Development of a Sex-Specific Marker for the Chinese Hooksnout Carp Opsariichthys bidens Günther, 1873 Based on Whole-Genome Resequencing and Bulked Segregant Analysis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample and DNA Extraction
2.2. Library Construction and BSA Sequencing
2.3. Initial Data Processing and Quality Assessment
2.4. Screening of Sex-Specific Regions
2.5. Design of Primers for Sex-Specific Regions and Their Screening Validation
3. Results
3.1. Sample Information
3.2. Sequencing Data Statistics
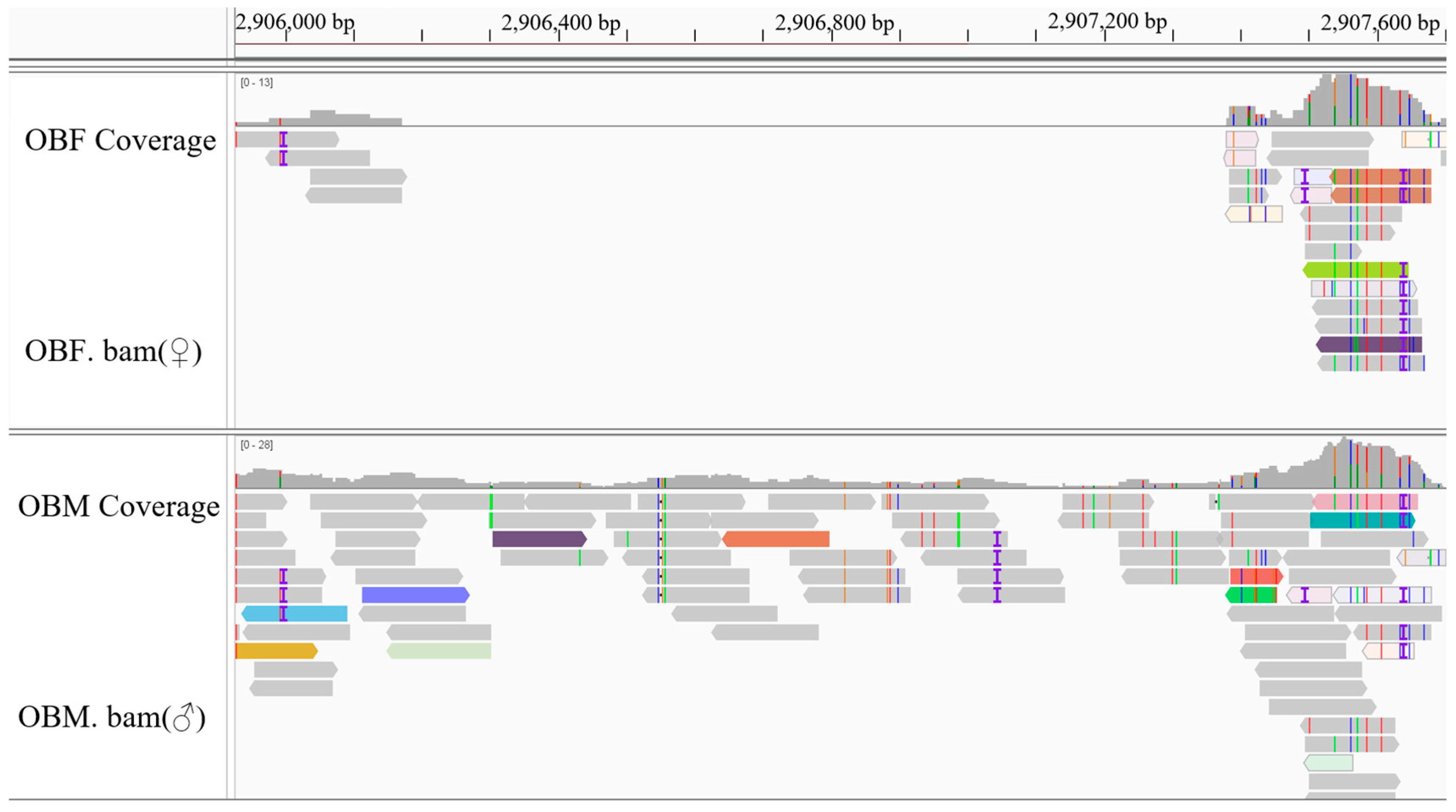
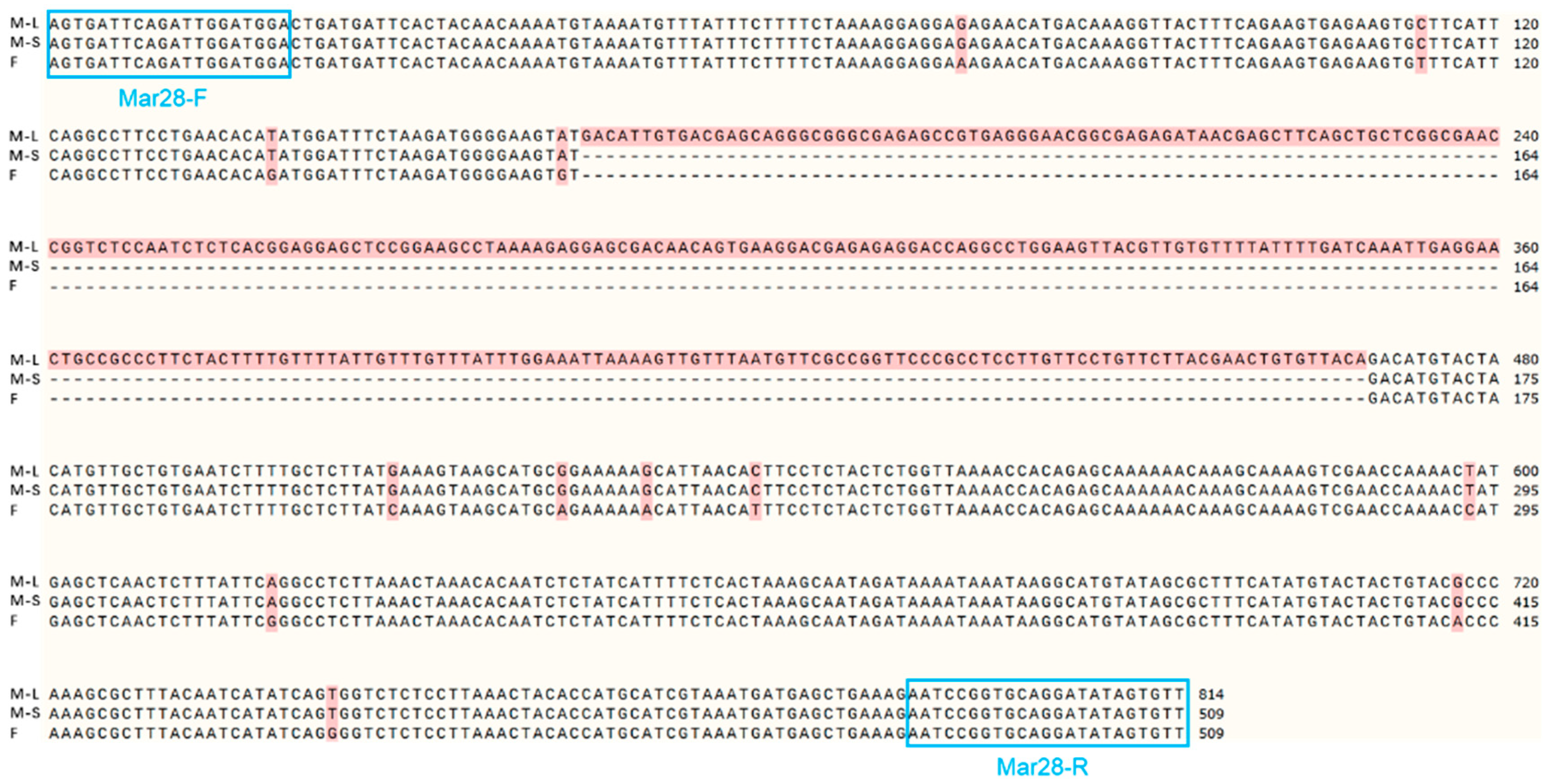
3.3. Screening and Validation of Sex-Specific Markers
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stelkens, R.B.; Wedekind, C. Environmental sex reversal, Trojan sex genes, and sex ratio adjustment: Conditions and population consequences. Mol. Ecol. 2010, 19, 627–646. [Google Scholar] [CrossRef]
- Anastasiadi, D.; Vandeputte, M.; Sánchez-Baizán, N.; Allal, F.; Piferrer, F. Dynamic epimarks in sex-related genes predict gonad phenotype in the European sea bass, a fish with mixed genetic and environmental sex determination. Epigenetics 2018, 13, 988–1011. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, X.; Bian, C.; Jiao, K.; Zhang, L.; Huang, Y.; Yang, W.; Li, Y.; Shi, G.; Huang, Y.; et al. Allelic variation and duplication of the dmrt1 were associated with sex chromosome turnover in three representative Scatophagidae fish species. Commun. Biol. 2025, 8, 627. [Google Scholar] [CrossRef]
- Xue, L.; Guo, X.; Zhou, Y.; Wang, Z.; Fan, H.; Li, D.; Gui, J. Screening and characterization of sex-specific markers by 2b-RAD sequencing in zig-zag eel (Mastacembelus armatus) with implication of XY sex determination system. Aquaculture 2020, 528, 735550. [Google Scholar] [CrossRef]
- Wang, L.; Xie, N.; Shen, Y.; Ye, B.; Yue, G.; Feng, X. Constructing high-density genetic maps and developing sexing markers in northern snakehead (Channa argus). Mar. Biotechnol. 2019, 21, 348–358. [Google Scholar] [CrossRef]
- Han, C.; Zhu, Q.; Lu, H.; Wang, C.; Zhou, X.; Peng, C.; Tang, L.; Han, L.; Chen, J.; Li, S.; et al. Screening and characterization of sex-specific markers developed by a simple NGS method in mandarin fish (Siniperca chuatsi). Aquaculture 2020, 527, 735495. [Google Scholar] [CrossRef]
- Li, Y.; Cui, Y.; Ye, X.; Wang, M.; Xu, H.; Wu, Y.; Jin, J.; Huang, H.; Lai, M.; Wang, Z.; et al. Identification of the sex-linked region of Siniperca scherzeri and development of sex-specific markers. Aquaculture 2025, 600, 742231. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, S. Fish sex-specific molecular marker screening and marker-assisted sex-controlled breeding: History, progress and prospect. J. Fish. China 2023, 47, 34–43. [Google Scholar]
- Qin, W.; Han, C.; Yang, J.; Yu, Z.; Feng, Y.; Wu, Y.; Lu, B.; Cui, M.; Shu, H. Development of genetic sex markers of zig-zag eel (Mastacembelus armatus) by a NGS method. Aquaculture 2023, 571, 739498. [Google Scholar] [CrossRef]
- Bentley, D.R. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006, 16, 545–552. [Google Scholar] [CrossRef]
- Wen, M.; Wang, S.; Zhu, C.; Zhang, Y.; Liu, Z.; Wu, C.; Wang, S.; Wang, Y.; Ren, L.; Tao, M.; et al. Identification of sex locus and a male-specific marker in blunt-snout bream (Megalobrama amblycephala) using a whole genome resequencing method. Aquaculture 2024, 582, 740559. [Google Scholar] [CrossRef]
- Fu, Y.; Luo, L.; Wang, S.; Yu, Y.; Wang, Y.; Gao, Z. Identification of sex-specific markers using genome re-sequencing in the blunt snout bream (Megalobrama amblycephala). BMC Genom. 2024, 25, 963. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Y.; Li, S.; Liu, Y.; Li, C.; Han, S.; Liu, K.; Doretto, L.B.; Liu, B.; Huang, H.; et al. Investigation of sex determination in starry flounder (Platichthys stellatus) reveals sex chromosome evolution in Pleuronectiformes and identifies a sex-specific marker. Zool. Res. 2024, 45, 1347–1356. [Google Scholar] [CrossRef]
- Michelmore, R.W.; Paran, I.; Kesseli, R. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Zhang, Y.; Wang, S.; Hu, F.; Tang, C.; Li, Q.; Qin, Q.; Tao, M.; Zhang, C.; Zhao, R.; et al. Sex locus and sex markers identification using whole genome pool-sequencing approach in the largemouth bass (Micropterus salmoides L.). Aquaculture 2022, 559, 738375. [Google Scholar] [CrossRef]
- He, P.; Wei, P.; Ma, Y.; Hu, S.; Yao, J.; Jiang, X.; Xu, Y.; Zhu, P.; Wei, M.; Jiang, W.; et al. Candidate sex-associated gene identification in Trachinotus ovatus (Carangidae) using an integrated SLAF-seq and bulked segregant analysis approach. Gene 2022, 809, 146026. [Google Scholar] [CrossRef]
- Liu, S.; Lian, Q.; Jia, Y.; Chi, M.; Li, F.; Jiang, J.; Liu, Y.; Zheng, J.; Cheng, S.; Gu, Z. Genetic diversity analysis of three Opsariichthys bidens populations in Zhejiang Province based on mitochondrial Cyt b gene sequences. Acta Agric. Zhejiangensis 2023, 35, 293–300. [Google Scholar]
- Xu, X.; Guan, W.; Niu, B.; Guo, D.; Xie, Q.; Zhan, W.; Yi, S.; Luo, B. Chromosome-level assembly of the Chinese hooksnout carp (Opsariichthys bidens) genome using PacBio sequencing and hi-C technology. Front. Genet. 2022, 12, 788547. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, Q.; Wang, W.; He, B.; Hou, Y.; Lin, Y.; Ye, J.; Ren, S.; Qin, Y.; Xiao, A.; et al. Transcriptomic analysis of colour dimorphism of Opsariichthys bidens provides insights into the mechanism of male colour. Aquac. Rep. 2023, 33, 101756. [Google Scholar] [CrossRef]
- Qiu, Q.; Luo, W. Apply RAPD to screen out sex differentiation related molecular marker in Opsariichthys bidens. J. Aquacult. 2012, 33, 38–42. [Google Scholar]
- Liu, D.; Gui, L.; Zhu, Y.; Xu, C.; Zhou, W.; Li, M. Chromosome-level assembly of male Opsariichthys bidens genome provides insights into the regulation of the GnRH signaling pathway and genome evolution. Biology 2022, 11, 1500. [Google Scholar] [CrossRef]
- Guan, W.; Xu, X.; Zhan, W.; Niu, B.; Luo, B. Induction of gynogenesis with ultra-violet irradiated Koi carp (Cyprinus carpio haematopterus) sperm demonstrates the XX/XY sex determination system in Opsariichthys bidens. Aquac. Rep. 2022, 26, 101286. [Google Scholar] [CrossRef]
- Xu, X.; Yu, J.; Ge, J.; Yi, S.; Weng, X.; Guan, W.; Niu, B.; Zhang, X.; Luo, B. A male-specific insert of Opsariichthys bidens identified based on genome-wide association analyses and comparative genomics. Aquac. Rep. 2024, 35, 101982. [Google Scholar] [CrossRef]
- Zhao, R.; Wang, R.; Deng, Y.; Wu, X.; Ouyang, S.; Lin, F.; Zhou, C. Development of SNP markers for sex identification of Opsariichthys bidens. J. Nanchang Univ. Nat. Sci. 2025, 49, 214–221. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Wang, Z.; He, C.; Yao, Z.; Li, J.; Chen, Z.; Deng, A.; Lai, Z.; Li, Y.; Dong, Z.; Guo, Y. Screening of Sex Genetic Markers in the Sanya Population of Oryzias curvinotus by Whole-Genome Resequencing. J. Guangdong Ocean Univ. 2023, 43, 69–75. [Google Scholar]
- Akkaya, M.S.; Bhagwat, A.A.; Cregan, P.B. Length polymorphisms of simple sequence repeat DNA in soybean. Genetics 1992, 132, 1131–1139. [Google Scholar] [CrossRef]
- Waldbieser, G.C.; Bosworth, B.G.; Nonneman, D.J.; Wolters, W.R. A microsatellite-based genetic linkage map for channel catfish, Ictalurus punctatus. Genetics 2001, 158, 727–734. [Google Scholar] [CrossRef]
- Felip, A.; Young, W.P.; Wheeler, P.A.; Thorgaard, G.H. An AFLP-based approach for the identification of sex-linked markers in rainbow trout (Oncorhynchus mykiss). Aquaculture 2001, 247, 35–43. [Google Scholar] [CrossRef]
- Han, C.; Zhou, X.; Lu, H.; Zhu, Q.; Han, L.; Li, S.; Lin, H.; Zhang, Y. A simple PCR-based genetic sex identification method in the blotched snakehead (Channa maculata) developed by high-throughput sequencing. Aquaculture 2021, 538, 736579. [Google Scholar] [CrossRef]
- Best, N.B.; McSteen, P. Mapping Maize Mutants Using Bulked-Segregant Analysis and Next-Generation Sequencing. Curr. Protoc. 2022, 2, e591. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, J.; Liu, C.; Yang, Y.; Wang, Y.; Mu, X. Identification of two effective sex-specific DNA markers in silver arowana (Osteoglossum bicirrhosum). Aquaculture 2025, 596, 741748. [Google Scholar] [CrossRef]
- Liu, G.; Chen, K.; Zheng, G.; Zhu, X.; Zhao, J.; Xu, P.; Sun, X. Screening and identification of female-specific DNA fragments in Channa argus using SSR-BSA. J. Fish. China 2011, 35, 170–175. [Google Scholar]
- Huang, W.; Lai, J.; Liang, W.; Ye, S.; Li, J.; Zhou, J.; Zhang, Y.; Peng, S.; Zhan, H.; Zheng, P.; et al. Identification of sex-specific DNA markers in the army fish (Spinibarbus hollandi) by whole genome re-sequencing method. Aquaculture 2024, 583, 740605. [Google Scholar] [CrossRef]
- Carvalho, C.M.; Lupski, J.R. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016, 17, 224–238. [Google Scholar] [CrossRef] [PubMed]
- Keane, T.M.; Goodstadt, L.; Danecek, P.; White, M.A.; Wong, K.; Yalcin, B.; Heger, A.; Agam, A.; Slater, G.; Goodson, M.; et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 2011, 477, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Xiao, Y.; Xiao, Z.; Li, J. Development of DNA insertion-specific markers based on the intergenic region of Oplegnathus punctatus Cdkn1/srsf3 for sex identification. Mar. Biotechnol. 2024, 26, 687–695. [Google Scholar] [CrossRef]
- Tang, R.; Zhu, Y.; Gan, W.; Zhang, Y.; Yao, Z.; Ren, J.; Li, M. De novo transcriptome analysis of gonads reveals the sex-associated genes in Chinese hook snout carp Opsariichthys bidens. Aquac. Rep. 2022, 23, 101068. [Google Scholar] [CrossRef]
- Zhou, C.; Lian, X.; Wang, R.; Wu, X.; Lin, F.; Ouyang, S.; Jian, S.; Hua, Q. Gonadal transcriptome analysis of Opsariichthys bidens reveals sex-associated genes. Comp. Biochem. Physiol. Part D Genom. Proteom. 2025, 54, 101379. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Tang, D.; Zhang, Y.; Gao, X.; Du, C.; Shen, W.; Jin, S.; Zhu, J. Transcriptomes of testes at different developmental stages in the Opsariichthys bidens predict key genes for testis development and spermatogenesis. Mar. Biotechnol. 2023, 25, 123–139. [Google Scholar] [CrossRef] [PubMed]




| Sample | Body Length | Body Weight | Date of Hatching | Age | Sample Size |
|---|---|---|---|---|---|
| Male for BSA | 13.02 ± 0.82 cm | 24.94 ± 4.69 g | September 2022 | 10th | 25 |
| Female for BSA | 12.24 ± 0.54 cm | 19.39 ± 1.69 g | September 2022 | 10th | 25 |
| Male from Jiangxi | 13.73 ± 0.46 cm | 27.95 ± 4.49 g | September 2022 | 12th | 20 |
| Female from Jiangxi | 12.72 ± 0.56 cm | 21.44 ± 3.00 g | September 2022 | 12th | 20 |
| Male from Fujian | 12.80 ± 1.06 cm | 35.26 ± 13.86 g | August 2023 | 12th | 20 |
| Female from Fujian | 11.85 ± 1.27 cm | 24.57 ± 11.77 g | August 2023 | 12th | 20 |
| Male from Zhejiang | 16.41 ± 0.65 cm | 74.08 ± 9.96 g | September 2023 | 10th | 20 |
| Female from Zhejiang | 13.71 ± 1.10 cm | 41.49 ± 9.30 g | September 2023 | 10th | 20 |
| Sample | Raw Bases | Raw Reads | Clean Bases | Clean Reads | Q20 (%) | Q30 (%) | GC (%) | Reads Passed Filters |
|---|---|---|---|---|---|---|---|---|
| male | 21,540,728,700 | 143,604,858 | 20,734,671,780 | 143,500,274 | 98.96 | 95.64 | 38.95 | 143,500,274 |
| female | 28,202,680,200 | 188,017,868 | 26,978,807,394 | 187,872,836 | 99.03 | 95.92 | 38.94 | 187,872,836 |
| Primer Name | Position | Primer Sequence (5′–3′) | Size (bp) | TM (°C) |
|---|---|---|---|---|
| Mar1 | Chr: 136,963,139–36,965,341 | F: GTCCTCCTCAGTGAAGTCATTATAC R: GCTCCTCCAGATGTTGTAGAGA | 201 | 57 |
| Mar2 | Chr1: 36,964,139–36,964,341 | F: TTCGAAAATCTCAGTGCAGC R: TTGACAGTGAAGCTCCTCCA | 386 | 57 |
| Mar3 | Chr1: 36,964,139–36,964,341 | F: CTCCACCACATTCTGCAGTT R: TTTGTGAATTGGCAGGAACA | 492 | 59 |
| Mar4 | Chr1: 36,964,139–36,964,341 | F: ACATGCAAGTGTGGCTTTGA R: ACATGCAAGTGTGGCTTTGA | 793 | 57 |
| Mar5 | Chr1: 36,964,145–36,964,340 | F: CTCACTCTTGTCATGTCCTCCTCA R: GAGAGTTGAAGGAAGACAGAAGGAC | 195 | 57 |
| Mar6 | Chr8: 33,584,367–33,586,928 | F: CAGGTTGGACTTCAGAACTTACA R: ACGCAGAACGAGCAGGAA | 267 | 58 |
| Mar7 | Chr8: 33,584,367–33,586,928 | F: AATGCGGAATCTAAGAGCCAAT R: CAACTGCCTCGGAAGAACAA | 530 | 58 |
| Mar8 | Chr8: 33,584,929–33,587,296 | F: TTCGTGTTGGCTCGGTCTG R: AGTTCATCTGTCATTATGGCTATCC | 554 | 58 |
| Mar9 | Chr8: 33,584,929–33,587,296 | F: CAGGTTGGACTTCAGAACTTACA R: ACGCAGAACGAGCAGGAA | 267 | 56 |
| Mar10 | Chr8: 33,585,297–33,587,717 | F: GGATAGCCATAATGACAGATGAACT R: AGCACCAGAACGACCAGAC | 499 | 58 |
| Mar11 | Chr8: 33,585,367–33,585,928 | F: TTACCAGCAGCTGCACAATC R: GGAATGCCAAAGTTATCGGA | 428 | 57 |
| Mar12 | Chr8: 33,585,367–33,585,928 | F: AAGGTTTTTGAGCCTGGGTT R: GGAATGCCAAAGTTATCGGA | 488 | 56 |
| Mar13 | Chr8: 33,585,407–33,586,707 | F: CCGATAACTTTGGCATTCCACTCG R: AAGGTATGCCCTGAGTTTCAGACC | 326 | 56 |
| Mar14 | Chr8: 33,585,407–33,586,707 | F: TGTAGTGTTCGTGTTGGCTCGG R: AGTGTCTGTGGGAGCAGTTCAT | 576 | 58 |
| Mar15 | Chr8: 33,585,929–33,586,296 | F: CCACTCGGTAAGTCTGCTCC R: CGACAAATGGGAGGAAACAG | 370 | 58 |
| Mar16 | Chr8: 33,585,929–33,586,296 | F: GGCATTCCACTCGGTAAGTC R: CGACAAATGGGAGGAAACAG | 376 | 57 |
| Mar17 | Chr8: 33,586,297–33,586,717 | F: CTGTTTCCTCCCATTTGTCG R: GTGTCTGTGGGAGCAGTTCA | 490 | 58 |
| Mar18 | Chr8: 33,586,297–33,586,717 | F: CTGTTTCCTCCCATTTGTCG R: TCATTATGGCTATCCCGGAC | 466 | 56 |
| Mar19 | Chr8: 33,625,278–33,627,696 | F: GAGCGATTCAGAGACCAGAGA R: CTTCCAGAGCACAACCATTACTT | 566 | 56 |
| Mar20 | Chr8: 33,625,278–33,627,696 | F: CTCTGGAAGCGTCTGAATGC R: CCAACCACAACTTAATCTGAACTAC | 231 | 57 |
| Mar21 | Chr8: 33,626,278–33,626,696 | F: AGCGCACTGCCTCTCATACT R: GCATTTCCCGACCTGTAAAA | 358 | 57 |
| Mar22 | Chr8: 33,626,278–33,626,696 | F: AGCGCACTGCCTCTCATACT R: CTTGTGAAATATGCCTCGCA | 375 | 56 |
| Mar23 | Chr8: 33,626,281–33,626,686 | F: AATGGTTGTGCTCTGGAAGCG R: CCTCGCATTTCCCGACCTGTAA | 274 | 58 |
| Mar24 | Chr8: 33,626,781–33,627,375 | F: CAAACACAAAAACGACACGC R: AAGCGCTTTGGGTGTACAGT | 453 | 58 |
| Mar25 | Chr8: 33,626,781–33,627,375 | F: TGTTTCATTCAGGCCTTCCT R: TGACTTTGACCTTTGACCCC | 500 | 57 |
| Mar26 | Chr8: 33,626,781–33,627,375 | F: GGATGGACTGATGATTCACTACAAC R: AAAGCGCTTTGGGTGTACAGT | 412 | 57 |
| Mar27 | Chr8: 33,626,781–33,627,375 | F: ACAGTTCCAGTTGTGTCTCCACTT R: TCTGATACGGCGCAATGAAGATCC | 405 | 56 |
| Mar28 | Chr8: 33,626,816–33,627,324 | F: AGTGATTCAGATTGGATGGACTGA R: AACACTATATCCTGCACCGGATT | 509 | 56 |
| Mar29 | Chr12: 6,143,157–6,143,771 | F: TGGAGATCTCAAGTGCTCTTCAAG R: ATGTGGTATTGCCTGGATGATCTC | 451 | 58 |
| Mar30 | Chr14: 16,536,659–16,536,680 | F: CCATAGCAACACCTTAGCAAACAC R: GGGCTTTGCAATGGTGGTTAAA | 320 | 59 |
| Mar31 | Chr14: 16,536,836–1,653,710 | F: CCAGTAACCACTCAGATCTTCTG R: TTAGTTGCTAGAGCATTGCTGTG | 220 | 56 |
| Mar32 | Chr14: 16,563,469–16,563,648 | F: TGGTTCATTCACACTGCCAGT R: TCCATGCGTGTTCACTATAGTCC | 204 | 58 |
| Mar33 | Chr18: 17,827,232–17,827,798 | F: TTGAACGACACTCTGCACCT R: TGAAGCTGAAAAAGCCACAA | 837 | 58 |
| Mar34 | Chr18: 17,827,232–17,827,798 | F: TTCTGCCAGTCTTTTCCTGG R: TTTGAACGACACTCTGCACC | 497 | 57 |
| Mar35 | Chr18: 17,827,812–17,828,407 | F: AAGCCATAGCCCCTTTTCAT R: ATCGGTGAGTCTGATACGGC | 466 | 58 |
| Mar36 | Chr18: 17,827,812–17,828,407 | F: ACAAAGCCATAGCCCCTTTT R: ATCGGTGAGTCTGATACGGC | 469 | 59 |
| Mar37 | Chr20: 2,906,171–2,906,720 | F: TCAATTTCTCATGCAGTTTGC R: AGCGTTGCACAAACACAAAG | 402 | 59 |
| Mar38 | Chr20: 2,906,224–2,907,256 | F: CTTTGTGTTTGTGCAACGCTGT R: ACACCATGTTGAAATGCTTCCAGT | 302 | 59 |
| Mar39 | Chr20: 2,906,721–2,907,377 | F: CATGCAGCATCTTCTCCAAA R: ACCAACAACCCAATAACCCA | 387 | 57 |
| Mar40 | Chr20: 2,906,721–2,907,377 | F: TCATTGAACCATCGGCTACA R: ACCAACAACCCAATAACCCA | 462 | 58 |
| Mar41 | Chr21: 10,711,992–10,712,371 | F: TGCACTTCTGTCAATAGATCCTCTT R: TCGGTCATCACCATCATCAGAT | 401 | 59 |
| Mar42 | Chr38: 4,802,762–4,802,784 | F: TCATCAGTCCACAGAACACTATCC R: ACCATAGGAAGCATCATTCGC | 350 | 56 |
| Mar43 | Chr38: 4,867,346–4,867,835 | F: ACATTCATTGGACAATCTTGTCGG R: AACAAGGAGTGGAATGAATTGCC | 469 | 57 |
| Mar44 | Chr38: 4,955,599–4,955,622 | F: TTATCTTGCTCCTTCATCCATCTCC R: ACTTAATGAAGACAGGAGGCAATGG | 248 | 57 |
| Mar45 | Chr39: 5,164,306–5,164,730 | F: AAGGAAACCGGTATGTGCTG R: TGTGCAAAAAGCAGTCTTGG | 396 | 58 |
| Mar46 | Chr39: 5,179,355–5,179,609 | F: TTTTGCAGCAAGAGCTTTCA R: TTGCCCTTGTGGTAATGACA | 423 | 58 |
| Mar47 | Chr39: 5,179,385–5,180,010 | F: GTGCCTTACAAGAATAATGCTCTGA R: CTTTGCCCTTGTGGTAATGACA | 325 | 58 |
| Mar48 | Chr39: 5,179,610–5,180,093 | F: TGTCATTACCACAAGGGCAA R: AGGTATGGGGAAGGTGGAGT | 371 | 56 |
| Mar49 | Chr39: 5,179,610–5,180,093 | F: TGTCATTACCACAAGGGCAA R: GGTGGAGTGCAGTGAGTCAG | 359 | 56 |
| Mar50 | Chr39: 5,180,173–5,180,725 | F: TGTCTTTGTTGCTGACCTTCA R: GGCCATCTCTTGTTGGAGTC | 399 | 58 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, F.; Zhao, R.; Miao, M.; Wang, Y.; Lei, N.; Ding, D.; Wang, R.; Ouyang, S.; Wu, X.; Zhou, C. Development of a Sex-Specific Marker for the Chinese Hooksnout Carp Opsariichthys bidens Günther, 1873 Based on Whole-Genome Resequencing and Bulked Segregant Analysis. Animals 2025, 15, 3164. https://doi.org/10.3390/ani15213164
Lin F, Zhao R, Miao M, Wang Y, Lei N, Ding D, Wang R, Ouyang S, Wu X, Zhou C. Development of a Sex-Specific Marker for the Chinese Hooksnout Carp Opsariichthys bidens Günther, 1873 Based on Whole-Genome Resequencing and Bulked Segregant Analysis. Animals. 2025; 15(21):3164. https://doi.org/10.3390/ani15213164
Chicago/Turabian StyleLin, Feng, Ruobing Zhao, Maosheng Miao, Yuchen Wang, Ning Lei, Dewen Ding, Rongrong Wang, Shan Ouyang, Xiaoping Wu, and Chunhua Zhou. 2025. "Development of a Sex-Specific Marker for the Chinese Hooksnout Carp Opsariichthys bidens Günther, 1873 Based on Whole-Genome Resequencing and Bulked Segregant Analysis" Animals 15, no. 21: 3164. https://doi.org/10.3390/ani15213164
APA StyleLin, F., Zhao, R., Miao, M., Wang, Y., Lei, N., Ding, D., Wang, R., Ouyang, S., Wu, X., & Zhou, C. (2025). Development of a Sex-Specific Marker for the Chinese Hooksnout Carp Opsariichthys bidens Günther, 1873 Based on Whole-Genome Resequencing and Bulked Segregant Analysis. Animals, 15(21), 3164. https://doi.org/10.3390/ani15213164





