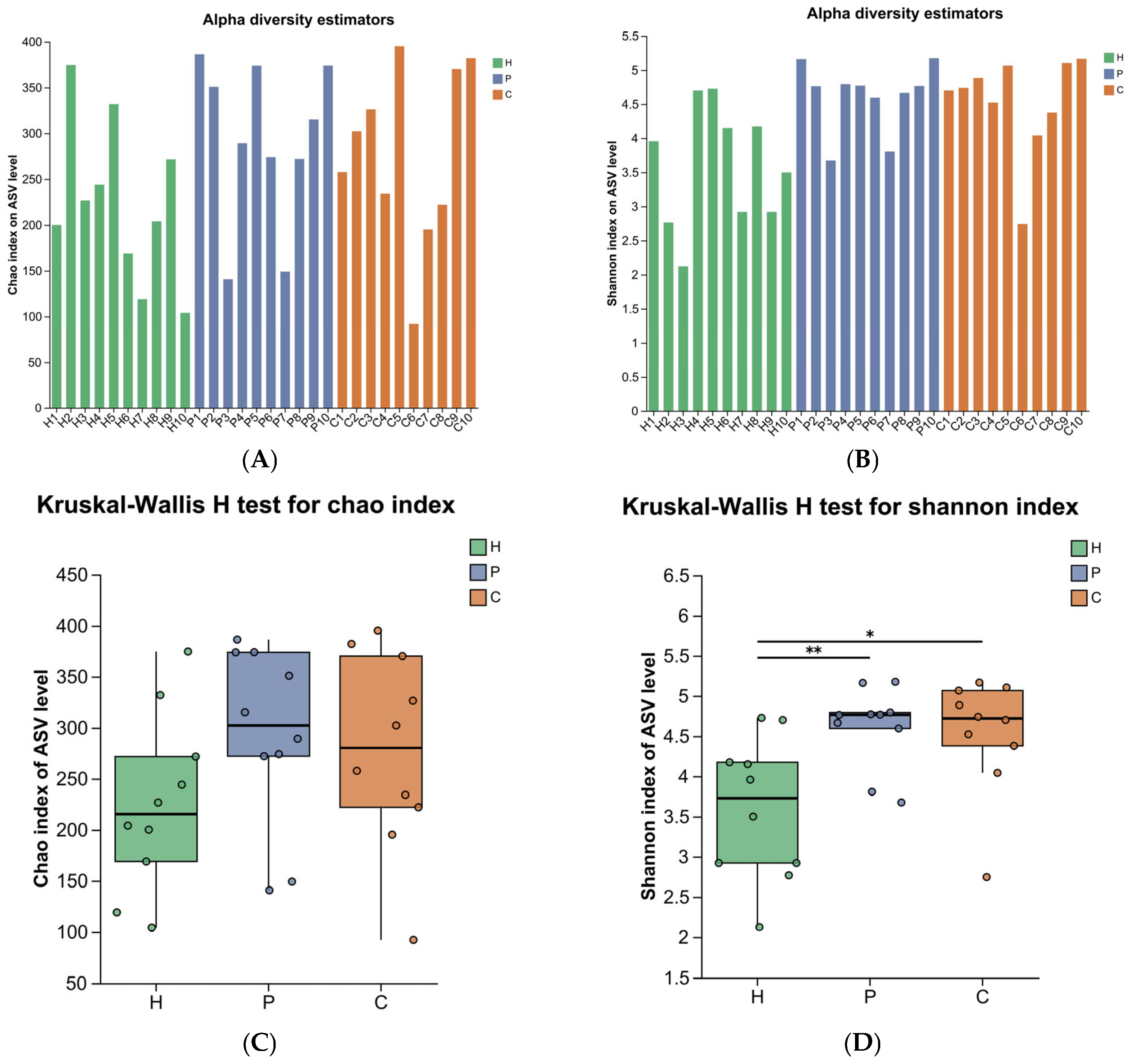
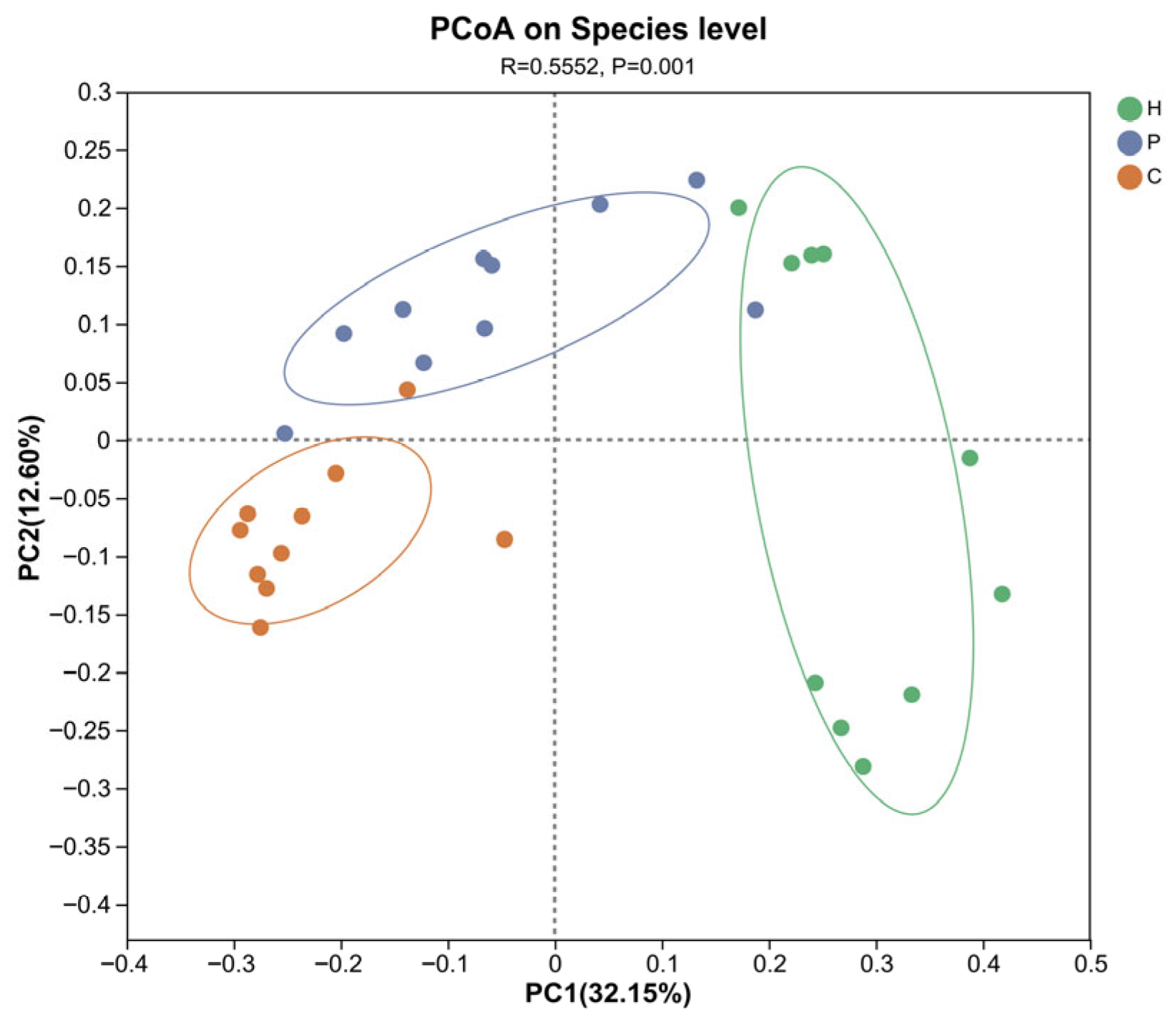
3.2. Analysis of Microbial Diversity During Canine Dental Calculus Formation
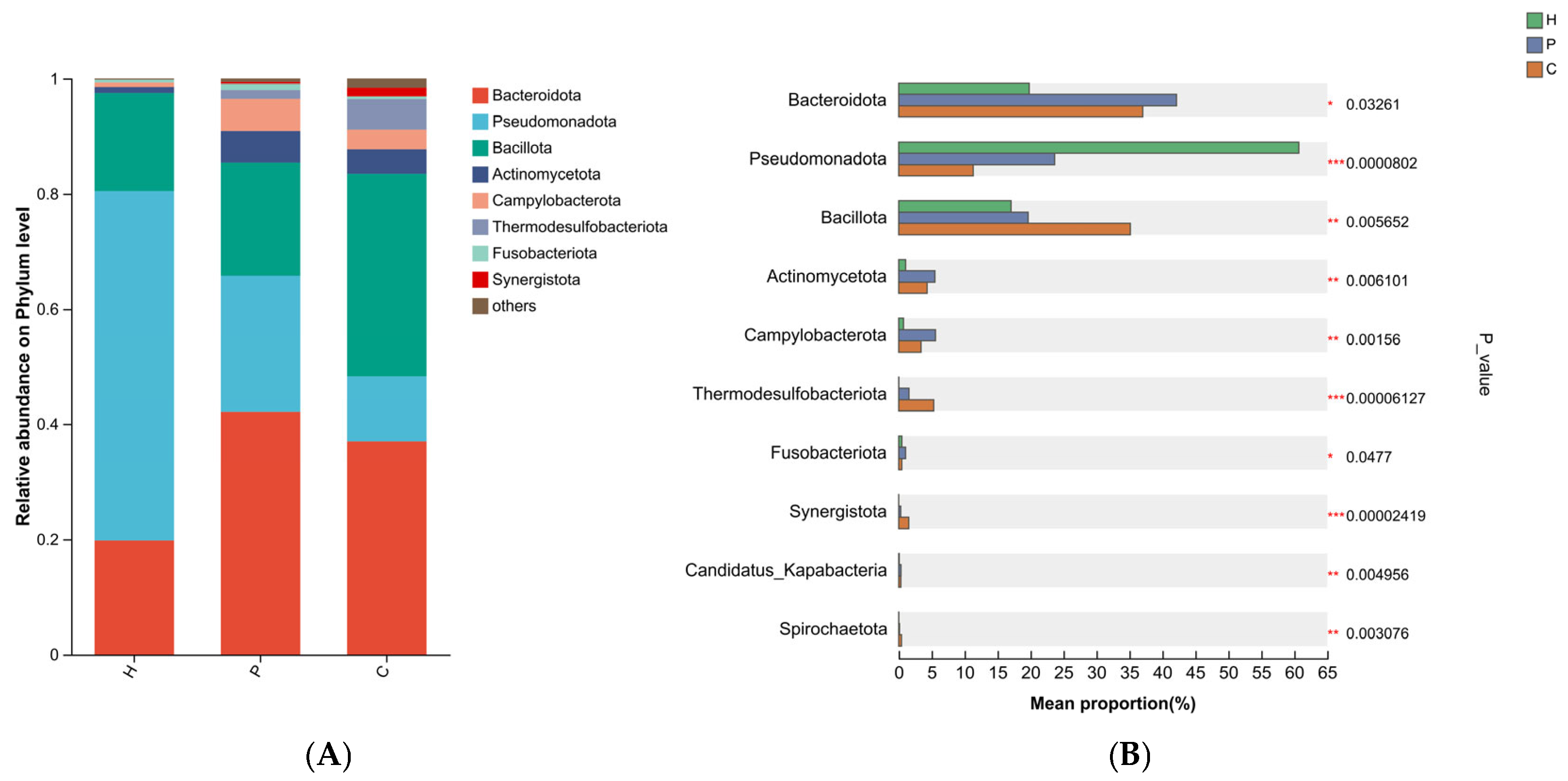
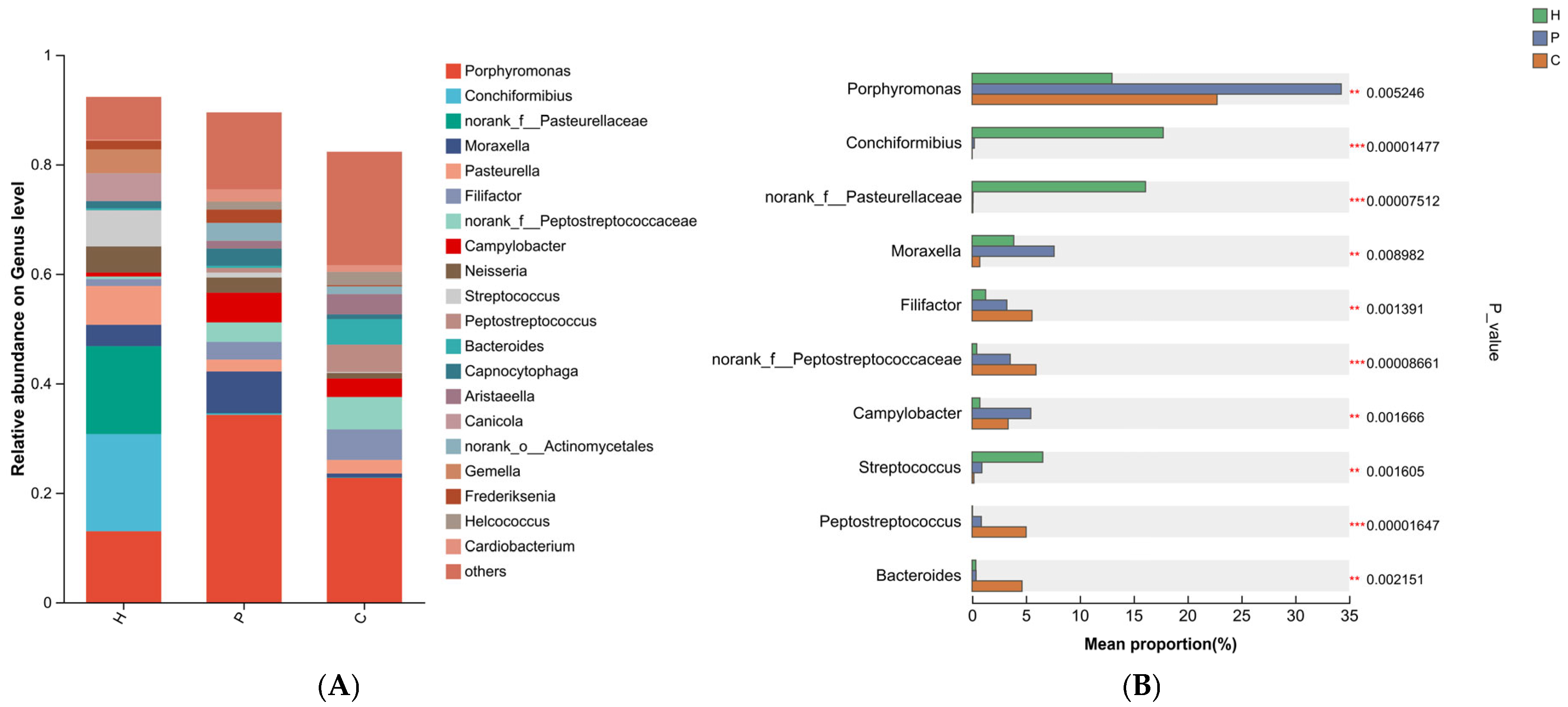
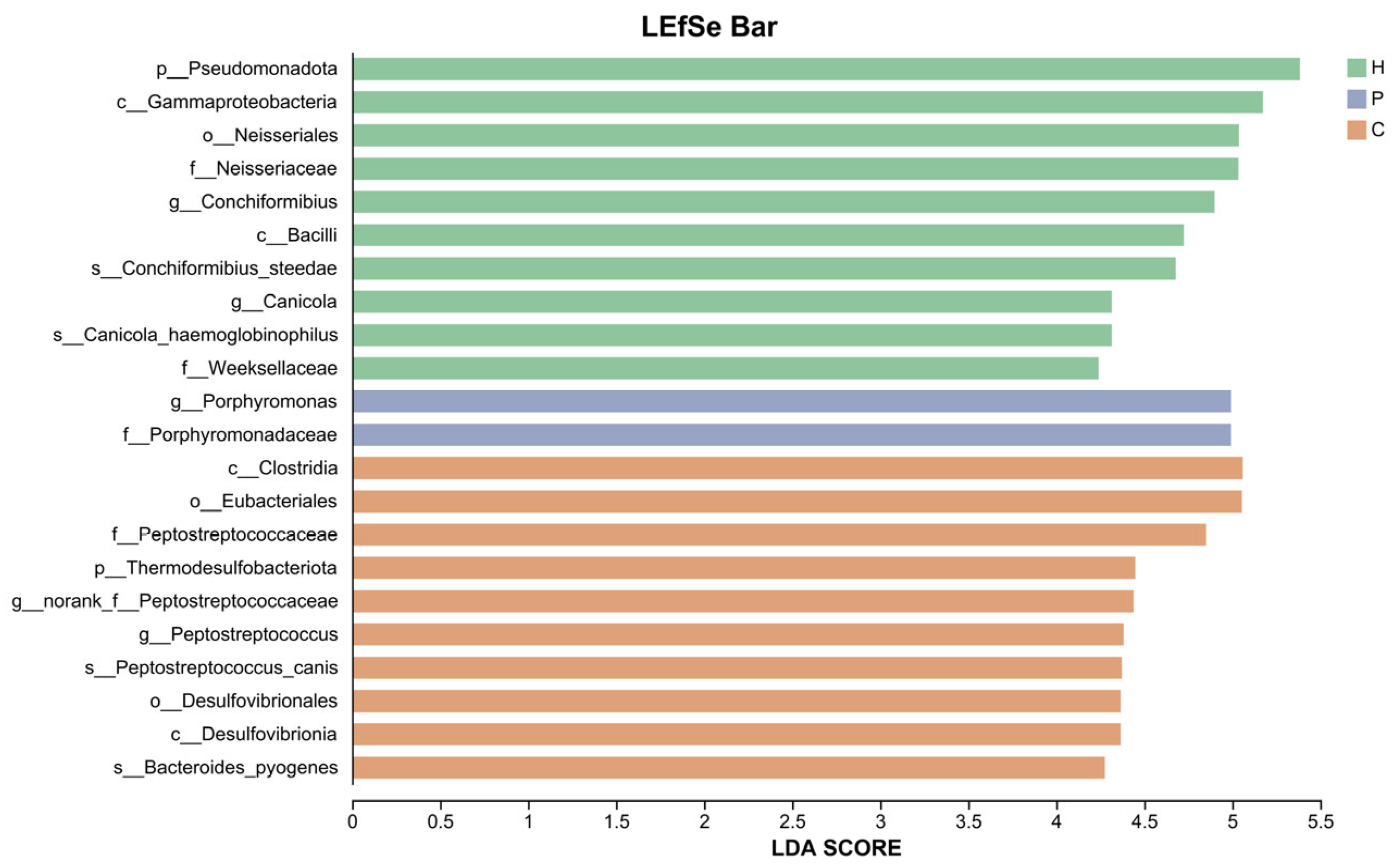
3.3. Analysis of Species Composition and Differences in the Microbiota During Canine Dental Calculus Formation
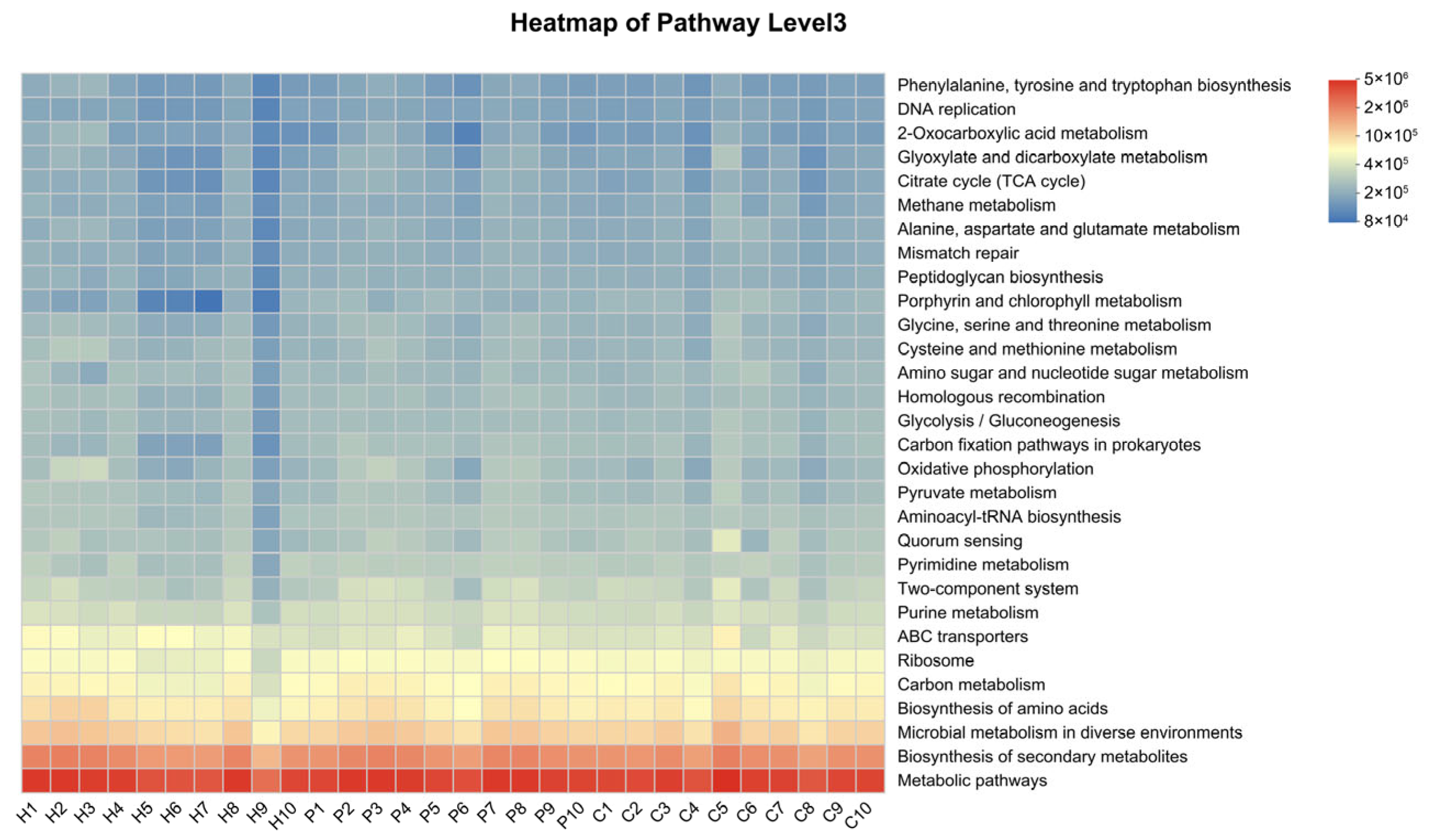
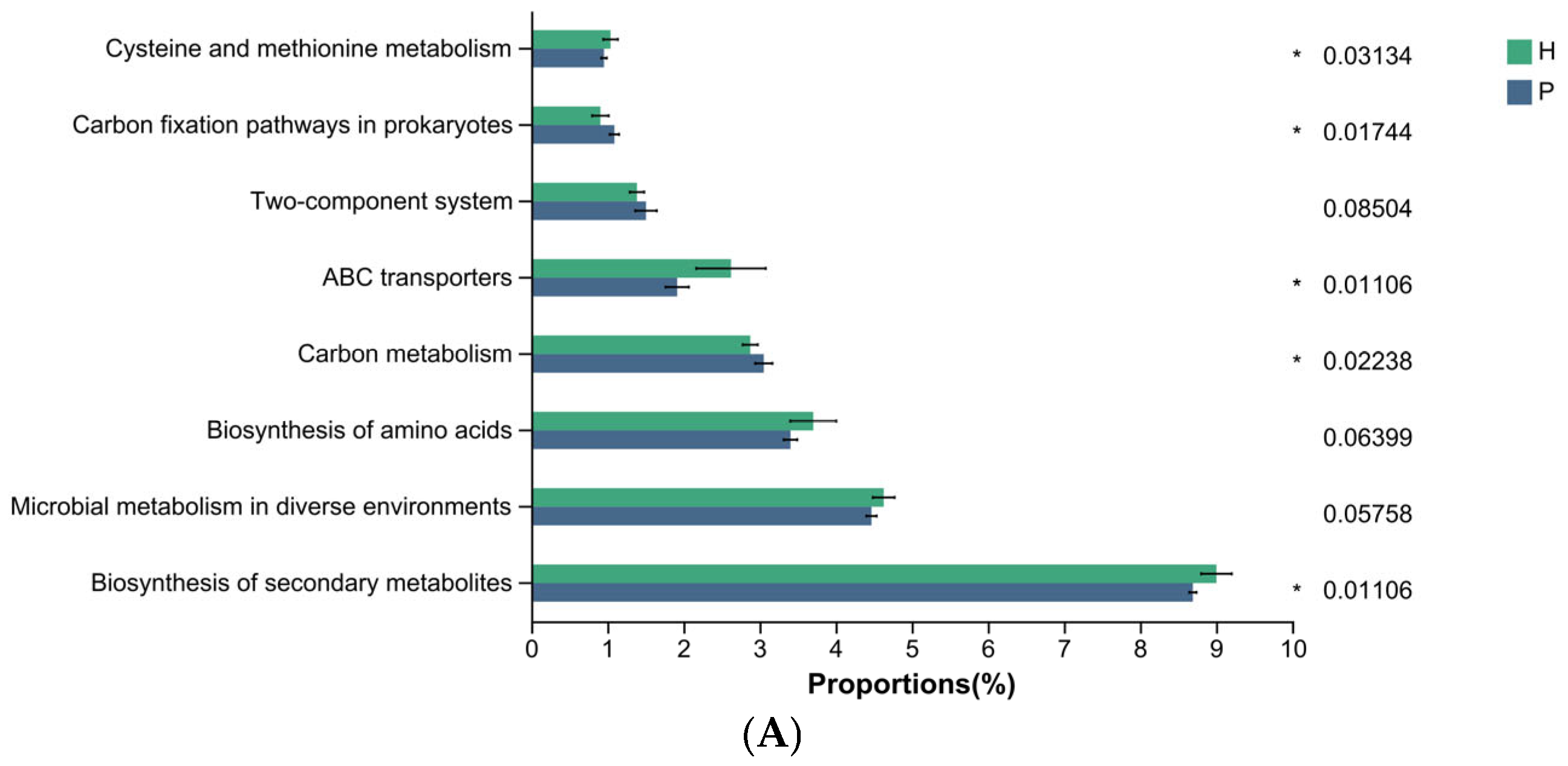
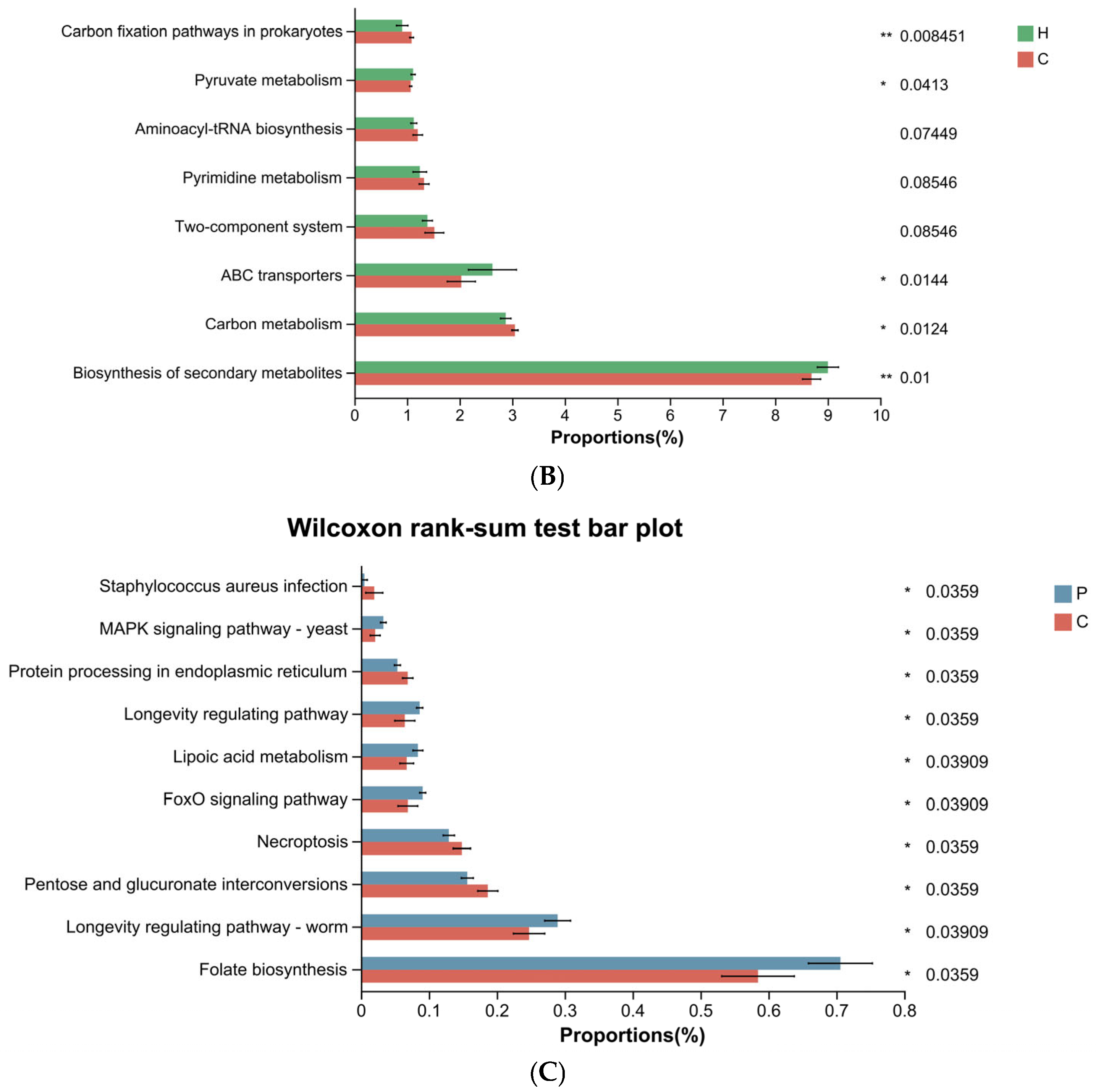
3.4. Differential Functional Analysis of Microbial Communities During Canine Dental Calculus Formation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PD | Periodontal disease |
| NGS | Next-generation sequencing |
| TGS | Third-generation sequencing |
| PacBio | Pacific biosciences |
| SMRT | Single molecule real-time |
| GI | Gingival index |
| PI | Plaque index |
| CI | Calculus index |
| BOP | Bleeding on probing |
| HiFi | High-fidelity |
| CCS | Circular consensus sequencing |
| QIIME2 | Quantitative insights into microbial 2 |
| ASVs | Amplicon sequence variants |
| PCoA | Principal co-ordinates analysis |
| LEfSe | Linear discriminant analysis effect size |
| LDA | Linear discriminant analysis |
| KEGG | Kyoto encyclopedia of genes and genomes |
| MuLDi | Multicellular longitudinal division |
| SRB | Sulfate-reducing bacteria |
| Ala | Alanine |
| Asp | Aspartate |
| Glu | Glutamate |
| SCFAs | Short-chain fatty acids |
References
- Perazzi, A.; Ricci, R.; Contiero, B.; Iacopetti, I. Evaluation of Salivary Biochemistry in Dogs with and without Plaque, Calculus, and Gingivitis: Preliminary Results. Animals 2022, 12, 1091. [Google Scholar] [CrossRef] [PubMed]
- AlRowis, R.; AlMoharib, H.S.; AlMubarak, A.; Bhaskardoss, J.; Preethanath, R.S.; Anil, S. Oral Fluid-Based Biomarkers in Periodontal Disease—Part 2. Gingival Crevicular Fluid. J. Int. Oral Health JIOH 2014, 6, 126–135. [Google Scholar] [PubMed]
- Buduneli, N.; Kinane, D.F. Host-derived Diagnostic Markers Related to Soft Tissue Destruction and Bone Degradation in Periodontitis. J. Clin. Periodontol. 2011, 38, 85–105. [Google Scholar] [CrossRef] [PubMed]
- Kačírová, J.; Sondorová, M.; Maďari, A.; Styková, E.; Mucha, R.; Nemcová, R.; Marečáková, N.; Farbáková, J.; Maďar, M. Detection of Periodontal Pathogens from Dental Plaques of Dogs with and without Periodontal Disease. Pathogens 2022, 11, 480. [Google Scholar] [CrossRef]
- Marsh, P.D. Dental Plaque as a Biofilm and a Microbial Community—Implications for Health and Disease. BMC Oral Health 2006, 6, S14. [Google Scholar] [CrossRef]
- Wei, Y.; Dang, G.; Ren, Z.; Wan, M.; Wang, C.; Li, H.; Zhang, T.; Tay, F.R.; Niu, L. Recent Advances in the Pathogenesis and Prevention Strategies of Dental Calculus. npj Biofilms Microbiomes 2024, 10, 56. [Google Scholar] [CrossRef]
- Belibasakis, G.N.; Manoil, D. Microbial Community-Driven Etiopathogenesis of Peri-Implantitis. J. Dent. Res. 2021, 100, 21–28. [Google Scholar] [CrossRef]
- Tatakis, D.N.; Kumar, P.S. Etiology and Pathogenesis of Periodontal Diseases. Dent. Clin. N. Am. 2005, 49, 491–516. [Google Scholar] [CrossRef]
- Radeerom, T.; Thongkorn, K.; Buddhachat, K.; Pradit, W.; Chomdej, S.; Siengdee, P.; Nganvongpanit, K. Köpek ve Kedi Diş Taşı Mikrobiyomunun Ileri Jenerasyon Sekanslama Kullanarak Araştırılması. Kafkas Univ. Vet. Fak. Derg. 2018, 24, 589–598. [Google Scholar] [CrossRef]
- Coignoul, E.; Cheville, N. Calcified Microbial Plaque. Dental Calculus of Dogs. Am. J. Pathol. 1984, 117, 499–501. [Google Scholar]
- Akcalı, A.; Lang, N.P. Dental Calculus: The Calcified Biofilm and Its Role in Disease Development. Periodontol. 2000 2018, 76, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Su, L.; Wang, Y.; Deng, S. Improved High-Throughput Sequencing of the Human Oral Microbiome: From Illumina to PacBio. Can. J. Infect. Dis. Med. Microbiol. 2020, 2020, 6678872. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, Z.D.; Müller, C.L.; Miraldi, E.R.; Littman, D.R.; Blaser, M.J.; Bonneau, R.A. Sparse and Compositionally Robust Inference of Microbial Ecological Networks. PLoS Comput. Biol. 2015, 11, e1004226. [Google Scholar] [CrossRef] [PubMed]
- Costea, P.I.; Hildebrand, F.; Arumugam, M.; Bäckhed, F.; Blaser, M.J.; Bushman, F.D.; de Vos, W.M.; Ehrlich, S.D.; Fraser, C.M.; Hattori, M.; et al. Enterotypes in the Landscape of Gut Microbial Community Composition. Nat. Microbiol. 2017, 3, 8–16. [Google Scholar] [CrossRef]
- Han, H.; Choi, Y.H.; Kim, S.Y.; Park, J.H.; Chung, J.; Na, H.S. Optimizing Microbiome Reference Databases with PacBio Full-Length 16S rRNA Sequencing for Enhanced Taxonomic Classification and Biomarker Discovery. Front. Microbiol. 2024, 15, 1485073. [Google Scholar] [CrossRef]
- Buetas, E.; Jordán-López, M.; López-Roldán, A.; D’Auria, G.; Martínez-Priego, L.; De Marco, G.; Carda-Diéguez, M.; Mira, A. Full-Length 16S rRNA Gene Sequencing by PacBio Improves Taxonomic Resolution in Human Microbiome Samples. BMC Genom. 2024, 25, 310. [Google Scholar] [CrossRef]
- Woese, C.R.; Fox, G.E. Phylogenetic Structure of the Prokaryotic Domain: The Primary Kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef]
- Starke, R.; Pylro, V.S.; Morais, D.K. 16S rRNA Gene Copy Number Normalization Does Not Provide More Reliable Conclusions in Metataxonomic Surveys. Microb. Ecol. 2021, 81, 535–539. [Google Scholar] [CrossRef]
- Athanasopoulou, K.; Boti, M.A.; Adamopoulos, P.G.; Skourou, P.C.; Scorilas, A. Third-Generation Sequencing: The Spearhead towards the Radical Transformation of Modern Genomics. Life 2021, 12, 30. [Google Scholar] [CrossRef]
- Meyer, M.; Stenzel, U.; Myles, S.; Prüfer, K.; Hofreiter, M. Targeted High-Throughput Sequencing of Tagged Nucleic Acid Samples. Nucleic Acids Res. 2007, 35, e97. [Google Scholar] [CrossRef]
- Hamady, M.; Walker, J.J.; Harris, J.K.; Gold, N.J.; Knight, R. Error-Correcting Barcoded Primers for Pyrosequencing Hundreds of Samples in Multiplex. Nat. Methods 2008, 5, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Mosher, J.J.; Bernberg, E.L.; Shevchenko, O.; Kan, J.; Kaplan, L.A. Efficacy of a 3rd Generation High-Throughput Sequencing Platform for Analyses of 16S rRNA Genes from Environmental Samples. J. Microbiol. Methods 2013, 95, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Mosher, J.J.; Bowman, B.; Bernberg, E.L.; Shevchenko, O.; Kan, J.; Korlach, J.; Kaplan, L.A. Improved Performance of the PacBio SMRT Technology for 16S rDNA Sequencing. J. Microbiol. Methods 2014, 104, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, J.; Chen, X.; Jiang, W.; Wang, S.; Xu, L.; Tu, Y.; Zheng, P.; Wang, Y.; Lin, X.; et al. Profiling of Oral Microbiota in Early Childhood Caries Using Single-Molecule Real-Time Sequencing. Front. Microbiol. 2017, 8, 2244. [Google Scholar] [CrossRef]
- Callahan, B.J.; Wong, J.; Heiner, C.; Oh, S.; Theriot, C.M.; Gulati, A.S.; McGill, S.K.; Dougherty, M.K. High-Throughput Amplicon Sequencing of the Full-Length 16S rRNA Gene with Single-Nucleotide Resolution. Nucleic Acids Res. 2019, 47, e103. [Google Scholar] [CrossRef]
- Guo, H.; Li, B.; Yao, H.; Liu, D.; Chen, R.; Zhou, S.; Ji, Y.; Zeng, L.; Du, M. Profiling the Oral Microbiomes in Patients with Alzheimer’s Disease. Oral Dis. 2023, 29, 1341–1355. [Google Scholar] [CrossRef]
- Ma, G.; Qiao, Y.; Shi, H.; Zhou, J.; Li, Y. Comparison of the Oral Microbiota Structure among People from the Same Ethnic Group Living in Different Environments. Biomed Res. Int. 2022, 2022, 6544497. [Google Scholar] [CrossRef]
- Löe, H.; Silness, J. Periodontal Disease in Pregnancy I. Prevalence and Severity. Acta Odontol. Scand. 1963, 21, 533–551. [Google Scholar] [CrossRef]
- Dale, S. Scherl.; Lori Coffman.; Misty Van Cleave.; Steve Lowry. Validation of a New Dental Plaque Quantification Method in Dogs. J. Vet. Dent. 2007, 24, 14–20. [Google Scholar] [CrossRef]
- Ramfjord, S.P. Indices for Prevalence and Incidence of Periodontal Disease. J. Periodontol. 1959, 30, 51–59. [Google Scholar] [CrossRef]
- Li, X.; Zhao, Z.; Guo, S.; Yang, C.; Gao, Y.; Li, L.; Ning, K.; Zhang, Q.; Zhou, N.; Zhang, H.; et al. Effects of Toothpaste Containing Inactivated Lacticaseibacillus Paracasei Probio-01 on Plaque-Induced Gingivitis and Dental Plaque Microbiota. Microb. Pathogen. 2024, 192, 106701. [Google Scholar] [CrossRef] [PubMed]
- Santibáñez, R.; Rodríguez-Salas, C.; Flores-Yáñez, C.; Garrido, D.; Thomson, P. Assessment of Changes in the Oral Microbiome That Occur in Dogs with Periodontal Disease. Vet. Sci. 2021, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Ruparell, A.; Inui, T.; Staunton, R.; Wallis, C.; Deusch, O.; Holcombe, L.J. The Canine Oral Microbiome: Variation in Bacterial Populations across Different Niches. BMC Microbiol. 2020, 20, 42. [Google Scholar] [CrossRef]
- Davis, I.J.; Wallis, C.; Deusch, O.; Colyer, A.; Milella, L.; Loman, N.; Harris, S. A Cross-Sectional Survey of Bacterial Species in Plaque from Client Owned Dogs with Healthy Gingiva, Gingivitis or Mild Periodontitis. PLoS ONE 2013, 8, e83158. [Google Scholar] [CrossRef]
- Wallis, C.; Milella, L.; Colyer, A.; O’Flynn, C.; Harris, S.; Holcombe, L.J. Subgingival Microbiota of Dogs with Healthy Gingiva or Early Periodontal Disease from Different Geographical Locations. BMC Vet. Res. 2021, 17, 7. [Google Scholar] [CrossRef]
- Pereira, A.M.; Clemente, A. Dogs’ Microbiome from Tip to Toe. Top. Companion Anim. Med. 2021, 45, 100584. [Google Scholar] [CrossRef]
- Nomura, R.; Inaba, H.; Yasuda, H.; Shirai, M.; Kato, Y.; Murakami, M.; Iwashita, N.; Shirahata, S.; Yoshida, S.; Matayoshi, S.; et al. Inhibition of Porphyromonas gulae and Periodontal Disease in Dogs by a Combination of Clindamycin and Interferon Alpha. Sci. Rep. 2020, 10, 3113. [Google Scholar] [CrossRef]
- Watanabe, A.; Okada, J.; Niwa, R.; Inui, Y.; Ito, K.; Shimokawa, Y.; Kihira, M. Bacterial Composition Changes in Canine Plaque over Periodontal Disease Severity and Daily Care Practices. Biorxiv 2023. [Google Scholar] [CrossRef]
- Niemiec, B.A.; Gawor, J.; Tang, S.; Prem, A.; Krumbeck, J.A. The Bacteriome of the Oral Cavity in Healthy Dogs and Dogs with Periodontal Disease. Am. J. Vet. Res. 2022, 83, 50–58. [Google Scholar] [CrossRef]
- Oba, P.M.; Carroll, M.Q.; Alexander, C.; Valentine, H.; Somrak, A.J.; Keating, S.C.J.; Sage, A.M.; Swanson, K.S. Microbiota Populations in Supragingival Plaque, Subgingival Plaque, and Saliva Habitats of Adult Dogs. Anim. Microbiome 2021, 3, 38. [Google Scholar] [CrossRef]
- Wallis, C.; Colyer, A.; Holcombe, L.J. Bacterial Associations with Periodontal Disease in Yorkshire Terriers. BMC Vet. Res. 2025, 21, 296. [Google Scholar] [CrossRef]
- Holcombe, L.J.; Patel, N.; Colyer, A.; Deusch, O.; O’Flynn, C.; Harris, S. Early Canine Plaque Biofilms: Characterization of Key Bacterial Interactions Involved in Initial Colonization of Enamel. PLoS ONE 2014, 9, e113744. [Google Scholar] [CrossRef]
- Man, S.M. The Clinical Importance of Emerging Campylobacter Species. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 669–685. [Google Scholar] [CrossRef]
- Arce, R.M.; Diaz, P.I.; Barros, S.P.; Galloway, P.; Bobetsis, Y.; Threadgill, D.; Offenbacher, S. Characterization of the Invasive and Inflammatory Traits of Oral Campylobacter Rectus in a Murine Model of Fetoplacental Growth Restriction and in Trophoblast Cultures. J. Reprod. Immunol. 2010, 84, 145–153. [Google Scholar] [CrossRef]
- Kato, Y.; Shirai, M.; Murakami, M.; Mizusawa, T.; Hagimoto, A.; Wada, K.; Nomura, R.; Nakano, K.; Ooshima, T.; Asai, F. Molecular Detection of Human Periodontal Pathogens in Oral Swab Specimens from Dogs in Japan. J. Vet. Dent. 2011, 28, 84–89. [Google Scholar] [CrossRef]
- O’Flynn, C.; Deusch, O.; Darling, A.E.; Eisen, J.A.; Wallis, C.; Davis, I.J.; Harris, S.J. Comparative Genomics of the Genus Porphyromonas Identifies Adaptations for Heme Synthesis within the Prevalent Canine Oral Species Porphyromonas cangingivalis. Genome Biol. Evol. 2015, 7, 3397–3413. [Google Scholar] [CrossRef] [PubMed]
- Iwashita, N.; Nomura, R.; Shirai, M.; Kato, Y.; Murakami, M.; Matayoshi, S.; Kadota, T.; Shirahata, S.; Ohzeki, L.; Arai, N.; et al. Identification and Molecular Characterization of Porphyromonas gulae fimA Types among Cat Isolates. Vet. Microbiol. 2019, 229, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Hamada, N.; Takahashi, Y.; Watanabe, K.; Kumada, H.; Oishi, Y.; Umemoto, T. Molecular and Antigenic Similarities of the Fimbrial Major Components between Porphyromonas gulae and P. Gingivalis. Vet. Microbiol. 2008, 128, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, Y.; Nomura, R.; Nakano, K.; Inaba, H.; Kuboniwa, M.; Hirai, N.; Shirai, M.; Kato, Y.; Murakami, M.; Naka, S.; et al. Distribution and Molecular Characterization of Porphyromonas gulae Carrying a New fimA Genotype. Vet. Microbiol. 2012, 161, 196–205. [Google Scholar] [CrossRef]
- Ito, N.; Itoh, N.; Kameshima, S. Volatile Sulfur Compounds Produced by the Anaerobic Bacteria Porphyromonas spp. Isolated from the Oral Cavities of Dogs. Vet. Sci. 2023, 10, 503. [Google Scholar] [CrossRef]
- Kislik, G.; Zhou, L.; Rubbi, L.; Pellegrini, M. Age-Correlated Changes in the Canine Oral Microbiome. Front. Microbiol. 2024, 15, 1426691. [Google Scholar] [CrossRef]
- Nyongesa, S.; Weber, P.M.; Bernet, È.; Pulido, F.; Nieves, C.; Nieckarz, M.; Delaby, M.; Viehboeck, T.; Krause, N.; Rivera-Millot, A.; et al. Evolution of Longitudinal Division in Multicellular Bacteria of the Neisseriaceae Family. Nat. Commun. 2022, 13, 4853. [Google Scholar] [CrossRef]
- Persson, S.; Edlund, M.; Claesson, R.; Carlsson, J. The Formation of Hydrogen Sulfide and Methyl Mercaptan by Oral Bacteria. Oral Microbiol. Immunol. 1990, 5, 195–201. [Google Scholar] [CrossRef]
- Polkowska, I.; Sobczyńska-Rak, A.; Gołyńska, M. Analysis of Gingival Pocket Microflora and Biochemical Blood Parameters in Dogs Suffering from Periodontal Disease. In Vivo 2014, 28, 1085–1090. [Google Scholar]
- Widdel, F. Microbiology and Ecology of Sulphateand Sulpur Reducing Bacteria. In Biology of Anaerobic Microorganisms; Zehnder, A.J.B., Ed.; Wiely: New York, NY, USA, 1988; pp. 469–585. [Google Scholar]
- Hansen, T.A. Carbon Metabolism of Sulfate-Reducing Bacteria. In The Sulfate-reducing Bacteria: Contemporary Perspectives; Odom, J.M., Singleton, R., Eds.; Springer: New York, NY, USA, 1993; pp. 21–40. ISBN 978-1-4613-9263-7. [Google Scholar]
- Kushkevych, I.; Coufalová, M.; Vítězová, M.; Rittmann, S.K.-M.R. Sulfate-Reducing Bacteria of the Oral Cavity and Their Relation with Periodontitis—Recent Advances. J. Clin. Med. 2020, 9, 2347. [Google Scholar] [CrossRef] [PubMed]
- Langendijk, P.S.; Kulik, E.M.; Sandmeier, H.; Meyer, J.; Van Der Hoeven, J.S. Isolation of Desulfomicrobium orale sp. Nov. and Desulfovibrio Strain NY682, Oral Sulfate-Reducing Bacteria Involved in Human Periodontal Disease. Int. J. Syst. Evol. Microbiol. 2001, 51, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Kushkevych, I.; Dordević, D.; Vítězová, M. Possible Synergy Effect of Hydrogen Sulfide and Acetate Produced by Sulfate-Reducing Bacteria on Inflammatory Bowel Disease Development. J. Adv. Res. 2021, 27, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-H.; Dong, Z.; Chu, L. Hydrogen Sulfide Induces Apoptosis in Human Periodontium Cells. J. Periodontal Res. 2010, 45, 71–78. [Google Scholar] [CrossRef]
- Murata, T.; Yaegaki, K.; Qian, W.; Herai, M.; Calenic, B.; Imai, T.; Sato, T.; Tanaka, T.; Kamoda, T.; Ii, H. Hydrogen Sulfide Induces Apoptosis in Epithelial Cells Derived from Human Gingiva. J. Breath Res. 2008, 2, 17007. [Google Scholar] [CrossRef]
- Wu, D.-D.; Ngowi, E.E.; Zhai, Y.-K.; Wang, Y.-Z.; Khan, N.H.; Kombo, A.F.; Khattak, S.; Li, T.; Ji, X.-Y. Role of Hydrogen Sulfide in Oral Disease. Oxid. Med. Cell. Longev. 2022, 2022, 1886277. [Google Scholar] [CrossRef]
- How, S.S.; Nathan, S.; Lam, S.D.; Chieng, S. ATP-Binding Cassette (ABC) Transporters: Structures and Roles in Bacterial Pathogenesis. J. Zhejiang Univ.-Sci. B 2025, 26, 58–75. [Google Scholar] [CrossRef]

| Evaluation Index | Scoring Criteria | Score |
|---|---|---|
| Gingival Index (GI) | Healthy gingiva, with no inflammation at the gingival margin and normal periodontal pocket depth (free gingiva) (<3 mm in dogs) | 0 |
| Mild gingivitis, with slight redness and swelling at the gingival margin, but no bleeding upon probing | 1 | |
| Moderate gingivitis, with redness and swelling at the gingival margin, and bleeding upon probing | 2 | |
| Severe gingivitis, with marked redness and swelling at the gingival margin, often accompanied by ulceration, and potentially spontaneous gingival bleeding | 3 | |
| Plaque Index (PI) | No visible plaque | 0 |
| Plaque covers less than 1/3 of the buccal tooth surface | 1 | |
| Plaque covers between 1/3 and 2/3 of the buccal tooth surface | 2 | |
| Plaque covers more than 2/3 of the buccal tooth surface | 3 | |
| Calculus Index (CI) | No visible calculus | 0 |
| Calculus covers less than 1/3 of the maxillofacial tooth surface | 1 | |
| Calculus covers between 1/3 and 2/3 of the maxillofacial tooth surface; little or no subgingival plaque accumulation | 2 | |
| Calculus covers more than 2/3 of the maxillofacial tooth surface and extends below the gingival margin | 3 |
| Healthy Group | Plaque Group | Calculus Group | p Value | H vs. P | H vs. C | P vs. C | |
|---|---|---|---|---|---|---|---|
| Age (months) | |||||||
| Mean ± SD | 10.8 ± 1.81 | 63.6 ± 3.92 | 120.2 ± 10.73 | <0.001 | <0.001 | <0.001 | <0.001 |
| Mix-Max | 8–14 | 58–70 | 107–140 | ||||
| Weight (kg) | |||||||
| Mean ± SD | 6.27 ± 3.46 | 6.75 ± 4.08 | 8.1 ± 5.22 | 0.554 | 0.739 | 0.353 | 0.393 |
| Mix-Max | 2.9–13.5 | 2.9–14 | 3.2–17 | ||||
| Sex | 0.873 | 0.653 | 0.435 | 0.653 | |||
| Males, n (%) | 4 (40%) | 5 (50%) | 4 (40%) | ||||
| Females, n (%) | 6 (60%) | 5 (50%) | 6 (60%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, L.; Shi, L.; Yang, Y.; Zheng, D.; Zhang, W.; Yang, J.; Qiao, M.; Shi, H. Compositional Changes and Comparative Analysis of Oral Microbial Community During the Formation of Canine Dental Calculus. Animals 2025, 15, 3335. https://doi.org/10.3390/ani15223335
Zeng L, Shi L, Yang Y, Zheng D, Zhang W, Yang J, Qiao M, Shi H. Compositional Changes and Comparative Analysis of Oral Microbial Community During the Formation of Canine Dental Calculus. Animals. 2025; 15(22):3335. https://doi.org/10.3390/ani15223335
Chicago/Turabian StyleZeng, Liwei, Lei Shi, Yufei Yang, Dongqiang Zheng, Wenkai Zhang, Jingyi Yang, Meilin Qiao, and Hao Shi. 2025. "Compositional Changes and Comparative Analysis of Oral Microbial Community During the Formation of Canine Dental Calculus" Animals 15, no. 22: 3335. https://doi.org/10.3390/ani15223335
APA StyleZeng, L., Shi, L., Yang, Y., Zheng, D., Zhang, W., Yang, J., Qiao, M., & Shi, H. (2025). Compositional Changes and Comparative Analysis of Oral Microbial Community During the Formation of Canine Dental Calculus. Animals, 15(22), 3335. https://doi.org/10.3390/ani15223335





