Development of Visual Detection of African Swine Fever Virus Using CRISPR/AapCas12b Lateral Flow Strip Based on Viral Major Capsid Protein Gene B646L
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Specimens
2.2. Cloning of AapCas12b Gene, Protein Expression, and Purification
2.3. Preparation of sgRNA
2.4. B646L-Mediated CRISPR/AapCas12b Reaction
2.5. Recombinase Polymerase Amplification (RPA)
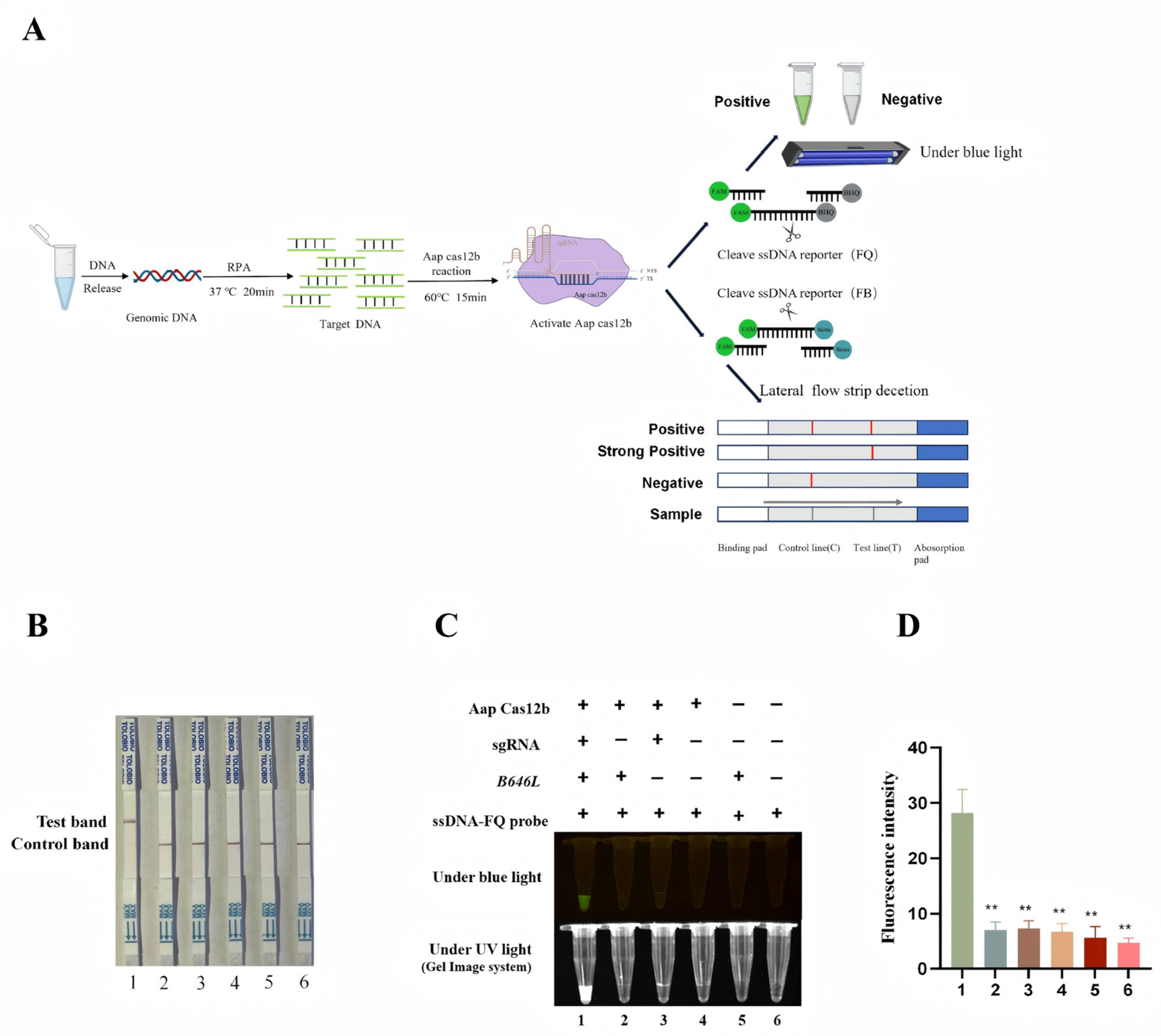
2.6. Establishment of RPA-CRISPR/AapCas12b-Lateral Flow Strip (LFS) Detection Method
2.7. Sensitivity and Specificity of B646L-RPA-AapCas12b-LFS Detection Method
2.8. Detection of Clinical Samples by RPA-AapCas12b-LFS
3. Results
3.1. The Expression and Purification of AapCas12b
3.2. Validation of the Cleavage Activities of the Purified AapCas12b Protein
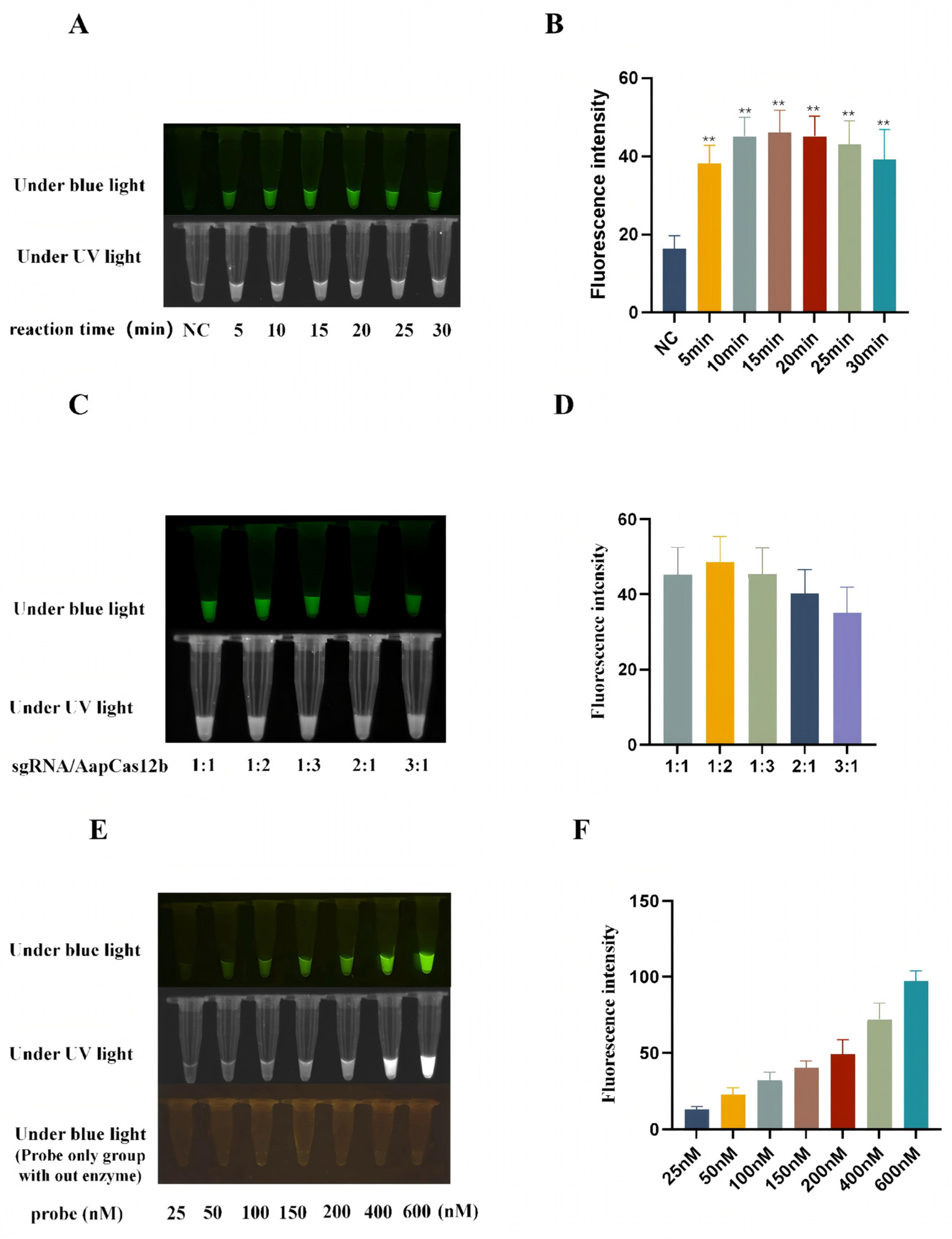
3.3. Optimization of Reaction Conditions of CRISPR/AapCas12b Assay
3.4. Establishment of CRISPR/AapCas12b Mediated Lateral Flow Strip (LFS) Method
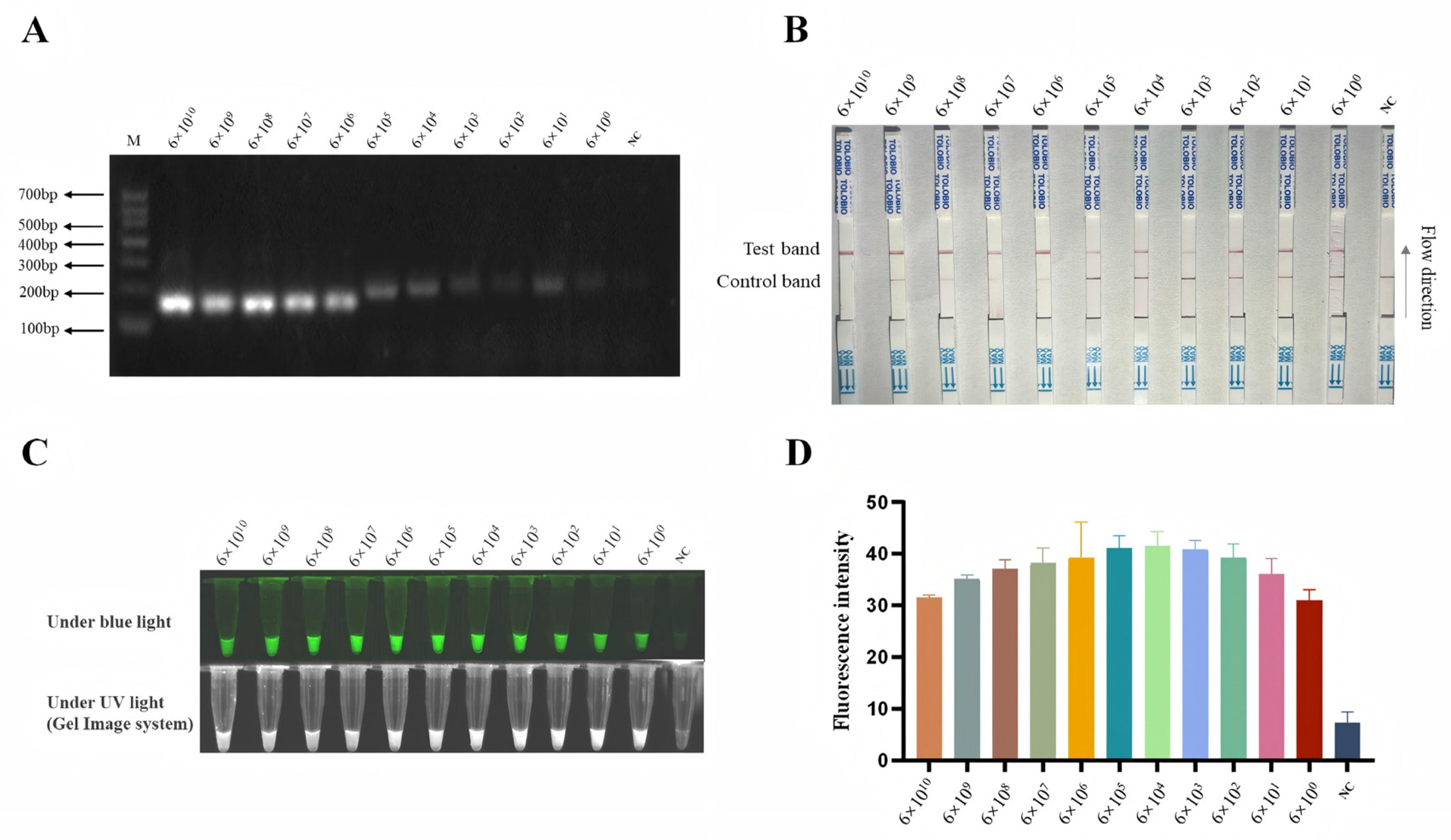
3.5. Sensitivity of B646L-RPA-CRISPR/AapCas12b-LFS Method
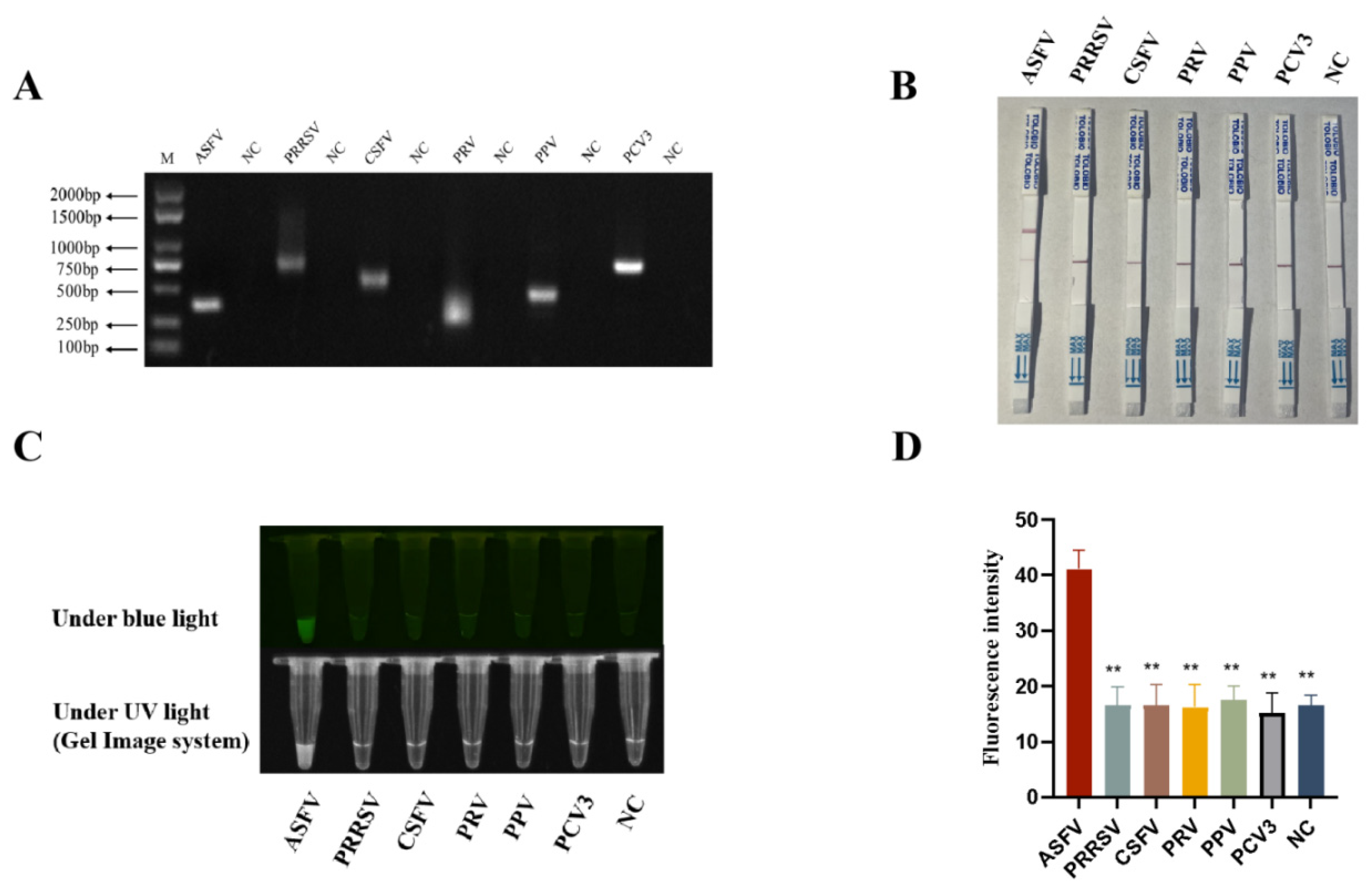
3.6. Specificity of RPA-CRISPR/AapCas12b-LFS Detection Method
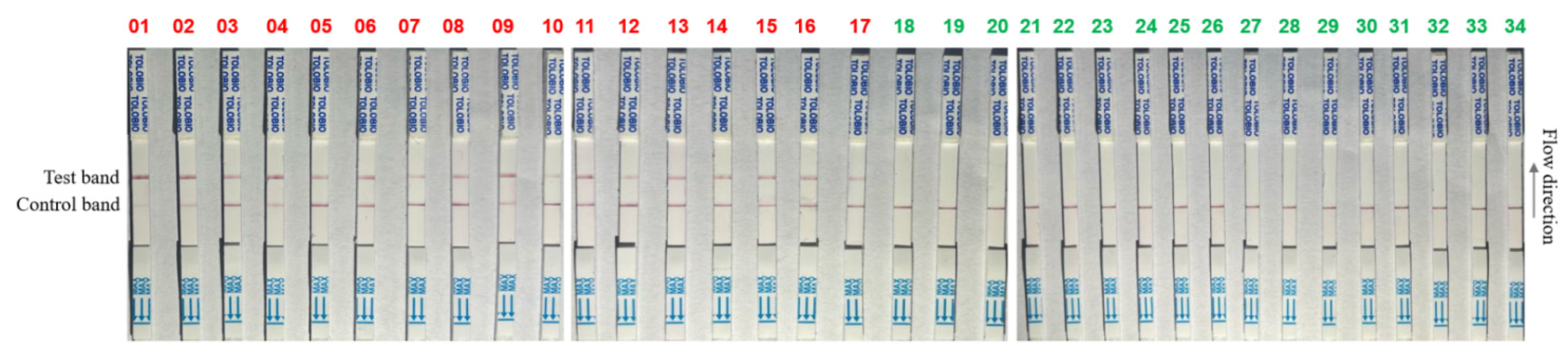
3.7. RPA-CRISPR/AapCas12b-LFS Detection of Clinical Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Solikhah, T.I.; Rostiani, F.; Nanra, A.F.P.; Dewi, A.D.P.P.; Nurbadri, P.H.; Agustin, Q.A.D.; Solikhah, G.P. African swine fever virus: Virology, pathogenesis, clinical impact, and global control strategies. Vet. World 2025, 18, 1599–1613. [Google Scholar] [CrossRef]
- Dixon, L.K.; Chapman, D.A.G.; Netherton, C.L.; Upton, C. African swine fever virus replication and genomics. Virus Res. 2013, 173, 3–14. [Google Scholar] [CrossRef]
- Zhu, G.L.; Xi, F.; Zeng, W.X.; Zhao, Y.F.; Cao, W.J.; Liu, C.; Yang, F.; Ru, Y.; Xiao, S.Q.; Zhang, S.L.; et al. Structural basis of RNA polymerase complexes in African swine fever virus. Nat. Commun. 2025, 16, 501. [Google Scholar] [CrossRef]
- Ruedas-Torres, I.; Nga, B.T.T.; Salguero, F.J. Pathogenicity and virulence of African swine fever virus. Virulence 2024, 15, 2375550. [Google Scholar] [CrossRef]
- Liu, C.X.; Li, T.T.; Huang, T.; Zhao, G.H.; Wang, X.; Li, J.N.; Huang, L.; Zhang, Z.X.; Zheng, J.; Weng, C.J. S5092 inhibit ASFV infection by targeting the pS273R protease activity in vitro. Vet. Microbiol. 2025, 304, 110473. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, L.; Ge, X.N.; Gao, P.; Zhou, Q.Q.; Han, J.; Guo, X.; Zhang, Y.N.; Yang, H.C. Advances in the diagnostic techniques of African swine fever. Virology 2025, 603, 110351. [Google Scholar] [CrossRef]
- Zhang, J.J.; Sun, Z.Y.; Sun, S.H.; Zhang, K.L.; Deng, D.F.; He, P.; Zhang, P.P.; Xia, N.W.; Jiang, S.; Zheng, W.L.; et al. The capsid protein p72 specific mAb and the corresponding novel epitope based ELISAs for detection of ASFV infection. Vet. Microbiol. 2025, 303, 110437. [Google Scholar] [CrossRef]
- Hu, Z.Q.; Tian, X.G.; Lai, R.R.; Wang, X.L.; Li, X.W. Current detection methods of African swine fever virus. Front. Vet. Sci. 2023, 10, 1289676. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.F.; Luo, S.H.; Li, W.B.; Su, W.T.; Chen, S.T.; Liu, C.C.; Pan, W.L.; Situ, B.; Zheng, L.; Li, L.; et al. Co-freezing localized CRISPR-Cas12a system enables rapid and sensitive nucleic acid analysis. J. Nanobiotechnol. 2024, 22, 602. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.J.; Fang, T.T.; Cai, Q.S.; Li, H.; Li, H.Y.; Sun, H.Q.; Zhu, M.Z.; Dai, L.S.; Shao, Y.Q.; Cai, L. Rapid detection of in sputum using CRISPR-Cas12b combined with cross-priming amplification in a single reaction. J. Clin. Microbiol. 2024, 62, e0092323. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Q.; Ye, X.F.; Liu, Y.F.; Li, W.T.; Zhang, X.; Zhang, W.; Yi, C.Q.; Liu, C.X. Regulating cleavage activity and enabling microRNA detection with split sgRNA in Cas12b. Nat. Commun. 2025, 16, 6392. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Macaluso, N.C.; Pizzano, B.L.M.; Cash, M.N.; Spacek, J.; Karasek, J.; Miller, M.R.; Lednicky, J.A.; Dinglasan, R.R.; Salemi, M.; et al. A thermostable Cas12b from leverages one-pot discrimination of SARS-CoV-2 variants of concern. Ebiomedicine 2022, 77, 103926. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, V.; Spinard, E.; Xu, L.Z.; Berninger, A.; Barrette, R.W.; Gladue, D.P.; Faburay, B. Full-Length ASFV B646L Gene Sequencing by Nanopore Offers a Simple and Rapid Approach for Identifying ASFV Genotypes. Viruses 2024, 16, 1522. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhao, D.M.; Wang, J.L.; Zhang, Y.L.; Wang, M.; Gao, Y.; Li, F.; Wang, J.F.; Bu, Z.G.; Rao, Z.H.; et al. Architecture of African swine fever virus and implications for viral assembly. Science 2019, 366, 640–644. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, S.; Balusamy, S.R.; Perumalsamy, H.; Ståhlberg, A.; Mijakovic, I. CRISPR-Cas target recognition for sensing viral and cancer biomarkers. Nucleic Acids Res. 2024, 52, 10040–10067. [Google Scholar] [CrossRef]
- Das, A.; Goswami, H.N.; Whyms, C.T.; Sridhara, S.; Li, H. Structural principles of CRISPR-Cas enzymes used in nucleic acid detection. J. Struct. Biol. 2022, 214, 107838. [Google Scholar] [CrossRef]
- Li, S.Y.; Cheng, Q.X.; Liu, J.K.; Nie, X.Q.; Zhao, G.P.; Wang, J. CRISPR-Cas12a has both and cleavage activities on single-stranded DNA. Cell Res. 2018, 28, 491–493. [Google Scholar] [CrossRef]
- Yoon, P.H.; Zhang, Z.Y.; Loi, K.J.; Adler, B.A.; Lahiri, A.; Vohra, K.; Shi, H.L.; Rabelo, D.B.; Trinidad, M.; Boger, R.S.; et al. Structure-guided discovery of ancestral CRISPR-Cas13 ribonucleases. Science 2024, 385, 538–543. [Google Scholar] [CrossRef]
- Teng, F.; Cui, T.T.; Feng, G.H.; Guo, L.; Xu, K.; Gao, Q.Q.; Li, T.D.; Li, J.; Zhou, Q.; Li, W. Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov. 2018, 4, 63. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, S.J.; Li, R.R.; Qiu, X.; Fan, T.Y.; Wang, B.; Zhang, B.; Zhang, L. Advances in application of CRISPR-Cas13a system. Front. Cell. Infect. Microbiol. 2024, 14, 1291557, Erratum in Front. Cell. Infect. Microbiol. 2024, 14, 1393044. https://doi.org/10.3389/fcimb.2024.1393044. [Google Scholar] [CrossRef]
- Zhang, Z.C.; Li, J.; Zhang, C.Q.; Bai, X.; Zhang, T. Rapid Detection of Feline Calicivirus Using Lateral Flow Dipsticks Based on CRISPR/Cas13a System. Animals 2024, 14, 3663. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Q.; Quan, X.Y.; Li, Y.C.; Guo, H.Y.; Kong, F.E.; Lu, J.H.; Teng, L.R.; Wang, J.S.; Wang, D. Visual detection of SARS-CoV-2 with a CRISPR/Cas12b-based platform. Talanta 2023, 253, 124093. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Rananaware, S.R.; Yang, L.G.; Macaluso, N.C.; Ocana-Ortiz, J.E.; Meister, K.S.; Pizzano, B.L.M.; Sandoval, L.S.W.; Hautamaki, R.C.; Fang, Z.R.; et al. Engineering highly thermostable Cas12b via.structural analyses for one-pot detection of nucleic acids. Cell Rep. Med. 2023, 4, 101037. [Google Scholar] [CrossRef]
- Zhang, D.S.; Jiang, S.; Xia, N.W.; Zhang, Y.W.; Zhang, J.J.; Liu, A.J.; Zhang, C.Y.; Chen, N.H.; Meurens, F.; Zheng, W.L.; et al. Rapid Visual Detection of African Swine Fever Virus with a CRISPR/Cas12a Lateral Flow Strip Based on Structural Protein Gene D117L. Animals 2023, 13, 3712. [Google Scholar] [CrossRef]
- Zhang, D.S.; Jiang, S.; Xia, N.W.; Zhang, J.J.; Liu, A.J.; Deng, D.F.; Zhang, C.Y.; Sun, Y.X.; Chen, N.H.; Kang, X.L.; et al. Development of visual detection of African swine fever virus using CRISPR/ LwCas13a lateral flow strip based on structural protein gene D117L. Vet. Microbiol. 2024, 293, 110073. [Google Scholar] [CrossRef]
- Han, H.J.; Zhang, D.S.; Hao, W.L.; Liu, A.J.; Xia, N.W.; Cui, M.; Luo, J.; Jiang, S.; Zheng, W.L.; Chen, N.H.; et al. Parallel and Visual Detections of ASFV by CRISPR-Cas12a and CRISPR-Cas13a Systems Targeting the Viral S273R Gene. Animals 2025, 15, 1902. [Google Scholar] [CrossRef]
- Tian, T.; Qiu, Z.Q.; Jiang, Y.Z.; Zhu, D.B.; Zhou, X.M. Exploiting the orthogonal CRISPR-Cas12a/Cas13a-cleavage for dual-gene virus detection using a handheld device. Biosens. Bioelectron. 2022, 196, 113701. [Google Scholar] [CrossRef]
- Bohorquez, J.A.; Lanka, S.; Rosell, R.; Pérez-Simó, M.; Alberch, M.; Rodriguez, F.; Ganges, L.; Maddox, C.W. Efficient detection of African Swine Fever Virus using minimal equipment through a LAMP PCR method. Front. Cell. Infect. Microbiol. 2023, 13, 1114772. [Google Scholar] [CrossRef]
- Petrovan, V.; Yuan, F.F.; Li, Y.H.; Shang, P.C.; Murgia, M.V.; Misra, S.; Rowland, R.R.R.; Fang, Y. Development and characterization of monoclonal antibodies against p30 protein of African swine fever virus. Virus Res. 2019, 269, 197632. [Google Scholar] [CrossRef] [PubMed]
- He, K.J.; Wu, Y.; Su, Z.P.; Zeng, Y.; Ye, G.S.; Wu, Q.; Li, L.; Zhang, A.D. Identification of a Conserved Linear Epitope on the p54 Protein of African Swine Fever Virus. Viruses 2025, 17, 823. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.L.; Bai, Y.L.; Zhang, S.; Zhao, X.Y.; Jin, J.X.; Zhu, X.J.; Wang, R.; Wu, Y.A.; Zhang, A.K.; Zhang, G.P.; et al. An Intracellular Epitope of ASFV CD2v Protein Elicits Humoral and Cellular Immune Responses. Animals 2023, 13, 1967. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.L.; Xu, M.H.; Luo, S.Y.; Yang, Y.; Zhong, J.Y.; Zhou, J.W.; Fan, H.Y.; Li, X.P.; Chen, Z. Advancements in the synergy of isothermal amplification and CRISPR-cas technologies for pathogen detection. Front. Bioeng. Biotech. 2023, 11, 1273988. [Google Scholar] [CrossRef] [PubMed]
- Azeez, S.S.; Hamad, R.S.; Hamad, B.K.; Shekha, M.S.; Bergsten, P. Advances in CRISPR-Cas technology and its applications: Revolutionising precision medicine. Front. Genome Ed. 2024, 6, 1509924. [Google Scholar] [CrossRef]
- Park, S.J.; Lee, G.E.; Cho, S.M.; Choi, E.H. Recent applications, future perspectives, and limitations of the CRISPR-Cas system. Mol. Ther. Nucleic Acids 2025, 36, 102634. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, W.P.; Wang, M.; Zhang, L.; Li, P.F. Nanozyme-assisted amplification-free CRISPR/Cas system realizes visual detection. Front. Bioeng. Biotech. 2024, 11, 1327498. [Google Scholar] [CrossRef] [PubMed]
- Ghouneimy, A.; Mahas, A.; Marsic, T.; Aman, R.; Mahfouz, M. CRISPR-Based Diagnostics: Challenges and Potential Solutions toward Point-of-Care Applications. ACS Synth. Biol. 2023, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]

| Primer Names | Primer Sequences (5′-3′) |
|---|---|
| pET28a-AapCas12b-F | ATGGGTCGCGGATCCGAATTCATGGCCGTAAAATCTATGAAAGTTAA |
| pET28a-AapCas12b-R | GTGGTGGTGGTGGTGCTCGAGTCAGATGTCCCCAGTGTTCTCA |
| B646L-sgDNA-F | GAAATTAATACGACTCACTATAGGGGTCTAGAGGACAGAATTTTTCAACGGGTGTGCCAATGGCCACTTTCCAGGTGGCAAAGCCCGTTGAGCTTCTCAAATCTGAGAAGTGGCACCGTATCCGATCACATTACCT |
| B646L-sgDNA-R | AGGTAATGTGATCGGATACGGTGCCACTTCTCAGATTTGAGAAGCTCAACGGGCTTTGCCACCTGGAAAGTGGCCATTGGCACACCCGTTGAAAAATTCTGTCCTCTAGACCCCTATAGTGAGTCGTATTAATTTC |
| ssDNA-probe | 5′-6-FAM-NNNNNNNNNNNN-BHQ1-3′ |
| RPA-B646L-F1 | CGCCATTATGCAGCCCACTCACCACGCAGA |
| RPA-B646L-R1 | GATAAGATTGATACCATGAGCAGTTACGGA |
| RPA-B646L-F2 | AGATTGGCACAAGTTCGGACATGTTGTTAACGCCA |
| RPA-B646L-R2 | TAGTGGAAGGGTATGTAAGAGCTGCAGAACTTTGA |
| PRRSV-UF | GCCCCTGCCCAYCACG |
| PRRSV-UR | TCGCCCTAATTGAATAGGTGA |
| CSFV-UF | CTGGGTGGTCTAAGTCCTGAGTA |
| CSFV-UR | GATTCAACTCCATGTGCCATGTA |
| PRV-gC-F1 | CGAGACCGAGGGCGTCTACAC |
| PRV-gC-R1 | GCCCATCATCAGCGCCTGC |
| PPV4-F1 | CTTTGCTTTGTCCAACGCAGA |
| PPV4-R1 | TAGATGTCCTGGCACAGATACTTGAC |
| PCV3-F1 | CTGTTATTTTGGATGATTTTTATG |
| PCV3-R1 | CACAGCCGTTACTTCACCC |
| B646L/p72-F | CGGGTGCGATGATGATTACC |
| B646L/p72-R | TCTCTTGCTCTGGATACGTTAATATGAC |
| B646L/p72-TaqMan | 5′-FAM-TCTCTTGCTCTGGATACGTTAATATGAC-BHQ1-3′ |
| β-actin-F: | ATGAAGATCAAGGTGAGTGCC |
| β-actin-R: | TCGTACTCCTGCTTGCTGATC |
| Sample Types | Samples Numbers | Detection Results (Positive Rates) | |
|---|---|---|---|
| RPA-AapCas12b-LFS | qPCR | ||
| Heart | 3 | 2/3 | 2/3 |
| Liver | 3 | 2/3 | 2/3 |
| Spleen | 3 | 2/3 | 2/3 |
| Lung | 3 | 2/3 | 2/3 |
| Kidney | 3 | 2/3 | 2/3 |
| Lymph node | 3 | 2/3 | 2/3 |
| serum | 5 | 1/5 | 1/5 |
| Blood | 5 | 2/5 | 2/5 |
| Oral swab | 6 | 2/6 | 2/6 |
| Total | 34 | 50% | 50% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, W.; Hao, W.; Chang, Y.; Zheng, W.; Lin, C.; Xu, Z.; Kang, X.; Chen, N.; Bai, J.; Zhu, J. Development of Visual Detection of African Swine Fever Virus Using CRISPR/AapCas12b Lateral Flow Strip Based on Viral Major Capsid Protein Gene B646L. Animals 2025, 15, 3274. https://doi.org/10.3390/ani15223274
Zheng W, Hao W, Chang Y, Zheng W, Lin C, Xu Z, Kang X, Chen N, Bai J, Zhu J. Development of Visual Detection of African Swine Fever Virus Using CRISPR/AapCas12b Lateral Flow Strip Based on Viral Major Capsid Protein Gene B646L. Animals. 2025; 15(22):3274. https://doi.org/10.3390/ani15223274
Chicago/Turabian StyleZheng, Wanglong, Weilin Hao, Yajing Chang, Wangli Zheng, Can Lin, Zijian Xu, Xilong Kang, Nanhua Chen, Jianfa Bai, and Jianzhong Zhu. 2025. "Development of Visual Detection of African Swine Fever Virus Using CRISPR/AapCas12b Lateral Flow Strip Based on Viral Major Capsid Protein Gene B646L" Animals 15, no. 22: 3274. https://doi.org/10.3390/ani15223274
APA StyleZheng, W., Hao, W., Chang, Y., Zheng, W., Lin, C., Xu, Z., Kang, X., Chen, N., Bai, J., & Zhu, J. (2025). Development of Visual Detection of African Swine Fever Virus Using CRISPR/AapCas12b Lateral Flow Strip Based on Viral Major Capsid Protein Gene B646L. Animals, 15(22), 3274. https://doi.org/10.3390/ani15223274








