Tracing Genetic Divergence and Phylogeographic Patterns of Gekko gecko Linnaeus, 1758 (Squamata: Gekkonidae) Across Southeast Asia Using RAG1 Sequence
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and PCR Amplification
2.3. DNA Sequence Analysis
2.4. Phylogenetic Tree Analysis
2.5. Species Delimitation Analysis
3. Results
3.1. Phylogenetic Tree and Species Delimitation
3.2. Haplotype Network
3.3. Spatial Analysis of Molecular Variance (SAMOVA)
3.4. RAG1 Sequence Variation
3.5. Genetic Differences and Genetic Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fieldsend, T.W.; Rösler, H.; Krysko, K.L.; Harman, M.E.A.; Mahony, S.; Collins, T.M. Genotypic and Phenotypic Evidence Reveals the Introduction of Two Distinct Forms of the Non-Native Reptile Gekko gecko to Southern Florida. Asain Herpetol. J. 2025, 35, 287–298. [Google Scholar] [CrossRef]
- Bauer, A. Geckos in Traditional Medicine: Forensic Implications. Appl. Herpetol. 2009, 6, 81–96. [Google Scholar] [CrossRef]
- Nijman, V.; Todd, M.; Shepherd, C.R. Wildlife Trade as an Impediment Wildlife trade as an impediment to conservation as exemplified by the trade in reptiles in Southeast Asia. In Biotic Evolution and Environmental Change in Southeast Asia; Gower, D.J., Johnson, K.G., Richardson, J.E., Rosen, B.R., Williams, S.T., Eds.; Cambridge University Press: Cambridge, UK, 2012; Volume 82, pp. 390–405. [Google Scholar] [CrossRef]
- Laoong, S.; Sribundit, W. Diet of Tokay Gecko (Gekko gecko) in Eastern and Northern Regions of Thailand. Wildl. Yearb. 2006, 7, 78–90. [Google Scholar]
- Kongbuntad, W.; Tantrawatpan, C.; Pilap, W.; Jongsomchai, K.; Chanaboon, T.; Laotongsan, P.; Petney, T.N.; Saijuntha, W. Genetic Diversity of the Red-Spotted Tokay Gecko (Gekko gecko Linnaeus, 1758) (Squamata: Gekkonidae) in Southeast Asia Determined with Multilocus Enzyme Electrophoresis. J. Asia-Pac. Biodivers. 2016, 9, 63–68. [Google Scholar] [CrossRef]
- Saijuntha, W.; Sedlak, S.; Agatsuma, T.; Jongsomchai, K.; Pilap, W.; Kongbuntad, W.; Tawong, W.; Suksavate, W.; Petney, T.N.; Tantrawatpan, C. Genetic Structure of the Red-Spotted Tokay Gecko, Gekko gecko (Linnaeus, 1758) (Squamata: Gekkonidae) from Mainland Southeast Asia. Asian Herpetol. Res. 2019, 10, 69–78. [Google Scholar]
- Caillabet, O. The Trade in Tokay Geckos in South-East Asia: A Case Study on Medicinal Claims in Peninsular Malaysia; TRAFFIC: Cambridge, UK, 2013; Available online: https://coilink.org/20.500.12592/0pqv61 (accessed on 6 August 2025).
- Qin, X.-M.; Li, H.-M.; Zeng, Z.-H.; Zeng, D.-L.; Guan, Q.-X. Genetic Variation and Differentiation of Gekko gecko from Different Populations Based on Mitochondrial Cytochrome b Gene Sequences and Karyotypes. Zool. Sci. 2012, 29, 384–389. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Mo, X.C.; Zeng, W.M.; Hu, D.N. A Molecular Phylogeny of Red Tokay and Black Tokay (Gekko gecko) Based on Mitochondrial 12S rRNA Gene Sequences. Guangxi Med. J. 2006, 28, 793–796. [Google Scholar]
- Yu, X.; Peng, Y.; Aowphol, A.; Ding, L.; Brauth, S.E.; Tang, Y.-Z. Geographic Variation in the Advertisement Calls of Gekko gecko in Relation to Variations in Morphological Features: Implications for Regional Population Differentiation. Ethol. Ecol. Evol. 2011, 23, 211–228. [Google Scholar] [CrossRef]
- Qin, X.; Liang, Y.; Huang, X. RAPD Analysis on Genetic Divergence and Phylogenesis of Gekko gecko from Different Areas. Chin. J. Zool. 2005, 40, 14. [Google Scholar]
- Wang, G.; Gong, S.; Jiang, L.; Peng, R.; Shan, X.; Zou, D.; Yang, C.; Zou, F. Genetic Variability of the Tokay Gecko Based on Mitochondrial and Nuclear DNA. Mitochondrial DNA 2013, 24, 518–527. [Google Scholar] [CrossRef]
- Fieldsend, T.W.; Krysko, K.L.; Sharp, P.; Collins, T.M. Provenance and Genetic Diversity of the Non-Native Geckos Phelsuma grandis Gray 1870 and Gekko Gecko (Linnaeus 1758) in Southern Florida, USA. Biol. Invasions 2021, 23, 1649–1662. [Google Scholar] [CrossRef]
- Shen, X.-X.; Liang, D.; Zhang, P. The Development of Three Long Universal Nuclear Protein-Coding Locus Markers and Their Application to Osteichthyan Phylogenetics with Nested PCR. PLoS ONE 2012, 7, e39256. [Google Scholar] [CrossRef]
- Bennett, D. Expedition Field Techniques: Reptiles and Amphibians, 1st ed.; Royal Geographical Society (with IBG): London, UK, 1999; pp. 1–100. [Google Scholar]
- Bennett, R.A. Reptile Anesthesia. Semin. Avian Exot. Pet. Med. 1998, 7, 30–40. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Dupanloup, I.; Schneider, S.; Excoffier, L. A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 2002, 11, 2571–2581. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModeltest v2. Program Distributed by the Author. Evolutionary Biology Centre, Uppsala University. Ampignons de l’Équateur (Pugillus IV). Bull. L’herbier Boissier 2004, 3, 53–74. [Google Scholar]
- Mesquite Project. Available online: https://www.mesquiteproject.org/ (accessed on 5 August 2025).
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A General Species Delimitation Method with Applications to Phylogenetic Placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef]
- Vences, M.; Miralles, A.; Brouillet, S.; Ducasse, J.; Fedosov, A.; Kharchev, V.; Kumari, S.; Patmanidis, S.; Puillandre, N.; Scherz, M.D.; et al. iTaxoTools 0.1: Kickstarting a specimen-based software toolkit for taxonomists. Megataxa 2021, 6, 77–92. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, C.; Li, L.; Zhao, C.; Chen, W.; Huang, Y. Insights from ecological niche modeling on the taxonomic status of the black- and red-spotted tokay (Gekko gecko). Ecol. Evol. 2014, 4, 4717–4727. [Google Scholar] [CrossRef]
- Brock, K.M.; McTavish, E.J.; Edwards, D.L. Color Polymorphism is a Driver of Diversification in the Lizard Family Lacertidae. Syst. Biol. 2021, 71, 24–39. [Google Scholar] [CrossRef]
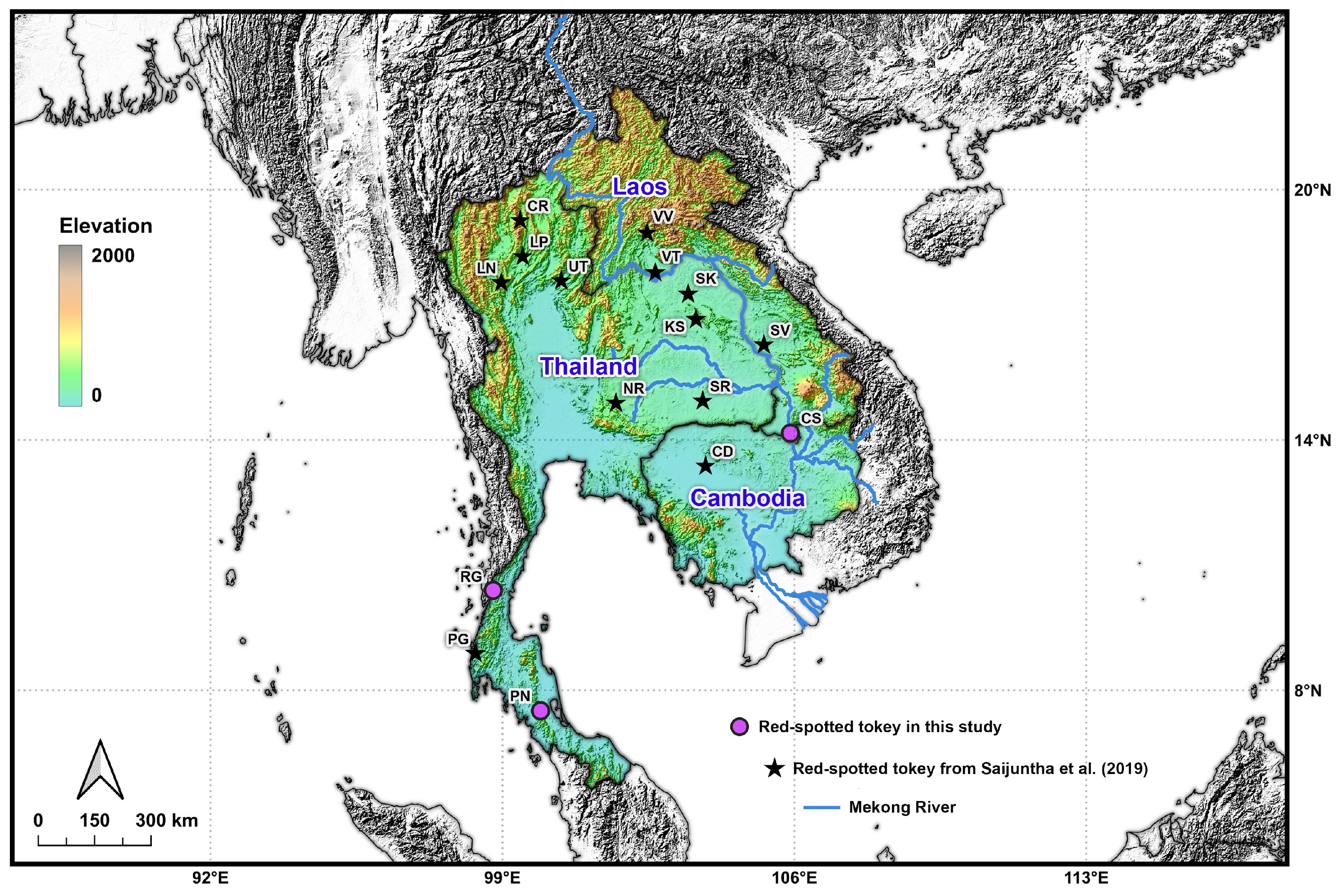

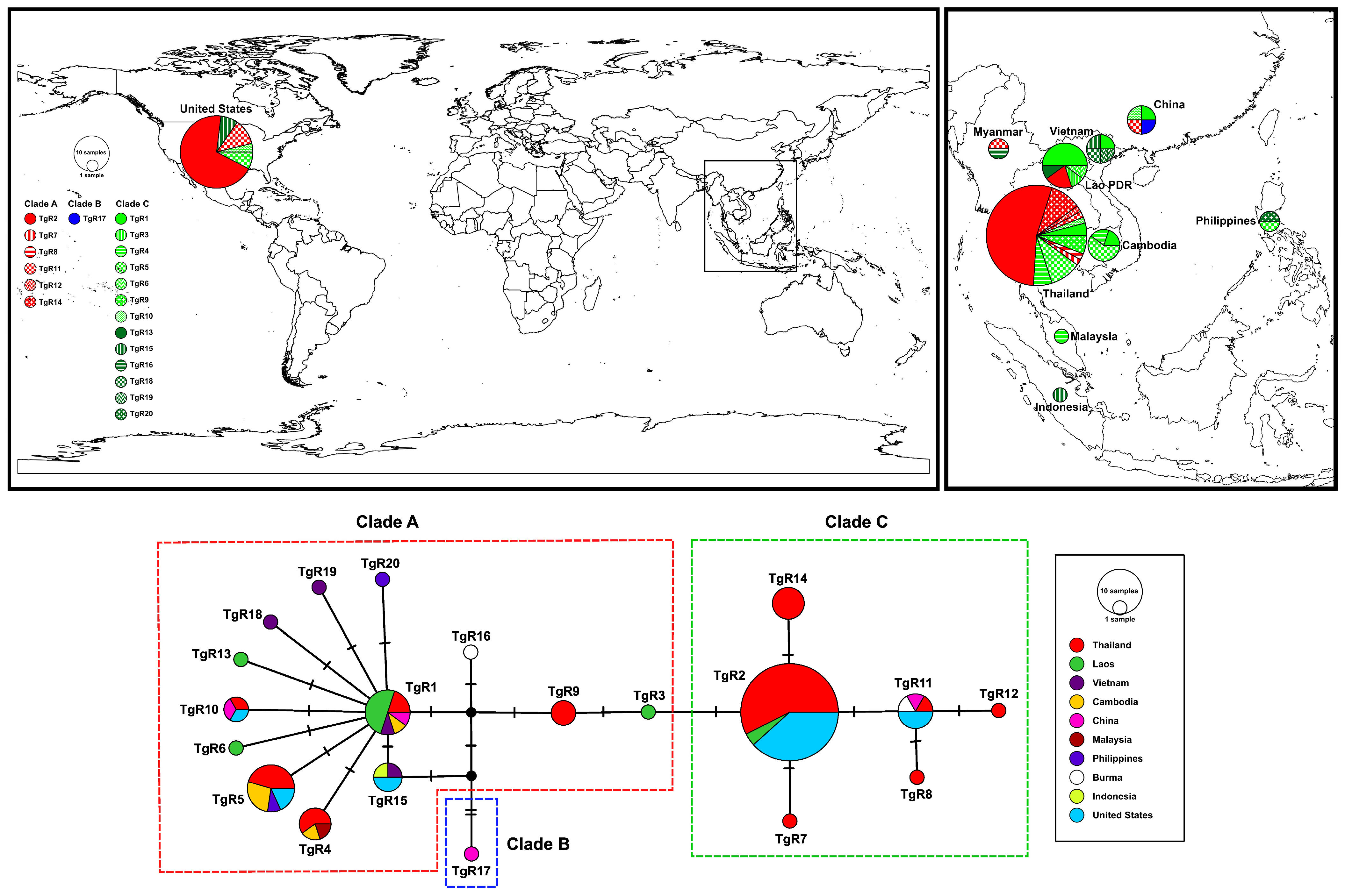
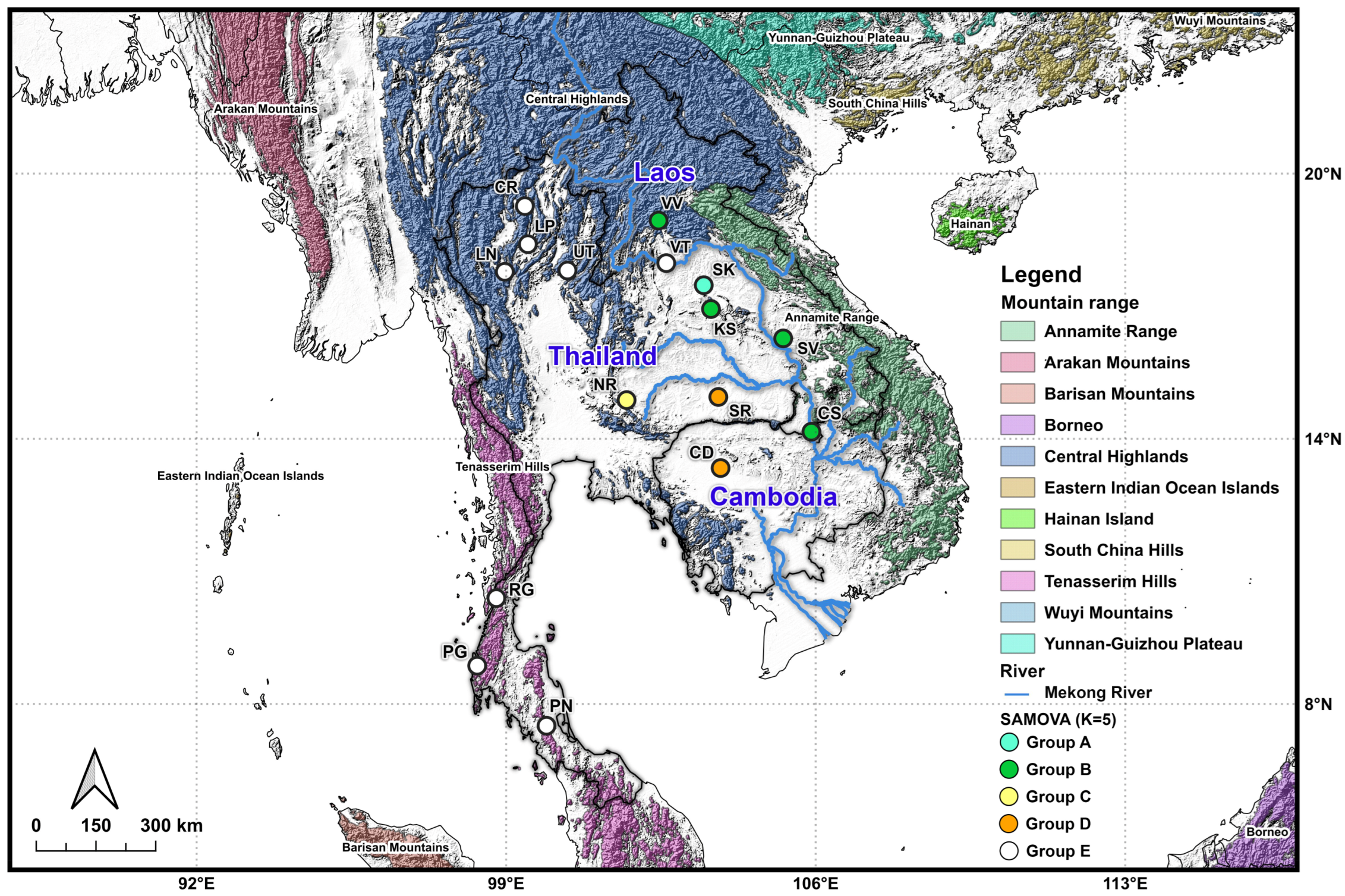
| Code | Province | Region | Country | Clade (Haplogroup) * | N | Coordinate |
|---|---|---|---|---|---|---|
| SV | Savannakhet | Central | Laos | D (G4) | 1 | 16.272846° N/105.266552° E |
| VT | Vientiane | North | Laos | B (G1) | 2 | 17.975828° N/102.621974° E |
| VV | Vang Viang (Laos) | North | Laos | B (G1) | 1 | 18.938490° N/102.448177° E |
| CS ** | Champasak | South | Laos | n/a | 6 | 13.954472° N/105.933777° E |
| LN | Lamphun | North | Thailand | E (G5) | 2 | 17.779125° N/98.970251° E |
| CR | Chiang Rai | North | Thailand | E (G5) | 1 | 19.265866° N/99.424861° E |
| UT | Uttaradit | North | Thailand | B (G2) | 6 | 17.809095° N/100.388379° E |
| LP | Lampang | North | Thailand | E (G5) | 2 | 18.403844° N/99.491655° E |
| KS | Kalasin | Northeast | Thailand | B (G1) | 2 | 16.936796° N/103.632850° E |
| NR | Nakhon Ratchasima | Northeast | Thailand | B (G1) | 4 | 14.876044° N/101.724654° E |
| SK | Sakon Nakhon | Northeast | Thailand | B (G1) | 1 | 17.475612° N/103.469636° E |
| SR | Surin | Northeast | Thailand | A (G3) | 8 | 14.947288° N/103.799448° E |
| RG ** | Ranong | South | Thailand | n/a | 5 | 10.395722° N/98.779416° E |
| PG | Phang-nga | South | Thailand | B (G6) | 6 | 8.866375° N/98.345836° E |
| PN ** | Pattalung | South | Thailand | n/a | 12 | 7.508305° N/99.913472° E |
| CD | Siem Reap | Cambodia | Cambodia | A (G3) | 5 | 13.338748° N/103.850614° E |
| Total | 64 |
| Populations | Molecular Diversity Indices | |||||
|---|---|---|---|---|---|---|
| n | S | H | Uh | Hd ± SD | π ± SD | |
| Clade A | 61 | 5 | 6 | 6 | 0.396 ± 0.075 | 0.0009 ± 0.0002 |
| Clade C | 43 * | 13 | 13 | 13 | 0.865 ± 0.030 | 0.0030 ± 0.0003 |
| Total | 104 * | 18 | 19 | 19 | 0.772 ± 0.039 | 0.0049 ± 0.0003 |
| Clade | A | B | C |
|---|---|---|---|
| A | – | 0.0110 | 0.0082 |
| B | 0.0111 | – | 0.0082 |
| C | 0.0082 | 0.0083 | – |
| Source of Variation | d.f. | Ss | Vc | %Va | Fi |
|---|---|---|---|---|---|
| Thailand, Laos, Cambodia, USA | |||||
| Among groups | 3 | 19.608 | 0.06634 | 5.26 | FCT = 0.05257 |
| Among populations within groups | 14 | 43.552 | 0.71146 | 56.39 | FSC = 0.53569 * |
| Within populations | 73 | 45.016 | 0.61666 | 48.87 | FST = 0.51128 * |
| Groups A–E | |||||
| Among groups | 4 | 54.55 | 1.34029 | 73.89 | FCT = 0.73889 * |
| Among populations within groups | 11 | 5.226 | 0.00054 | 0.03 | FSC = 0.00115 * |
| Within populations | 48 | 22.708 | 0.47309 | 26.08 | FST = 0.73919 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laotongsan, P.; Pilap, W.; Jaroenchaiwattanachote, C.; Pasorn, P.; Saijuntha, J.; Tawong, W.; Kongbuntad, W.; Wongpakam, K.; Inkhavilay, K.; Sithirith, M.; et al. Tracing Genetic Divergence and Phylogeographic Patterns of Gekko gecko Linnaeus, 1758 (Squamata: Gekkonidae) Across Southeast Asia Using RAG1 Sequence. Animals 2025, 15, 3004. https://doi.org/10.3390/ani15203004
Laotongsan P, Pilap W, Jaroenchaiwattanachote C, Pasorn P, Saijuntha J, Tawong W, Kongbuntad W, Wongpakam K, Inkhavilay K, Sithirith M, et al. Tracing Genetic Divergence and Phylogeographic Patterns of Gekko gecko Linnaeus, 1758 (Squamata: Gekkonidae) Across Southeast Asia Using RAG1 Sequence. Animals. 2025; 15(20):3004. https://doi.org/10.3390/ani15203004
Chicago/Turabian StyleLaotongsan, Panida, Warayutt Pilap, Chavanut Jaroenchaiwattanachote, Pattana Pasorn, Jatupon Saijuntha, Wittaya Tawong, Watee Kongbuntad, Komgrit Wongpakam, Khamla Inkhavilay, Mak Sithirith, and et al. 2025. "Tracing Genetic Divergence and Phylogeographic Patterns of Gekko gecko Linnaeus, 1758 (Squamata: Gekkonidae) Across Southeast Asia Using RAG1 Sequence" Animals 15, no. 20: 3004. https://doi.org/10.3390/ani15203004
APA StyleLaotongsan, P., Pilap, W., Jaroenchaiwattanachote, C., Pasorn, P., Saijuntha, J., Tawong, W., Kongbuntad, W., Wongpakam, K., Inkhavilay, K., Sithirith, M., Tantrawatpan, C., & Saijuntha, W. (2025). Tracing Genetic Divergence and Phylogeographic Patterns of Gekko gecko Linnaeus, 1758 (Squamata: Gekkonidae) Across Southeast Asia Using RAG1 Sequence. Animals, 15(20), 3004. https://doi.org/10.3390/ani15203004







