Simple Summary
Dairy cows frequently suffer from mastitis, or inflammation of the udder, and that causes major economic loss to farmers. If farmers could breed cows with better natural resistance to mastitis, they could thus achieve an important goal for sustainable milk production. In this study, we investigated data on genetics and milk from Japanese Holstein cows over a ten-year period, to see if we could identify any links in the data related to udder health. We found cows with one particular genetic variation were statistically at much greater risk of developing mastitis. This information on dairy cow genetics may help cattle breeders raise healthier, and more productive herds.
Abstract
We evaluated four candidate SNPs (GC-NPFFR2 rs137147462, GC-NPFFR2 rs109452259, BRCA1 rs134817801, and DGAT1 p.K232A) previously reported in relation to mastitis or milk production traits, using 10,729 test-day phenotypic records collected over 10 years from 269 Japanese Holstein cows (Bos taurus) enrolled in the national Dairy Herd Improvement (DHI) program. Linear mixed models were used to estimate genotypic effects on somatic cell score (SCS) and to test multiple inheritance models. To assess clinical relevance, mastitis severity was further analyzed using categories defined by somatic cell counts (SCC). Among the SNPs tested, GC-NPFFR2 rs137147462 showed the clearest and most consistent association with SCS under a recessive model, with GG cows exhibiting higher SCS throughout lactation. Ordinal logistic regression confirmed a higher probability of progression to severe mastitis in GG cows. DGAT1 p.K232A showed additive effects, with the A allele increasing milk yield while lowering fat and protein percentages. AA cows also showed higher SCS under a modest recessive effect. BRCA1 rs134817801 and GC-NPFFR2 rs109452259 had minimal effects. These findings support GC-NPFFR2 rs137147462 as a promising marker for mastitis resistance and indicate the importance of considering not only additive but also recessive genetic models in genomic selection strategies.
1. Introduction
Bovine mastitis presents a major challenge for the dairy industry, as it leads to decreased milk yield, discarded milk, higher veterinary costs, and increased culling [1,2,3]. It is a complex condition to which environmental factors, herd management practices, and genetic predisposition all may contribute [4,5]. Losses to farmers may be substantial whether the mastitis is clinical or subclinical. Cases of the latter are particularly difficult to detect, although subclinical mastitis can reportedly be responsible for elevated bulk-tank somatic cell count (SCC) and degraded milk quality [1,2,5]. As well as causing economic losses, mastitis has an adverse impact on animal welfare and drives increased antimicrobial use at the herd level, making it an important challenge for the sustainability of dairy production [3,5,6,7].
Genetic variation in mastitis resistance is well established, although the heritability of clinical mastitis is generally low, ranging from 0.01 to 0.10 [8,9]. In contrast, somatic cell score (SCS), which is the logarithmic transformation of SCC, exhibits moderate heritability (0.15 to 0.20), and is widely used as an indicator trait for improving udder health through selective breeding [10]. SCCs are routinely recorded in testing under the Japanese national Dairy Herd Improvement (DHI) program, and reflect leukocyte infiltration during intramammary infection. As base-2 logarithmic transformation stabilizes any variance present in SCC data, and yields an approximately normal distribution of data for genetic evaluation, SCS has been widely adopted as a proxy indicator for udder health and predictions of milk loss and increased risk of clinical mastitis [5,10,11,12,13,14]. However, despite the success of genomic selection for production traits, the identification of robust and biologically relevant markers for mastitis resistance remains a challenge as shown in previous studies including TLR4 [15,16]. Single nucleotide polymorphisms (SNPs) represent a potentially fruitful line of research for establishing such genetic markers.
The GC-NPFFR2 region on BTA6 has been identified as a major source of signals associated with SCS in a large-scale, genome-wide association study (GWAS) in North American Holsteins [17]. That study involved two complementary approaches: one with an approximate generalized least squares (AGLS) method, and the other with a Bayesian linear mixed model implemented via BOLT-LMM (version 2.3.2) software [18,19]. GC encodes the vitamin D–binding protein, while NPFFR2 encodes a neuropeptide receptor implicated in neuro-immune and inflammatory regulation [20,21,22,23]. Within the GC-NPFFR2 region, the rs137147462 and rs109452259 SNPs appear to be the most promising for further investigation. Accordingly, we opted to target them for validation as independent genetic markers for SCS (and hence mastitis) in a Japanese Holstein population. We further opted to evaluate BRCA1 rs134817801 as a useful comparator SNP, because of its essential roles in DNA repair and immune function and its previously reported association with SCS in cattle [24]. We also targeted DGAT1 as a major quantitative trait locus (QTL) for milk fat synthesis, focusing on the well-known causal SNP (p.K232A) in that QTL. DGAT1 p.K232A is known to exert a strong effect on milk composition [25,26,27].
Against this background, in the current study, we aimed to evaluate the effects of the four candidate SNPs (GC-NPFFR2 rs137147462, GC-NPFFR2 rs109452259, BRCA1 rs134817801, and DGAT1 p.K232A) initially identified for their potential association with SCS, on test-day traits in Japanese Holstein cows. Using longitudinal records from the Japanese national DHI program, we investigated the associations of the four candidate SNPs with SCS and major milk composition traits. The two analytical frameworks we applied were a main-effect model, and an interaction model including genotype-by-lactation-stage effects. This approach was designed to identify reliable genetic markers for mastitis resistance and to determine whether the potential effects of these SNPs are consistent or changeable across different stages of lactation. Although this study provides detailed longitudinal insights, it was conducted using records from a single commercial herd, and the number of cows carrying certain rare genotypes was limited; therefore, the findings should be interpreted within this population context.
2. Materials and Methods
2.1. Animals and Phenotypic Records
Data were obtained from a commercial Holstein dairy farm in Japan participating in the national DHI program. All milk testing was conducted by the Hokkaido Dairy Milk Recording & Testing Association (Sapporo, Japan) under this program. SCC and milk composition were analyzed using a CombiFoss 7 or FT+ (Foss, Hillerød, Denmark). Milk samples were collected once daily from individual cows during either the morning or evening milking on each test day. A total of 10,729 test-day records from 269 cows, collected between 2010 and 2025, were analyzed. Records included milk yield, fat content (fat%), protein content (protein%), solids-not-fat content (SNF%), and SCC, which was converted to SCS for evaluation [12]. Each record was linked to cow identity, parity, and days in milk (DIM) for use in longitudinal models. Ear tissue samples were collected during routine ear tagging carried out in accordance with Japan’s national cattle identification system, and animals were subject to no experimental procedures; therefore, ethical approval was waived for this study. The 269 cows in the current study population included 256 individuals we had targeted in a previous, separate evaluation of linkage disequilibrium for another SNP [28].
2.2. DNA Extraction and Genotyping
Genomic DNA was extracted with the GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA). Four SNPs (GC-NPFFR2 rs137147462, GC-NPFFR2 rs109452259, BRCA1 rs134817801, and DGAT1 p.K232A) were genotyped with Custom TaqMan assays on a StepOne Plus Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA). Genotype classification followed the ARS-UCD2.0 reference genome, with details summarized in Table 1.

Table 1.
Target SNPs and genotype classification.
2.3. Evaluation of Genotypic Distributions
We evaluated Hardy–Weinberg equilibrium (HWE) and pairwise linkage disequilibrium (r2 and D′) across the four loci and computed exact 95% confidence intervals for allele frequencies with the Clopper–Pearson method, using R software (version 4.3).
2.4. Data Classification
Test-day records were classified into categories of DIM stage, parity, and season, and calving year. Detailed class definitions and descriptive statistics for all traits are provided in Supplementary Table S1.
2.5. Statistical Analysis
All primary analyses were conducted under a codominant framework, in which the three genotype classes (Wild, Heterozygote, Mutant) were treated as distinct fixed levels. For each SNP, two linear mixed models were constructed: the first estimated the main effect of genotype, and the second included genotype × DIM stage interactions to evaluate lactation-stage specific effects. In both models, the response variable was one of the test-day traits: milk yield, fat percentage, protein percentage, SNF percentage, or SCS. Fixed effects were DIM stage, parity, season, and calving year, and cow identity was included as a random effect to account for repeated measures. The models were specified as:
- Main effect model:
- Interaction model:where represents the test-day phenotype, is the fixed effect of genotype, is DIM stages, is the genotype-by-DIM interaction, is parity group, is season, is calving year, is the random effect of cowID and n denotes repeated test-day records within each cow, and is the residual error.
Least-squares means (LSMs) were estimated for each genotype, and multiple comparisons were adjusted using Tukey’s honestly significant difference (HSD) test at α = 0.05. Genotype effects were evaluated by Type III ANOVA, with significance levels denoted as * p < 0.05, ** p < 0.01, and *** p < 0.001. For the main-effect model, SNPs showing significant genotype effects were additionally subjected to false discovery rate (FDR) correction across SNP–trait combinations to confirm the robustness of the results. FDR correction was applied only within the subset of SNP–trait combinations that showed significant genotype effects in the main-effect codominant model, rather than across all tests performed.
In addition to the codominant framework, additive, dominant, recessive, and overdominant inheritance models were also evaluated for each SNP–trait combination. For a bi-allelic locus with alleles A (wild-type) and B (mutant) (as defined in Table 1), the codominant model treated genotype as a categorical variable with three levels (AA vs. AB vs. BB), representing each genotype separately; additive used B-allele dosage (AA = 0, AB = 1, BB = 2); dominant compared AA vs. AB+BB; recessive compared AA+AB vs. BB; and overdominant compared AB vs. AA+BB. Genotypes were coded numerically as (0/1/2 for additive; 0/1 for dominant, recessive, or overdominant), with definitions consistent with standard practice. The same fixed-and-random-effects structure was applied across all models.
To complement the mixed-model analyses, a simplified auxiliary analysis was performed to appraise clinical relevance based on the highest SCC during each lactation. An ordinal logistic regression was fitted for mastitis severity, defined as corresponding to healthy cattle (<200,000 cells/mL), subclinical cases (≥200,000 and <400,000), or clinical cases (≥400,000). Genotypes were modeled as categorical variables with AA as the reference. The proportional-odds assumption was evaluated with the Brant test and was not violated (Omnibus p = 0.388). These SCC cut-offs followed established individual-cow criteria and recent practice in udder-health research [29,30,31].
2.6. Software
All statistical analyses were performed in R (version 4.3) [32] using HardyWeinberg (version 1.7.8) for HWE tests [33], genetics (version 1.3.8.13) for LD analyses [34], lme4 (version 1.1-37) and lmerTest (version 3.1-3)for linear mixed models [35,36], emmeans (version 1.11.2) for least-squares means [37], MASS (version 7.3-65) for ordinal logistic regression [38], and broom (version 1.0.9) and brant (version 0.3.0) for regression diagnostics [39]. Figures were generated with ggplot2 (version 3.4.2) [40].
3. Results
3.1. Genetic Variation and Independence of Candidate SNPs
All four loci were polymorphic with adequate minor allele frequencies. Exact 95% confidence intervals for allele frequencies are included in Table 2. Pairwise LD across loci was weak, justifying separate analyses (Table 3).

Table 2.
Genotype and allele frequencies with HWE test results.

Table 3.
Genotype cross-tabulation and LD test results.
3.2. Main Effects of Genotype on Test-Day Milk Production and Somatic Cell Score
SCS was significantly affected only by GC-NPFFR2 rs137147462, among the four candidate SNPs (p = 0.0099), with GG cows consistently exhibiting the highest values; Tukey’s HSD showed a significant difference between GG and AA, with AG intermediate (Figure 1; Supplementary Table S2). The overall SNP effect remained significant after FDR correction (FDR-adjusted p = 0.040), confirming its robustness. For the same SNP, SNF% showed a small but significant overall difference (p = 0.047), although it became non-significant after FDR correction (FDR-adjusted p = 0.14), and pairwise contrasts were not significant. Four production traits (milk yield, fat%, protein%, and SNF) were significantly affected by DGAT1 p.K232A (all p < 0.001; FDR-adjusted p < 0.001), reproducing the established trade-off between lower yield and higher component content in KK cows. BRCA1 rs134817801 was associated with fat% (p = 0.0296), but this association became non-significant after FDR correction (FDR-adjusted p = 0.099). CC cows had the highest values, and similar non-additive tendencies were observed, for protein% (p = 0.073) and SNF% (p = 0.079), and AC cows showed lower values than both homozygotes for these parameters. GC-NPFFR2 rs109452259 showed no significant effects on any trait.
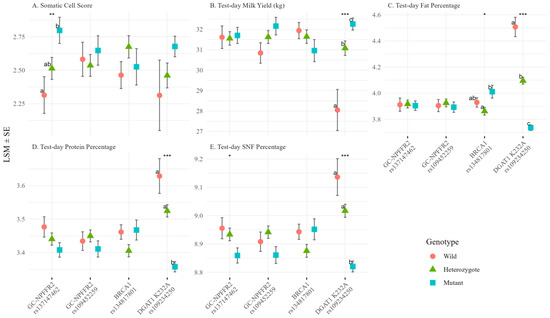
Figure 1.
Genotype effects on test-day traits. (A) Somatic Cell Score, (B) Test-day Milk Yield (kg), (C) Test-day Fat Percentage, (D) Test-day Protein Percentage, and (E) Test-day Solids-not-fat (SNF) Percentages are shown for four candidate SNPs. Least squares means (LSMs ± SE) were estimated using linear mixed models including DIM stage, parity group, and season as fixed effects and cow ID as a random effect. For genotype main effects, significance was assessed by ANOVA (* p < 0.05; ** p < 0.01, *** p < 0.001). Different letters indicate statistically significant differences between genotypes (Tukey’s HSD test, p < 0.05).
3.3. Genotype-by-DIM Interactions Affecting Milk Traits and SCS
Results are summarized in Figure 2 (multi-panel A–T) and Supplementary Table S3.
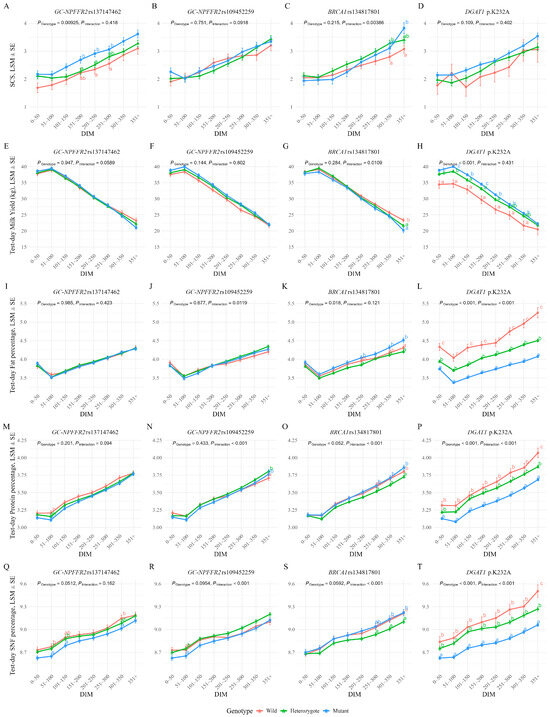
Figure 2.
LSM ± SE of test-day SCS and milk production traits across lactation stages by genotype. Panels (A–D): SCS; (E–H): milk yield; (I–L): fat%; (M–P): protein%; (Q–T): SNF. Each row corresponds to one SNP: GC-NPFFR2 rs137147462 (A,E,I,M,Q), GC-NPFFR2 rs109452259 (B,F,J,N,R), BRCA1 rs134817801 (C,G,K,O,S), and DGAT1 p.K232A (D,H,L,P,T). DIM were grouped by 50-day intervals up to 350 d, with recorded values after 350 d combined. Different letters indicate statistically significant differences between genotypes (Tukey’s HSD test, p < 0.05).
3.3.1. GC-NPFFR2 rs137147462
Cows with the GG genotype consistently showed higher SCS throughout lactation (overall p = 0.00925). At 151–200 DIM and 201–250 DIM, GG cows had significantly higher SCS than AG cows, while AA cows showed intermediate values (Tukey, p < 0.05; Figure 2A). A similar pattern was observed at 251–300 DIM, where GG cows showed significantly higher SCS than AA cows. These results provide supporting evidence for a recessive effect of the G allele. This SNP showed no significant main effects on protein% or SNF%, but the order of mean values by genotype (AA > AG > GG) remained stable throughout lactation. For protein%, the overall effect was not significant (p = 0.201), but AA cows tended to have higher values than GG cows between 51 and 150 DIM, with AG cows showing intermediate values. For SNF%, the overall effect approached significance (p = 0.0512). At 51–100 DIM, both AA and AG cows showed significantly higher SNF than GG cows (p < 0.05), and similar trends were observed at 101–150 DIM and 301–350 DIM. Taken together, these results indicate that although effect sizes were small, the descending order of AA > AG > GG was consistently maintained across lactation for both protein% and SNF% (Figure 2M,Q).
3.3.2. GC-NPFFR2 rs109452259
No significant effects were observed on SCS, but significant genotype-by-DIM interactions occurred for fat% (p = 0.0119), protein% (p < 0.001), and SNF% (p < 0.001). CC cows showed a marked decline in these three parameters from 201 to 250 DIM.
For protein%, significant pairwise differences were detected only in late lactation (351 DIM and after), where CA cows showed the lowest values, while AA and CC cows showed higher levels. For SNF%, significant differences were observed only at 51–100 DIM, with CA cows showing significantly lower values than AA and CC cows (p < 0.05; Figure 2J,N,R).
3.3.3. BRCA1 rs134817801
We detected no main SNP effect on SCS, but a significant interaction was detected (overall p = 0.00386). AC cows generally showed higher SCS than AA cows, while CC cows showed a sharply rising SCS from 150 DIM and their SCS exceeded that in AA cows in late lactation. At 301–350 DIM, AC cows showed a significantly higher SCS than AA cows, and CC cows showed a significantly higher SCS than AA cows at 351 DIM and after (p < 0.05; Figure 2C). For milk yield, AA cows had significantly higher values than AC and CC cows in late lactation (351 DIM and after; p < 0.05). CC cows consistently showed the highest fat%, protein%, and SNF%, with differences becoming more pronounced in mid-to-late stages of lactation (Figure 2G,K,O,S).
3.3.4. DGAT1 p.K232A
No significant effects on SCS were observed, but strong effects were found for other production traits (all p < 0.001). Milk yield descended (by genotype) in the order of AA > KA > KK, whereas fat%, protein%, and SNF% all showed values in a descending order of KK > KA > AA. Fat% showed significant pairwise differences between each of the three genotypes at every stage of lactation (p < 0.05; Figure 2L), and protein% and SNF%, differed significantly between the three genotypes at 351 DIM and after (p < 0.05; Figure 2P,T).
3.4. Evaluation of Genetic Inheritance Models
Four inheritance models (additive, dominant, recessive, and overdominant) were compared for each SNP–trait combination. The results for SCS evaluations, with significant effects (p < 0.05), are summarized in Table 4, and the comprehensive results for all traits are provided in Supplementary Table S4.

Table 4.
Summary of inheritance-model effects on SCS.
In these inheritance models, SCS was significantly affected by GC-NPFFR2 rs137147462 and DGAT1 p.K232A. For GC-NPFFR2 rs137147462, significant effects were detected under both the dominant (p = 0.037) and recessive (p = 0.0054) models, with the latter showing the strongest association; GG cows consistently exhibited higher SCS values than AA or AG cows. DGAT1 p.K232A also showed a significant recessive effect (p = 0.044), with AA cows having slightly higher SCS than KA and KK cows.
These findings support a predominantly recessive mode of inheritance for GC-NPFFR2 rs137147462 and indicate that the DGAT1 locus exerted a minor but statistically significant effect on SCS, whereas other loci showed no meaningful associations.
3.5. Supporting Analysis of SCC-Based Mastitis Severity Associated with GC-NPFFR2 rs137147462
Given that GC-NPFFR2 rs137147462 showed significant associations with SCS in multiple models, we further examined its clinical relevance using ordinal logistic regression with mastitis severity classified as healthy, subclinical mastitis (SCM), or clinical mastitis (CM). GG cows were likely to show a progression to more severe mastitis than AA cows (OR = 1.63; 95% CI 1.18–2.28; p = 0.0035), whereas AG cows showed no significance (Table 5). The proportional-odds assumption was not violated (Brant omnibus p = 0.39).

Table 5.
Association of GC-NPFFR2 rs137147462 with mastitis severity.
4. Discussion
4.1. Effects of Two Loci in the GC-NPFFR2 Region on SCS and SCC-Based Mastitis Traits
We targeted two SNPs in the GC-NPFFR2 region on BTA6 (rs137147462 and rs109452259) for investigation in this study, based on significant signals reported in a large GWAS in North American Holsteins [17]. In our study population, GC-NPFFR2 rs137147462 showed a robust association with SCS, detected primarily under a recessive model (p = 0.0054), with weaker support under a dominant model (p = 0.037). Furthermore, GG cows showed consistently higher SCS values than AA or AG cows (overall p = 0.00925). Mastitis-severity analyses provided further evidence for this recessive pattern, which occurred without clear penalties on milk yield, fat%, or protein%; however, we noted a non-significant trend toward lower SNF%. Within the DIM-adjusted framework, the GG genotype showed persistently higher SCS values across the lactation period, with differences versus other genotypes reaching significance at 151–200 and 201–250 DIM (vs. AG) and again at 251–300 DIM (vs. AA). These results indicate a persistent recessive effect, rather than a stage-restricted fluctuation. For milk-composition traits, genotype rankings were small and stable across stages. Milk lactose percentage is negatively associated with SCC in dairy cattle and declines during or after intramammary inflammation; similar patterns are reported in buffaloes and small ruminants [41,42,43,44]. Because lactose is the principal milk osmole, epithelial damage and leaky tight junctions reduce the amount of lactose retained in the alveolar lumen, leading to lower milk yield, and altered mineral balance (mediated by increased sodium and potassium concentrations) [45,46,47,48,49,50]. Considering this phenomenon, lactose is a promising predictor of subclinical mastitis, and genomic studies show overlap between lactose-associated regions and loci for mastitis/udder-health traits [42,51,52,53,54]. In our data, protein% did not differ consistently by genotype, whereas GG cows (the higher-SCS genotype) showed a slightly lower SNF (overall effect approaching significance). Given that lactose dominates SNF, this pattern is most plausibly explained by a modest reduction in lactose rather than protein, aligning with the established SCC–lactose antagonism [41,42].
The results for GC-NPFFR2 rs109452259 showed a contrasting pattern. This SNP exerted no sustained effect on SCS, but it exhibited stage-dependent associations with milk composition, consistent with significant genotype-by-DIM interactions for fat%, protein%, and SNF%. Based on these findings, we suggest that rs109452259 reflects a haplotypic structure in the specific chromosomal region rather than an independent causal variant.
Although mastitis resistance is regarded as generally polygenic [15,55], our findings indicate that a single SNP, GC-NPFFR2 rs137147462, can exert a consistent influence on SCS when non-additive inheritance is considered. Trade-offs between mastitis resistance and milk yield have previously been reported [56,57], whereas a copy number variant (CNV) in the GC region has been to shown reduce resistance but increase milk production [58]. Our results suggest that the effect of GC-NPFFR2 rs137147462 may not be governed by such trade-offs, and raise the possibility that this SNP is related to structural variation or has a functional role in itself. Such a marker could play a valuable role in selective breeding, but validation in larger and more diverse populations is essential. GC encodes vitamin D-binding protein, regulating vitamin D metabolism and immune function [20], and NPFFR2 is linked to anti-inflammatory macrophage activation [21,22,23].
4.2. Effects of BRCA1 rs134817801 on SCS and Milk Composition Traits
BRCA1 is a tumor-suppressor gene involved in DNA repair and cell-cycle control [59,60]. In cattle, it lies on BTA19 within a QTL for SCS and milk composition traits [61,62,63], and its missense variant, rs134817801, is reportedly associated with SCS in Chinese Holsteins [24].
In our Japanese Holstein population, we detected no main BRCA1 rs134817801 effect on SCS; however, we did detect a significant interaction effect for genotype-by-DIM, with CC cows showing sharply increasing SCS in late lactation and AC cows showing generally elevated SCS values. Milk yield also showed a stage-dependent reversal, with higher values in AA than AC or CC cows in later lactation. In contrast, this SNP had clear effects on milk composition: CC cows consistently showed the highest fat%, protein%, and SNF% values, whereas heterozygotes showed the lowest. Model comparisons supported recessive and overdominant inheritance, consistent with a pattern of heterozygote disadvantage.
Compared with GC-NPFFR2 rs109452259, which showed heterozygote advantage for SNF%, BRCA1 rs134817801 exhibited the opposite pattern, underscoring the diversity of non-additive effects among mastitis-associated loci. Although the additive signal of this SNP is limited and it may be overlooked in conventional GWAS, its detection through candidate-gene approaches highlights the value of longitudinal and non-additive models in uncovering hidden genetic effects.
4.3. Effects of DGAT1 p.K232A on Milk Composition and Mastitis-Related Traits
DGAT1 encodes a key enzyme for triglyceride synthesis, and its p.K232A polymorphism is a well-characterized causal mutation located in a major QTL on BTA14. This variant exerts a substantial effect on milk fat content and smaller antagonistic effects on milk yield [25,64], findings that have been consistently replicated across breeds and confirmed in meta-analyses [27].
In our study population, DGAT1 p.K232A showed a significant association with SCS under a recessive model (p = 0.044), with AA cows exhibiting higher SCS than KA or KK cows. Given that the K-homozygote was rare in our cohort (KK 4.8%, n = 13; see genotype counts in Table 2), estimates for contrasts involving KK and made in stratified analyses carry wider uncertainty and should be interpreted with caution. Our results provide interesting evidence in areas where previous reports are inconsistent: some reports have suggested favorable effects of the K allele [65,66], whereas others have found no meaningful relationship [67,68]. Taking our data and the previous reports collectively, we suggest that any link between DGAT1 and udder health is likely minor and population specific.
In contrast, we confirmed strong additive effects on milk production traits for this SNP: KK cows produced less milk but showed higher fat, protein, and SNF percentages, patterns that intensified in later lactation. Model comparisons suggested additive inheritance for milk yield and milk fat content, and recessive effects for protein and SNF, consistent with prior meta-analyses [27].
According to the inheritance–model comparisons summarized in Supplementary Table S4, fat% followed an additive pattern, whereas both protein% and SNF% showed their most markedly statistically significant results under a recessive coding (lowest p-values), with AA cows being lowest for both traits. Notably, the proportional decline in SNF for AA cows appeared larger than that in protein% (visually evident in Figure 2), which is unlikely to be explained by reduced protein alone. Given that lactose constitutes the major component of SNF, this pattern may indicate a concurrent reduction in lactose, consistent with substrate reallocation under DGAT1 p.K232A. Taking the relevant results together, we suggest that p.K232A exerted clear additive effects on fat and milk yield, while minor recessive decreases in protein and SNF were observed in AA cows. Such a phenomenon may be best interpreted as a metabolic trade-off within mammary synthesis rather than a change in udder-health status. Such an interpretation is consistent with the SCS–SNF antagonism noted earlier for GC-NPFFR2 rs137147462.
4.4. Limitations, Perspectives, and Practical Implications
This study has several limitations that should be acknowledged. The sample size was moderate (269 cows), and although genotype frequencies were generally well balanced, some less frequent genotypes, particularly p.K232A KK, were underrepresented. Consequently, the statistical power to detect recessive effects was limited when the recessive homozygote class was rare, and precision further declined in stratified analyses. The dataset originated from a single commercial herd, which may limit the generalizability of the findings to wider Holstein populations. Generalizability thus needs to be confirmed with analysis beyond this single-herd setting, to confirm reproducibility in independent herds. However, in the mixed model framework, parity, season, calving year, and DIM stage were included as fixed effects, and cow identity was included as a random effect, thereby accounting for variation related to management, environment, and repeated measurements within cows. These model structures help mitigate the potential effects of farm-level management or temporal factors such as feed changes and climate fluctuations. From a broader perspective, this work provides a targeted validation of SNPs previously identified by GWAS, demonstrating that candidate variants can yield consistent phenotypic associations when evaluated under appropriate genetic models. The use of mixed models across DIM stages allowed us to detect genotype effects that would likely be masked in cross-sectional or purely additive analyses. While future research using high-density SNP arrays or whole-genome sequencing will be essential to pinpoint causal variants and resolve the surrounding haplotype structures, the present study highlights the continuing value of hypothesis-driven validation studies in bridging large-scale genomic signals with practical genetic markers. Such markers can support genomic selection strategies that improve udder health without compromising milk productivity, thereby enhancing the sustainability of dairy production.
5. Conclusions
In conclusion, we suggest that non-additive genetic signals relevant to udder health can be elucidated through analyses targeting longitudinal test-day records with explicit inheritance-model comparisons. Going forward, research in the near future should involve validation in larger multi-herd and international populations, and functional studies to elucidate the relevant biological mechanism. In the longer term, integrating validated markers into genomic selection programs and clarifying their physiological roles may enhance udder health and milk productivity through sustainable dairy breeding. The expected DGAT1 p.K232A associations served as an internal positive control, supporting the robustness of our analytical framework.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ani15223239/s1: Table S1: Descriptive statistics; Table S2: Summary of genotype main effects on test-day traits; Table S3: LSM ± SE, Tukey-adjusted groupings, and p-values of genotype effects by DIM stage for five milk traits; Table S4: Full results of inheritance-model comparisons for all SNP-trait combinations.
Author Contributions
Conceptualization, Y.A., T.A. and N.M.; methodology, Y.A. and N.M.; data curation, Y.A., N.N., M.A., Y.I. and S.W.; formal analysis, Y.A.; investigation, Y.A., N.N., M.A., Y.I. and S.W.; visualization, Y.A.; writing—original draft preparation, Y.A.; writing—review and editing, T.A. and N.M.; supervision, T.A. and N.M.; funding acquisition, N.M. All authors have read and agreed to the published version of the manuscript.
Funding
Financial support was provided through the Livestock Promotional Subsidy of the Japan Racing Association (JRA). The funding agency had no influence on the study design, data handling, interpretation, manuscript writing, or publication decision.
Institutional Review Board Statement
Ethical review and approval were waived for this study because it used ear tissue collected during routine ear tagging under Japan’s national cattle identification program.
Informed Consent Statement
Written informed consent was obtained from the owner of the animals involved in this study.
Data Availability Statement
The datasets analyzed during the current study are publicly available at Figshare: https://doi.org/10.6084/m9.figshare.30071734.
Acknowledgments
We thank SLC farm for their cooperation and for providing DHI records, and the staff of the Hokkaido Dairy Milk Recording & Testing Association for their helpful explanation regarding the milk testing equipment used in this study. We also thank Henry Smith for English language editing, and Yu-Chang Lai, Md Mahfuzur Rahman, Hui-Wen Chen, Al Asmaul Husna, and Md Nazmul Hasan for their technical assistance. We acknowledge the contributions of graduate students who assisted with DNA sampling and PCR experiments. During the preparation of this manuscript, the authors used ChatGPT (OpenAI, San Francisco, CA; versions GPT-4o and GPT-5) for language editing only (grammar and phrasing). The authors reviewed and edited all text and take full responsibility for the final content.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Seegers, H.; Fourichon, C.; Beaudeau, F.O. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet. Res. 2003, 34, 475–491. [Google Scholar] [CrossRef]
- Halasa, T.; Huijps, K.; Østerås, O.; Hogeveen, H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q. 2007, 29, 18–31. [Google Scholar] [CrossRef]
- Hogeveen, H.; Huijps, K.; Lam, T.J. Economic aspects of mastitis: New developments. N. Z. Vet. J. 2011, 59, 16–23. [Google Scholar] [CrossRef]
- Bradley, A. Bovine mastitis: An evolving disease. Vet. J. 2002, 164, 116–128. [Google Scholar] [CrossRef]
- Ruegg, P.L. A 100-Year Review: Mastitis detection, management, and prevention. J. Dairy Sci. 2017, 100, 10381–10397. [Google Scholar] [CrossRef]
- McCubbin, K.D.; de Jong, E.; Lam, T.J.G.M.; Kelton, D.F.; Middleton, J.R.; McDougall, S.; De Vliegher, S.; Godden, S.; Rajala-Schultz, P.J.; Rowe, S.; et al. Invited review: Selective use of antimicrobials in dairy cattle at drying-off. J. Dairy Sci. 2022, 105, 7161–7189. [Google Scholar] [CrossRef]
- de Jong, E.; McCubbin, K.D.; Speksnijder, D.; Dufour, S.; Middleton, J.R.; Ruegg, P.L.; Lam, T.J.G.M.; Kelton, D.F.; McDougall, S.; Godden, S.M.; et al. Invited review: Selective treatment of clinical mastitis in dairy cattle. J. Dairy Sci. 2023, 106, 3761–3778. [Google Scholar] [CrossRef]
- Zwald, N.R.; Weigel, K.A.; Chang, Y.M.; Welper, R.D.; Clay, J.S. Genetic selection for health traits using producer-recorded data. I. Incidence rates, heritability estimates, and sire breeding values. J. Dairy Sci. 2004, 87, 4287–4294. [Google Scholar] [CrossRef] [PubMed]
- Bloemhof, S.; Dejong, G.; Dehaas, Y. Genetic parameters for clinical mastitis in the first three lactations of Dutch Holstein cattle. Vet. Microbiol. 2009, 134, 165–171. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rupp, R.; Boichard, D. Genetics of resistance to mastitis in dairy cattle. Vet. Res. 2003, 34, 671–688. [Google Scholar] [CrossRef] [PubMed]
- Schutz, M.M. Genetic evaluation of somatic cell scores for United States dairy cattle. J. Dairy Sci. 1994, 77, 2113–2129. [Google Scholar] [CrossRef]
- Wiggans, G.R.; Shook, G.E. A Lactation Measure of Somatic Cell Count. J. Dairy Sci. 1987, 70, 2666–2672. [Google Scholar] [CrossRef]
- Miglior, F.; Fleming, A.; Malchiodi, F.; Brito, L.F.; Martin, P.; Baes, C.F. A 100-Year Review: Identification and genetic selection of economically important traits in dairy cattle. J. Dairy Sci. 2017, 100, 10251–10271. [Google Scholar] [CrossRef]
- Ali, A.K.A.; Shook, G.E. An Optimum Transformation for Somatic Cell Concentration in Milk1. J. Dairy Sci. 1980, 63, 487–490. [Google Scholar] [CrossRef]
- Sahana, G.; Cai, Z.; Sanchez, M.P.; Bouwman, A.C.; Boichard, D. Invited review: Good practices in genome-wide association studies to identify candidate sequence variants in dairy cattle. J. Dairy Sci. 2023, 106, 5218–5241. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, M.; Kumar, H.; Nayak, S.S.; Rajawat, D.; Parida, S.; Bhushan, B.; Sharma, A.; Dutt, T. Molecular characterization of CRBR2 fragment of TLR4 gene in association with mastitis in Vrindavani cattle. Microb. Pathog. 2022, 165, 105483. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, L.; Prakapenka, D.; Vanraden, P.M.; Cole, J.B.; Da, Y. A Large-Scale Genome-Wide Association Study in U.S. Holstein Cattle. Front. Genet. 2019, 10, 412. [Google Scholar] [CrossRef]
- Loh, P.R.; Tucker, G.; Bulik-Sullivan, B.K.; Vilhjalmsson, B.J.; Finucane, H.K.; Salem, R.M.; Chasman, D.I.; Ridker, P.M.; Neale, B.M.; Berger, B.; et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015, 47, 284–290. [Google Scholar] [CrossRef]
- Loh, P.R.; Kichaev, G.; Gazal, S.; Schoech, A.P.; Price, A.L. Mixed-model association for biobank-scale datasets. Nat. Genet. 2018, 50, 906–908. [Google Scholar] [CrossRef] [PubMed]
- Bouillon, R.; Schuit, F.; Antonio, L.; Rastinejad, F. Vitamin D Binding Protein: A Historic Overview. Front. Endocrinol. 2020, 10, 910. [Google Scholar] [CrossRef]
- Cikos, S.; Gregor, P.; Koppel, J. Sequence and tissue distribution of a novel G-protein-coupled receptor expressed prominently in human placenta. Biochem. Biophys. Res. Commun. 1999, 256, 352–356. [Google Scholar] [CrossRef]
- Mollereau, C.; Mazarguil, H.; Marcus, D.; Quelven, I.; Kotani, M.; Lannoy, V.; Dumont, Y.; Quirion, R.; Detheux, M.; Parmentier, M.; et al. Pharmacological characterization of human NPFF(1) and NPFF(2) receptors expressed in CHO cells by using NPY Y(1) receptor antagonists. Eur. J. Pharmacol. 2002, 451, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Waqas, S.F.H.; Hoang, A.C.; Lin, Y.-T.; Ampem, G.; Azegrouz, H.; Balogh, L.; Thuróczy, J.; Chen, J.-C.; Gerling, I.C.; Nam, S.; et al. Neuropeptide FF increases M2 activation and self-renewal of adipose tissue macrophages. J. Clin. Investig. 2017, 127, 2842–2854. [Google Scholar] [CrossRef]
- Yuan, Z.; Li, J.; Li, J.; Zhang, L.; Gao, X.; Gao, H.J.; Xu, S. Investigation on BRCA1 SNPs and its effects on mastitis in Chinese commercial cattle. Gene 2012, 505, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Grisart, B.; Coppieters, W.; Farnir, F.; Karim, L.; Ford, C.; Berzi, P.; Cambisano, N.; Mni, M.; Reid, S.; Simon, P.; et al. Positional candidate cloning of a QTL in dairy cattle: Identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 2002, 12, 222–231. [Google Scholar] [CrossRef]
- Thaller, G.; Krämer, W.; Winter, A.; Kaupe, B.; Erhardt, G.; Fries, R. Effects of DGAT1 variants on milk production traits in German cattle breeds. J. Anim. Sci. 2003, 81, 1911–1918. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, P.; Rashidi, A. Strong evidence for association between K232A polymorphism of the DGAT1 gene and milk fat and protein contents: A meta-analysis. J. Dairy Sci. 2023, 106, 2573–2587. [Google Scholar] [CrossRef]
- Akiyama, Y.; Ando, T.; Nozaki, N.; Arif, M.; Ide, Y.; Wang, S.; Miura, N. Strong Linkage Disequilibrium and Proxy Effect of PPP1R16A rs109146371 for DGAT1 K232A in Japanese Holstein Cattle. Genes 2025, 16, 1000. [Google Scholar] [CrossRef]
- Lipkens, Z.; Piepers, S.; De Visscher, A.; De Vliegher, S. Evaluation of test-day milk somatic cell count information to predict intramammary infection with major pathogens in dairy cattle at drying off. J. Dairy Sci. 2019, 102, 4309–4321. [Google Scholar] [CrossRef]
- Costa, A.; Bovenhuis, H.; Egger-Danner, C.; Fuerst-Waltl, B.; Boutinaud, M.; Guinard-Flament, J.; Obritzhauser, W.; Visentin, G.; Penasa, M. Mastitis has a cumulative and lasting effect on milk yield and lactose content in dairy cows. J. Dairy Sci. 2025, 108, 635–650. [Google Scholar] [CrossRef]
- Smith, K.L.; Hillerton, J.E.; Harmon, R.J. Guidelines on Normal and Abnormal Raw Milk Based on Somatic Cell Counts and Signs of Clinical Mastitis; National Mastitis Council: Madison, WI, USA, 2001. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2025; Available online: https://www.R-project.org (accessed on 1 August 2025).
- Graffelman, J. Exploring Diallelic Genetic Markers: The HardyWeinberg Package. J. Stat. Softw. 2015, 64, 1–23. [Google Scholar] [CrossRef]
- Warnes, G.R.; Bolker, B.; Lumley, T. R Package, version 1.3.8.1.2. Genetics: Population Genetics. R Foundation for Statistical Computing: Vienna, Austria, 2022. Available online: https://CRAN.R-project.org/package=genetics (accessed on 1 August 2025).
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Kuznetsova, A.; Brockhoff, P.B.; Christensen, R.H.B. lmerTest Package: Tests in Linear Mixed Effects Models. J. Stat. Softw. 2017, 82, 1–26. [Google Scholar] [CrossRef]
- Lenth, R.V. R Package, version 1.8.8. emmeans: Estimated Marginal Means, aka Least-Squares Means. R Foundation for Statistical Computing: Vienna, Austria, 2023. Available online: https://CRAN.R-project.org/package=emmeans (accessed on 1 August 2025).
- Venables, W.N.; Ripley, B.D. Random and Mixed Effects. In Modern Applied Statistics with S; Venables, W.N., Ripley, B.D., Eds.; Springer: New York, NY, USA, 2002; pp. 271–300. [Google Scholar]
- Brant, R. Assessing proportionality in the proportional odds model for ordinal logistic regression. Biometrics 1990, 46, 1171–1178. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Costa, A.; Egger-Danner, C.; Mészáros, G.; Fuerst, C.; Penasa, M.; Sölkner, J.; Fuerst-Waltl, B. Genetic associations of lactose and its ratios to other milk solids with health traits in Austrian Fleckvieh cows. J. Dairy Sci. 2019, 102, 4238–4248. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.; Lopez-Villalobos, N.; Sneddon, N.W.; Shalloo, L.; Franzoi, M.; De Marchi, M.; Penasa, M. Invited review: Milk lactose—Current status and future challenges in dairy cattle. J. Dairy Sci. 2019, 102, 5883–5898. [Google Scholar] [CrossRef]
- Stelwagen, K.; Farr, V.C.; McFadden, H.A. Alteration of the Sodium to Potassium Ratio in Milk and the Effect on Milk Secretion in Goats. J. Dairy Sci. 1999, 82, 52–59. [Google Scholar] [CrossRef]
- Carta, S.; Cesarani, A.; Correddu, F.; Macciotta, N.P.P. Understanding the phenotypic and genetic background of the lactose content in Sarda dairy sheep. J. Dairy Sci. 2023, 106, 3312–3320. [Google Scholar] [CrossRef]
- Zhao, X.; Lacasse, P. Mammary tissue damage during bovine mastitis: Causes and control. J. Anim. Sci. 2008, 86, 57–65. [Google Scholar] [CrossRef]
- Herve, L.; Quesnel, H.; Veron, M.; Portanguen, J.; Gross, J.J.; Bruckmaier, R.M.; Boutinaud, M. Milk yield loss in response to feed restriction is associated with mammary epithelial cell exfoliation in dairy cows. J. Dairy Sci. 2019, 102, 2670–2685. [Google Scholar] [CrossRef]
- Wellnitz, O.; Bruckmaier, R.M. Invited review: The role of the blood–milk barrier and its manipulation for the efficacy of the mammary immune response and milk production. J. Dairy Sci. 2021, 104, 6376–6388. [Google Scholar] [CrossRef]
- Visentin, G.; Penasa, M.; Niero, G.; Cassandro, M.; De Marchi, M. Phenotypic characterisation of major mineral composition predicted by mid-infrared spectroscopy in cow milk. Ital. J. Anim. Sci. 2018, 17, 549–556. [Google Scholar] [CrossRef]
- Costa, A.; Lopez-Villalobos, N.; Visentin, G.; De Marchi, M.; Cassandro, M.; Penasa, M. Heritability and repeatability of milk lactose and its relationships with traditional milk traits, somatic cell score and freezing point in Holstein cows. Animal 2019, 13, 909–916. [Google Scholar] [CrossRef]
- Singh, A.; Kumar, A.; Thakur, M.S.; Khare, V.; Jain, A.; Tiwari, S.P. Genetic analysis of milk minerals in dairy cattle: A review. J. Appl. Genet. 2024, 65, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.; Schwarzenbacher, H.; Mészáros, G.; Fuerst-Waltl, B.; Fuerst, C.; Sölkner, J.; Penasa, M. On the genomic regions associated with milk lactose in Fleckvieh cattle. J. Dairy Sci. 2019, 102, 10088–10099. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimie, E.; Ebrahimi, F.; Ebrahimi, M.; Tomlinson, S.; Petrovski, K.R. A large-scale study of indicators of sub-clinical mastitis in dairy cattle by attribute weighting analysis of milk composition features: Highlighting the predictive power of lactose and electrical conductivity. J. Dairy Res. 2018, 85, 193–200. [Google Scholar] [CrossRef]
- Meredith, B.; Lynn, D.; Berry, D.; Kearney, F.; Bradley, D.; Finlay, E.; Fahey, A. A genome-wide association study for somatic cell score using the Illumina high-density bovine beadchip identifies several novel QTL potentially related to mastitis susceptibility. Front. Genet. 2013, 4, 229. [Google Scholar] [CrossRef]
- Tiezzi, F.; Parker-Gaddis, K.L.; Cole, J.B.; Clay, J.S.; Maltecca, C. A Genome-Wide Association Study for Clinical Mastitis in First Parity US Holstein Cows Using Single-Step Approach and Genomic Matrix Re-Weighting Procedure. PLoS ONE 2015, 10, e0114919. [Google Scholar] [CrossRef]
- Sahana, G.; Guldbrandtsen, B.; Thomsen, B.; Holm, L.E.; Panitz, F.; Brøndum, R.F.; Bendixen, C.; Lund, M.S. Genome-wide association study using high-density single nucleotide polymorphism arrays and whole-genome sequences for clinical mastitis traits in dairy cattle. J. Dairy Sci. 2014, 97, 7258–7275. [Google Scholar] [CrossRef]
- Olsen, H.G.; Knutsen, T.M.; Lewandowska-Sabat, A.M.; Grove, H.; Nome, T.; Svendsen, M.; Arnyasi, M.; Sodeland, M.; Sundsaasen, K.K.; Dahl, S.R.; et al. Fine mapping of a QTL on bovine chromosome 6 using imputed full sequence data suggests a key role for the group-specific component (GC) gene in clinical mastitis and milk production. Genet. Sel. Evol. 2016, 48, 79. [Google Scholar] [CrossRef]
- Cai, Z.; Guldbrandtsen, B.; Lund, M.S.; Sahana, G. Prioritizing candidate genes post-GWAS using multiple sources of data for mastitis resistance in dairy cattle. BMC Genom. 2018, 19, 656. [Google Scholar] [CrossRef]
- Lee, Y.-L.; Takeda, H.; Costa Monteiro Moreira, G.; Karim, L.; Mullaart, E.; Coppieters, W.; Appeltant, R.; Veerkamp, R.F.; Groenen, M.A.M.; Georges, M.; et al. A 12 kb multi-allelic copy number variation encompassing a GC gene enhancer is associated with mastitis resistance in dairy cattle. PLoS Genet. 2021, 17, e1009331. [Google Scholar] [CrossRef]
- Miki, Y.; Swensen, J.; Shattuck-Eidens, D.; Futreal, P.A.; Harshman, K.; Tavtigian, S.; Liu, Q.; Cochran, C.; Bennett, L.M.; Ding, W.; et al. A Strong Candidate for the Breast and Ovarian Cancer Susceptibility Gene BRCA1. Science 1994, 266, 66–71. [Google Scholar] [CrossRef]
- Roy, R.; Chun, J.; Powell, S.N. BRCA1 and BRCA2: Different roles in a common pathway of genome protection. Nat. Rev. Cancer 2011, 12, 68–78. [Google Scholar] [CrossRef]
- Bennewitz, J.; Reinsch, N.; Grohs, C.; Levéziel, H.; Malafosse, A.; Thomsen, H.; Xu, N.; Looft, C.; Kühn, C.; Brockmann, G.A.; et al. Combined analysis of data from two granddaughter designs: A simple strategy for QTL confirmation and increasing experimental power in dairy cattle. Genet. Sel. Evol. 2003, 35, 319. [Google Scholar] [CrossRef] [PubMed]
- Bennewitz, J.R.; Reinsch, N.; Guiard, V.; Fritz, S.; Thomsen, H.; Looft, C.; Kühn, C.; Schwerin, M.; Weimann, C.; Erhardt, G.; et al. Multiple Quantitative Trait Loci Mapping With Cofactors and Application of Alternative Variants of the False Discovery Rate in an Enlarged Granddaughter Design. Genetics 2004, 168, 1019–1027. [Google Scholar] [CrossRef]
- Daetwyler, H.D.; Schenkel, F.S.; Sargolzaei, M.; Robinson, J.A.B. A Genome Scan to Detect Quantitative Trait Loci for Economically Important Traits in Holstein Cattle Using Two Methods and a Dense Single Nucleotide Polymorphism Map. J. Dairy Sci. 2008, 91, 3225–3236. [Google Scholar] [CrossRef] [PubMed]
- Winter, A.; Kramer, W.; Werner, F.A.; Kollers, S.; Kata, S.; Durstewitz, G.; Buitkamp, J.; Womack, J.E.; Thaller, G.; Fries, R. Association of a lysine-232/alanine polymorphism in a bovine gene encoding acyl-CoA:diacylglycerol acyltransferase (DGAT1) with variation at a quantitative trait locus for milk fat content. Proc. Natl. Acad. Sci. USA 2002, 99, 9300–9305. [Google Scholar] [CrossRef]
- Barbosa Da Silva, M.V.G.; Sonstegard, T.S.; Thallman, R.M.; Connor, E.E.; Schnabel, R.D.; Van Tassell, C.P. Characterization of DGAT1 Allelic Effects in a Sample of North American Holstein Cattle. Anim. Biotechnol. 2010, 21, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Manga, I.; Říha, H. The DGAT1 gene K232A mutation is associated with milk fat content, milk yield and milk somatic cell count in cattle (Short Communication). Arch. Anim. Breed. 2011, 54, 257–263. [Google Scholar] [CrossRef]
- Bovenhuis, H.; Visker, M.H.P.W.; Van Valenberg, H.J.F.; Buitenhuis, A.J.; Van Arendonk, J.A.M. Effects of the DGAT1 polymorphism on test-day milk production traits throughout lactation. J. Dairy Sci. 2015, 98, 6572–6582. [Google Scholar] [CrossRef] [PubMed]
- Bobbo, T.; Tiezzi, F.; Penasa, M.; De Marchi, M.; Cassandro, M. Short communication: Association analysis of diacylglycerol acyltransferase (DGAT1) mutation on chromosome 14 for milk yield and composition traits, somatic cell score, and coagulation properties in Holstein bulls. J. Dairy Sci. 2018, 101, 8087–8091. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).