Metabolomics Reveals Abnormal Citrate Cycle and Phenylalanine Metabolism in Testes from Infertile Hybrid Dzo
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Widely Targeted Metabolomics Analysis
2.2.1. Sample Preparation and Extraction
2.2.2. HPLC Conditions
2.2.3. ESI-QTRAP-MS/MS
2.2.4. Data Preprocessing and Quality Control
2.2.5. Qualitative and Quantitative Analysis of Metabolites
2.3. Bioinformatics Analysis of Widely Targeted Metabolomics
2.4. Integrative Metabolomics and Transcriptomics Pathway Analysis in Testes
2.5. m6A and Reactive Oxygen Species Content Analysis
2.6. LDHC mRNA Expression and Association with Phenyl Lactate Content
2.7. miRNA-Seq and Target miRNA Prediction for IDH Genes
2.8. DNA Methylation Levels and Chromatin Accessibility in PDHA2 Promoters
2.9. Statistical Analysis
3. Results
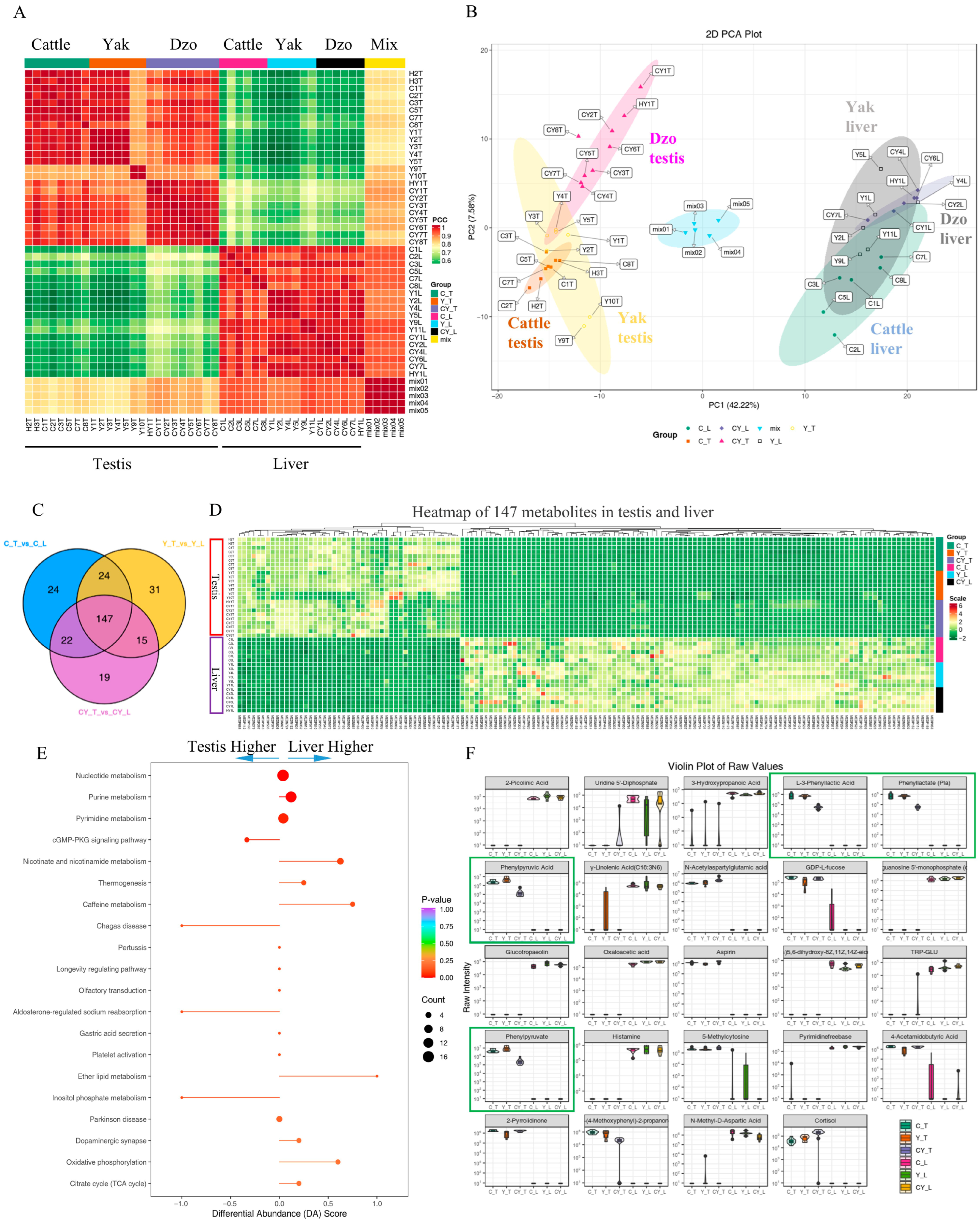
3.1. Global View of Metabolites Data
3.2. Differentially Abundant Metabolites Between Liver and Testis Tissues in Bovine
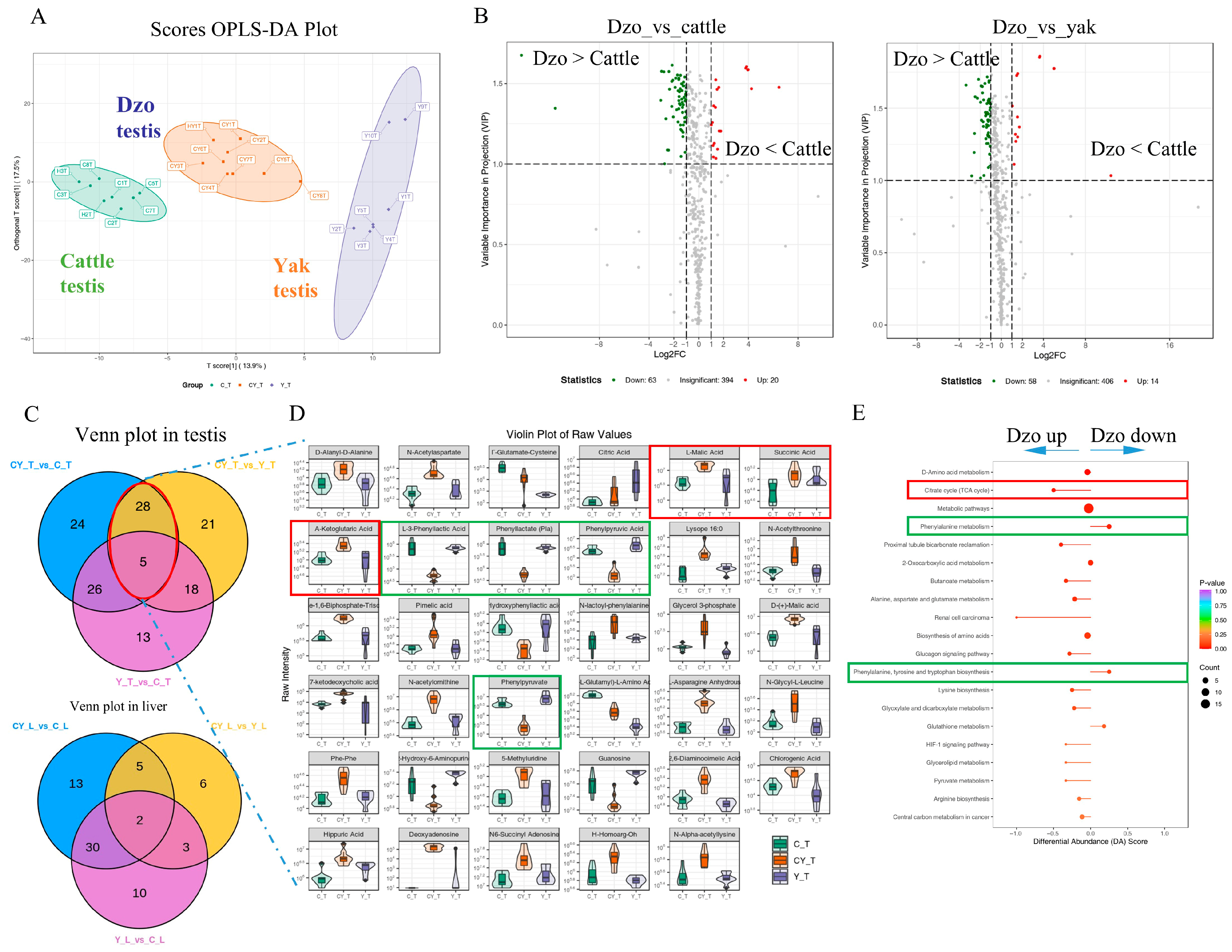
3.3. Differentially Abundant Metabolites Between Fertile and Infertile Testis Tissues
3.4. α-Ketoglutarate-Dependent Oxygenase mRNA Expression Upregulated in Dzo Testes
3.5. Transcriptomic and Metabolomics Integrative Analysis Reveals IDH and PDHA2 Genes Were Associated with Citrate Cycle Disorder in Dzo Testes
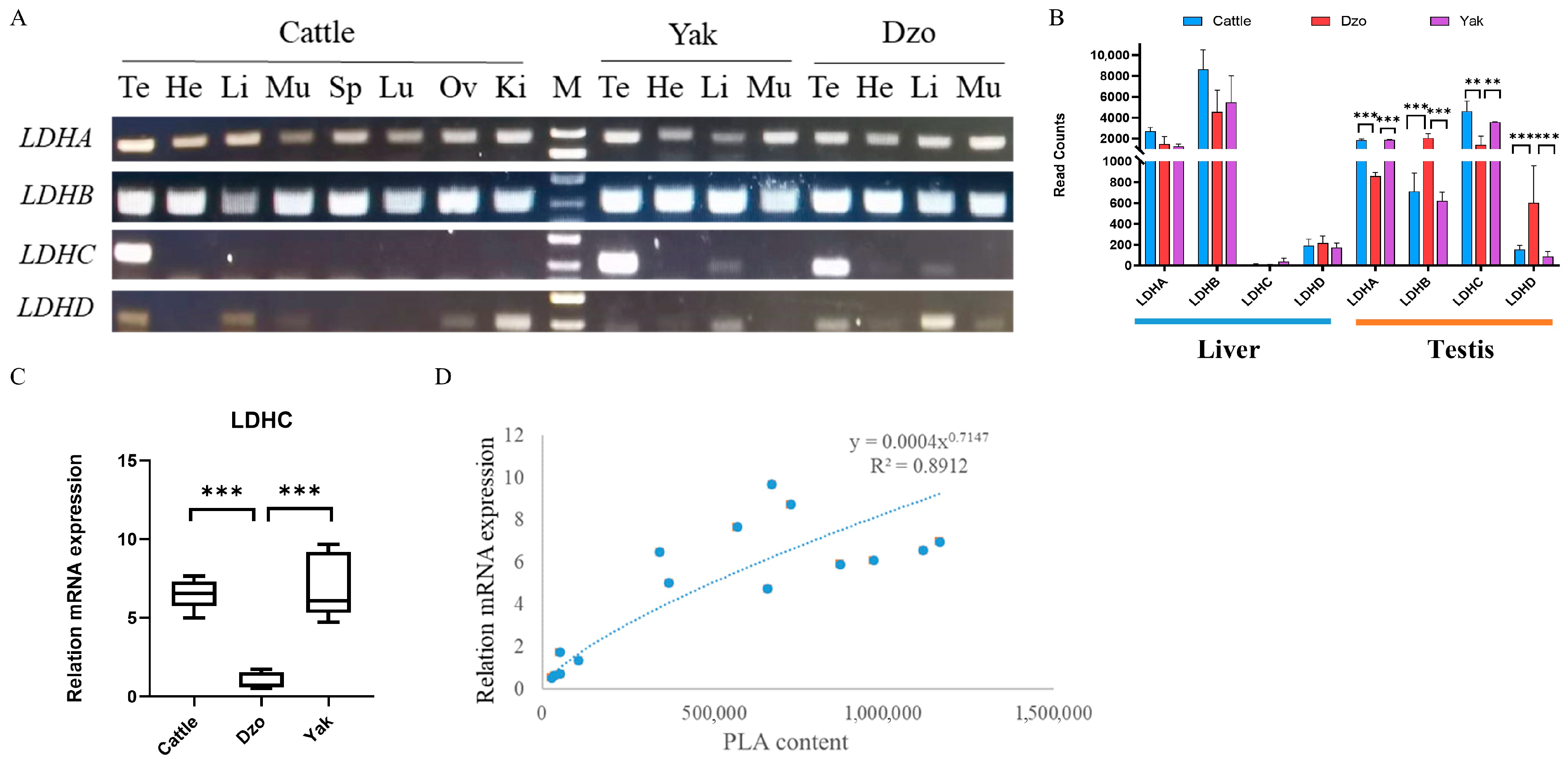
3.6. Testis-Specific LDHC mRNA Downregulation Associated with Lower Phenylalanine Metabolism in Dzo Testes
3.7. miRNA and DNA Methylation Associated with Regulating Citrate Cycle Disordered in Dzo Testes

4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TCA | citrate cycle |
| 2OGX | 2OG-dependent oxygenases |
| KDM | lysine demethylases |
| HIF | hypoxia inducible factor |
| ALKBH | AlkB homolog family |
| DEGs | differentially expressed genes |
| Pla | phenyl lactate |
| ROS | reactive oxygen species |
| OGDH | oxoglutarate dehydrogenase |
| 2OG | α-ketoglutarate or 2-oxoglutarate |
| PDHA | pyruvate dehydrogenase E1 alpha |
| LDH | lactate dehydrogenase |
| IDH3A | isocitrate dehydrogenase 3 [NAD (+)] alpha |
| LZ | leptotene or zygotene spermatocytes |
| PS | pachytene spermatocytes |
| OXPHOS | oxidative phosphorylation |
| piRNA | piwi-interacting RNA |
| PCA | principal component analysis |
| OPLS-DA | orthogonal partial least squares discriminant analysis |
| VIP | variable importance in projection |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Chang, X.; Xu, Y.; Cheng, L.; Yi, K.; Gu, X.; Luo, Z.; Zhang, J.; Wang, J.; Geng, F. Quantitative proteomic analysis of cattle-yak and yak longissimus thoracis provides insights into the differential mechanisms of meat quality. Food Res. Int. 2023, 173, 113253. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Z.; Hu, R.; Peng, Q.; Xue, B.; Wang, L. Comparison of carcass characteristics and meat quality between Simmental crossbred cattle, cattle-yaks and Xuanhan yellow cattle. J. Sci. Food Agric. 2021, 101, 3927–3932. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Ma, X.-M.; Xiong, L.; Wu, X.-Y.; Liang, C.-N.; Bao, P.-J.; Yu, Q.-L.; Yan, P. Effects of Intensive Fattening with Total Mixed Rations on Carcass Characteristics, Meat Quality, and Meat Chemical Composition of Yak and Mechanism Based on Serum and Transcriptomic Profiles. Front. Vet. Sci. 2021, 7, 599418. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.; Gao, Y.; Zhao, X.; Liang, J.; Pei, L.; Yao, Y.; Tang, D. Bacterial community structure and metabolomic profiles of yak milk and cattle-yak milk during refrigeration in Gannan region: Analysis of interspecific differences in milk spoilage. Food Chem. 2025, 463, 141022. [Google Scholar] [CrossRef]
- Zhang, G.W.; Wu, Y.; Luo, Z.; Guan, J.; Wang, L.; Luo, X.; Zuo, F. Comparison of Y-chromosome-linked TSPY, TSPY2, and PRAMEY genes in Taurus cattle, yaks, and interspecific hybrid bulls. J. Dairy Sci. 2019, 102, 6263–6275. [Google Scholar] [CrossRef]
- Takase, H.; Tumennasan, K.; Hiratsuka, K.; Chandley, A.; Hotta, Y. Fertility Investigation in F1 Hybrid and Backcross Progeny of Cattle (Bos taurus) and Yak (B. gruniens) in Mongolia. Niigata J. Health Welf. 2002, 2, 42–52. [Google Scholar]
- Zhang, G.W.; Wang, L.; Wu, J.; Ye, Y.; Zhao, J.; Du, Y.; Tu, Y.; Luo, Z.; Fu, S.; Zuo, F. Evaluation of MYBL1 as the master regulator for pachytene spermatocyte genes dysregulated in interspecific hybrid dzo. J. Dairy. Sci. 2023, 106, 4366–4379. [Google Scholar] [CrossRef]
- Cai, X.; Yu, S.; Mipam, T.; Yang, F.; Zhao, W.; Liu, W.; Cao, S.; Shen, L.; Zhao, F.; Sun, L.; et al. Comparative analysis of testis transcriptomes associated with male infertility in cattleyak. Theriogenology 2017, 88, 28–42. [Google Scholar] [CrossRef]
- Cai, X.; Wu, S.; Mipam, T.; Luo, H.; Yi, C.; Xu, C.; Zhao, W.; Wang, H.; Zhong, J. Testis transcriptome profiling identified lncRNAs involved in spermatogenic arrest of cattleyak. Funct. Integr. Genom. 2021, 21, 665–678. [Google Scholar] [CrossRef]
- Zhang, G.W.; Wang, L.; Chen, H.; Guan, J.; Wu, Y.; Zhao, J.; Luo, Z.; Huang, W.; Zuo, F. Promoter hypermethylation of PIWI/piRNA pathway genes associated with diminished pachytene piRNA production in bovine hybrid male sterility. Epigenetics 2020, 15, 914–931. [Google Scholar] [CrossRef]
- Wu, S.-X.; Wan, R.-d.; Wang, G.-W.; Zhang, Y.-W.; Yang, Q.-E. Comparative proteomic analysis identifies differentially expressed proteins associated with meiotic arrest in cattle-yak hybrids. Proteomics 2023, 23, e2300107. [Google Scholar] [CrossRef]
- Li, Y.C.; Wang, G.W.; Xu, S.R.; Zhang, X.N.; Yang, Q.E. The expression of histone methyltransferases and distribution of selected histone methylations in testes of yak and cattle-yak hybrid. Theriogenology 2020, 144, 164–173. [Google Scholar] [CrossRef]
- Wang, X.; Pei, J.; Guo, S.; Cao, M.; Kang, Y.; Xiong, L.; La, Y.; Bao, P.; Liang, C.; Yan, P.; et al. Characterization of N6-methyladenosine in cattle-yak testis tissue. Front. Vet. Sci. 2022, 9, 971515. [Google Scholar] [CrossRef]
- Hong, R.; Wu, J.; Chen, X.; Zhang, Z.; Liu, X.; Li, M.; Zuo, F.; Zhang, G.-W. mRNA-Seq of testis and liver tissues reveals a testis-specific gene and alternative splicing associated with hybrid male sterility in dzo. J. Anim. Sci. 2024, 102, skae091. [Google Scholar] [CrossRef]
- Schvartzman, J.M.; Thompson, C.B.; Finley, L.W.S. Metabolic regulation of chromatin modifications and gene expression. J. Cell Biol. 2018, 217, 2247–2259. [Google Scholar] [CrossRef]
- Losman, J.A.; Kaelin, W.G., Jr. What a difference a hydroxyl makes: Mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes. Dev. 2013, 27, 836–852. [Google Scholar] [CrossRef]
- Islam, M.S.; Leissing, T.M.; Chowdhury, R.; Hopkinson, R.J.; Schofield, C.J. 2-Oxoglutarate-Dependent Oxygenases. Annu. Rev. Biochem. 2018, 87, 585–620. [Google Scholar] [CrossRef]
- Klose, R.J.; Kallin, E.M.; Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006, 7, 715–727. [Google Scholar] [CrossRef]
- Tsukada, Y.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006, 439, 811–816. [Google Scholar] [CrossRef]
- Aas, P.A.; Otterlei, M.; Falnes, P.O.; Vågbø, C.B.; Skorpen, F.; Akbari, M.; Sundheim, O.; Bjørås, M.; Slupphaug, G.; Seeberg, E.; et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 2003, 421, 859–863. [Google Scholar] [CrossRef]
- Martinez-Reyes, I.; Diebold, L.P.; Kong, H.; Schieber, M.; Huang, H.; Hensley, C.T.; Mehta, M.M.; Wang, T.; Santos, J.H.; Woychik, R.; et al. TCA Cycle and Mitochondrial Membrane Potential Are Necessary for Diverse Biological Functions. Mol. Cell 2016, 61, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.T.; Sun, X.; Tsai, T.S.; Johnson, J.L.; Gould, J.A.; Garama, D.J.; Gough, D.J.; McKenzie, M.; Trounce, I.A.; St John, J.C. Mitochondrial DNA haplotypes induce differential patterns of DNA methylation that result in differential chromosomal gene expression patterns. Cell Death Discov. 2017, 3, 17062. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhou, L.A.; Chen, J.H.; Li, Z.Z.; Li, X.X.; Hu, X.D.; Huang, Y.; Zhao, X.K.; Liang, C.Z.; Wang, Y.; Sun, L.A.; et al. Integrated Profiling of MicroRNAs and mRNAs: MicroRNAs Located on Xq27.3 Associate with Clear Cell Renal Cell Carcinoma. PLoS ONE 2010, 5, e15224. [Google Scholar] [CrossRef]
- Martínez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef]
- Zhang, Z.; Miao, J.; Wang, Y. Mitochondrial regulation in spermatogenesis. Reproduction 2022, 163, R55–R69. [Google Scholar] [CrossRef]
- Yang, X.; Li, J.; Shi, G.; Zeng, M.; Liu, Z. Improving 3-phenyllactic acid production of Lactobacillus plantarum AB-1 by enhancing its quorum-sensing capacity. J. Food Sci. Technol. 2019, 56, 2605–2610. [Google Scholar] [CrossRef]
- Coonrod, S.; Vitale, A.; Duan, C.W.; Bristol-Gould, S.; Herr, J.; Goldberg, E. Testis-specific lactate dehydrogenase (LDH-C.; Ldh3) in murine oocytes and preimplantation embryos. J. Androl. 2006, 27, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Curry, E.; Safranski, T.J.; Pratt, S.L.J.T. Differential expression of porcine sperm microRNAs and their association with sperm morphology and motility. Theriogenology 2011, 76, 1532–1539. [Google Scholar] [CrossRef]
- Reza, A.M.M.T.; Choi, Y.J.; Han, S.G.; Song, H.; Park, C.; Hong, K.; Kim, J.H.J.B.R. Roles of microRNAs in mammalian reproduction: From the commitment of germ cells to peri-implantation embryos. Biol. Rev. 2019, 94, 415–438. [Google Scholar] [CrossRef]
- Tang, H.; Goldberg, E. A-MYB (MYBL1) stimulates murine testis-specific Ldhc expression via the cAMP-responsive element (CRE) site. Biol. Reprod. 2012, 86, 30. [Google Scholar] [CrossRef] [PubMed]
- da Cruz, I.; Rodríguez-Casuriaga, R.; Santiñaque, F.F.; Farías, J.; Curti, G.; Capoano, C.A.; Folle, G.A.; Benavente, R.; Sotelo-Silveira, J.R.; Geisinger, A.J.B.g. Transcriptome analysis of highly purified mouse spermatogenic cell populations: Gene expression signatures switch from meiotic-to postmeiotic-related processes at pachytene stage. BMC Genom. 2016, 17, 294. [Google Scholar] [CrossRef]
- Liu, X.; Liu, W.; Lenstra, J.A.; Zheng, Z.; Wu, X.; Yang, J.; Li, B.; Yang, Y.; Qiu, Q.; Liu, H.; et al. Evolutionary origin of genomic structural variations in domestic yaks. Nat. Commun. 2023, 14, 5617. [Google Scholar] [CrossRef]
- Zhang, M.; Wu, S.; Mipam, T.; Xu, C.; Zhao, W.; Shah, M.A.; Yi, C.; Luo, H.; Cai, X.; Zhong, J. Testis transcriptome profiling identified genes involved in spermatogenic arrest of cattleyak. PLoS ONE 2020, 15, e0229503. [Google Scholar] [CrossRef]
- Juárez-Rojas, L.; Casillas, F.; López, A.; Betancourt, M.; Ommati, M.M.; Retana-Márquez, S. Physiological role of reactive oxygen species in testis and epididymal spermatozoa. Andrologia 2022, 54, e14367. [Google Scholar] [CrossRef] [PubMed]
- McBride, H.M.; Neuspiel, M.; Wasiak, S. Mitochondria: More than just a powerhouse. Curr. Biol. 2006, 16, R551–R560. [Google Scholar] [CrossRef]
- Floyd, B.J.; Wilkerson, E.M.; Veling, M.T.; Minogue, C.E.; Xia, C.; Beebe, E.T.; Wrobel, R.L.; Cho, H.; Kremer, L.S.; Alston, C.L.J.M.c. Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol. Cell 2016, 63, 621–632. [Google Scholar] [CrossRef]
- Bayona-Bafaluy, M.P.; Müller, S.; Moraes, C.T. Fast adaptive coevolution of nuclear and mitochondrial subunits of ATP synthetase in orangutan. Mol. Biol. Evol. 2005, 22, 716–724. [Google Scholar] [CrossRef]
- Wang, J.; Xiang, H.; Liu, L.; Kong, M.; Yin, T.; Zhao, X. Mitochondrial haplotypes influence metabolic traits across bovine inter- and intra-species cybrids. Sci. Rep. 2017, 7, 4179. [Google Scholar] [CrossRef]
- Shen, Z.H.; Huang, L.; Jin, S.Y.; Zheng, Y.C. Cloning and Expression Analysis of Two Kdm Lysine Demethylases in the Testes of Mature Yaks and Their Sterile Hybrids. Animals 2020, 10, 521. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, J.; Yan, Y.; Zhu, C.; Wang, L.; Fu, S.; Zuo, F.; Zhang, G.W. N6-methyladenosine RNA demethylase ALKBH5 is testis-specifically downregulated in hybrid male sterile dzo and is a target gene of bta-miR-200a. Theriogenology 2022, 187, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Iannello, R.C.; Young, J.; Sumarsono, S.; Tymms, M.J.; Dahl, H.H.; Gould, J.; Hedger, M.; Kola, I. Regulation of Pdha-2 expression is mediated by proximal promoter sequences and CpG methylation. Mol. Cell. Biol. 1997, 17, 612–619. [Google Scholar] [CrossRef]
- Aravin, A.A.; Sachidanandam, R.; Bourc’his, D.; Schaefer, C.; Pezic, D.; Toth, K.F.; Bestor, T.; Hannon, G.J. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 2008, 31, 785–799. [Google Scholar] [CrossRef]
- Pezic, D.; Manakov, S.A.; Sachidanandam, R.; Aravin, A.A. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes. Dev. 2014, 28, 1410–1428. [Google Scholar] [CrossRef]
- Li, X.; Jiang, B.; Pan, B.; Mu, W.; Zhang, T. Purification and partial characterization of Lactobacillus species SK007 lactate dehydrogenase (LDH) catalyzing phenylpyruvic acid (PPA) conversion into phenyllactic acid (PLA). J. Agric. Food Chem. 2008, 56, 2392–2399. [Google Scholar] [CrossRef] [PubMed]
- Bolcun-Filas, E.; Bannister, L.A.; Barash, A.; Schimenti, K.J.; Hartford, S.A.; Eppig, J.J.; Handel, M.A.; Shen, L.S.; Schimenti, J.C. A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development 2011, 138, 3319–3330. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, J.; Dao, Y.; Liang, L.; Hong, R.; Chen, H.; Yan, Y.; Wang, L.; Zuo, F.; Zhang, G. Metabolomics Reveals Abnormal Citrate Cycle and Phenylalanine Metabolism in Testes from Infertile Hybrid Dzo. Animals 2025, 15, 3023. https://doi.org/10.3390/ani15203023
Ding J, Dao Y, Liang L, Hong R, Chen H, Yan Y, Wang L, Zuo F, Zhang G. Metabolomics Reveals Abnormal Citrate Cycle and Phenylalanine Metabolism in Testes from Infertile Hybrid Dzo. Animals. 2025; 15(20):3023. https://doi.org/10.3390/ani15203023
Chicago/Turabian StyleDing, Jiaojiao, Yan Dao, Lingqian Liang, Rui Hong, Huiyou Chen, Yi Yan, Ling Wang, Fuyuan Zuo, and Gongwei Zhang. 2025. "Metabolomics Reveals Abnormal Citrate Cycle and Phenylalanine Metabolism in Testes from Infertile Hybrid Dzo" Animals 15, no. 20: 3023. https://doi.org/10.3390/ani15203023
APA StyleDing, J., Dao, Y., Liang, L., Hong, R., Chen, H., Yan, Y., Wang, L., Zuo, F., & Zhang, G. (2025). Metabolomics Reveals Abnormal Citrate Cycle and Phenylalanine Metabolism in Testes from Infertile Hybrid Dzo. Animals, 15(20), 3023. https://doi.org/10.3390/ani15203023



