Genomic Selection for Cashmere Traits in Inner Mongolian Cashmere Goats Using Random Forest, Gradient Boosting Decision Tree, Extreme Gradient Boosting and Light Gradient Boosting Machine Methods
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Experimental Methods
2.2.1. Environment Configuration
2.2.2. Random Forest
2.2.3. Gradient Boosting Decision Tree
2.2.4. Extreme Gradient Boosting
2.2.5. Light Gradient Boosting Machine
2.2.6. GBLUP
2.2.7. Genomic Prediction Accuracy Evaluation
3. Results
3.1. Results of Descriptive Statistical Analysis of Phenotypic Data
3.2. Optimal Hyperparameters of Machine Learning Models
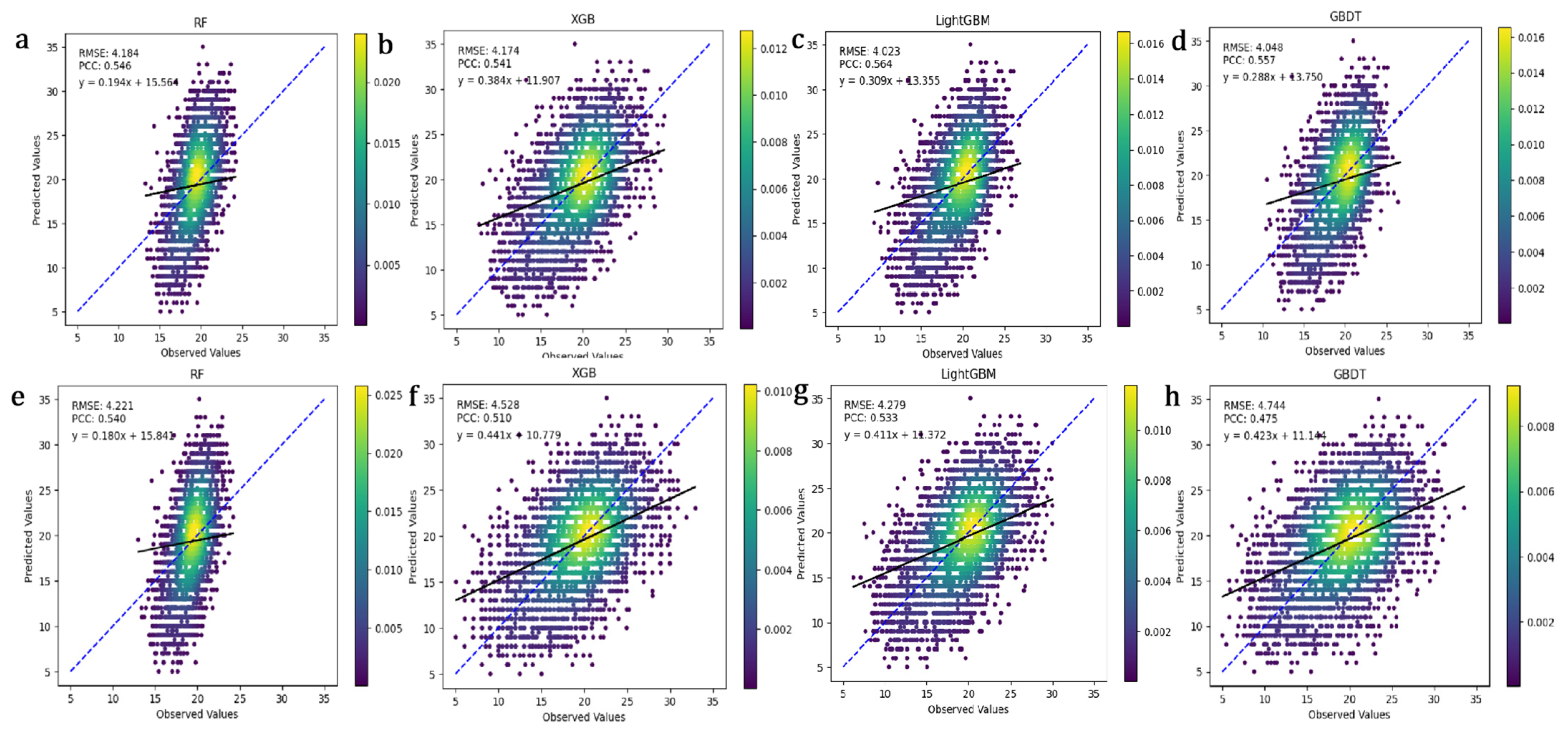
3.3. Genomic Selection Accuracy Evaluation
3.4. Evaluation of Error Metrics for Five Method
3.5. Computational Efficiency of Four Machine Learning Algorithms
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Han, W.; Yang, F.; Wu, Z.; Guo, F.; Zhang, J.; Hai, E.; Shang, F.; Su, R.; Wang, R.; Wang, Z.; et al. Inner Mongolian Cashmere Goat Secondary Follicle Development Regulation Research Based on mRNA-miRNA Co-analysis. Sci. Rep. 2020, 10, 4519. [Google Scholar] [CrossRef]
- Gong, W.; Liu, J.; Mu, Q.; Chahaer, T.; Liu, J.; Ding, W.; Bou, T.; Wu, Z.; Zhao, Y. Melatonin promotes proliferation of Inner Mongolia cashmere goat hair follicle papilla cells through Wnt10b. Genomics 2024, 116, 110844. [Google Scholar] [CrossRef]
- Li, X.; Hao, F.; Hu, X.; Wang, H.; Dai, B.; Wang, X.; Liang, H.; Cang, M.; Liu, D. Generation of Tβ4 knock-in Cashmere goat using CRISPR/Cas9. Int. J. Biol. Sci. 2019, 15, 1743–1754. [Google Scholar] [CrossRef]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2002, 157, 1819–1829. [Google Scholar] [CrossRef]
- Clark, S.A.; van der Werf, J. Genomic best linear unbiased prediction (gBLUP) for the estimation of genomic breeding values. Methods Mol. Biol. 2013, 1019, 321–330. [Google Scholar]
- Forni, S.; Aguilar, I.; Misztal, I. Different genomic relationship matrices for single-step analysis using phenotypic, pedigree and genomic information. Genet. Sel. Evol. 2011, 43, 1. [Google Scholar] [CrossRef]
- Gianola, D.; van Kaam, J.B. Reproducing kernel hilbert spaces regression methods for genomic assisted prediction of quantitative traits. Genetics 2008, 178, 2289–2303. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ishwaran, H. Random forests for genomic data analysis. Genomics 2012, 99, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Howard, J.T.; Rathje, T.A.; Bruns, C.E.; Wilson-Wells, D.F.; Kachman, S.D.; Spangler, M.L. The Impact of Selective Genotyping on the Response to Selection Using Single-step Genomic Best Linear Unbiased Prediction. J. Anim. Sci. 2018, 96, 4532–4542. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Gao, L.; Zhou, B.; Xu, Q.; Liu, Y.; Li, J.; Lv, Q.; Zhang, Y.; Wang, R.; Su, R.; et al. Genomic Selection for Early Growth Traits in Inner Mongolian Cashmere Goats Using ABLUP, GBLUP, and ssGBLUP Methods. Animals 2025, 15, 1733. [Google Scholar] [CrossRef]
- Negro, A.; Cesarani, A.; Cortellari, M.; Bionda, A.; Fresi, P.; Macciotta, N.P.P.; Grande, S.; Biffani, S.; Crepaldi, P. A comparison of genetic and genomic breeding values in Saanen and Alpine goats. Animal 2024, 18, 101118. [Google Scholar] [CrossRef]
- Alves, A.A.C.; Espigolan, R.; Bresolin, T.; Costa, R.M.; Fernandes Júnior, G.A.; Ventura, R.V.; Carvalheiro, R.; Albuquerque, L.G. Genome-enabled prediction of reproductive traits in Nellore cattle using parametric models and machine learning methods. Anim. Genet. 2021, 52, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Lai, X.S.; Liu, D.Y.; Zhang, Z.Y.; Ma, P.P.; Wang, Q.S.; Zhang, Z.; Pan, Y.C. Applications of support vector machine in genomic prediction in pig and maize populations. Front. Genet. 2020, 11, 598318. [Google Scholar] [CrossRef]
- Montesinos López, O.A.; Montesinos López, A.; Crossa, J. Random Forest for Genomic Prediction. In Multivariate Statistical Machine Learning Methods for Genomic Prediction; Springer International Publishing: Cham, Switzerland, 2022; pp. 633–681. [Google Scholar]
- Mahesh, B. Machine Learning Algorithms—A Review. Int. J. Sci. Res. 2020, 9, 381–386. [Google Scholar] [CrossRef]
- Liang, M.; Miao, J.; Wang, X.; Chang, T.; An, B.; Duan, X.; Xu, L.; Gao, X.; Zhang, L.; Li, J.; et al. Application of ensemble learning to genomic selection in chinese simmental beef cattle. J. Anim. Breed. Genet. 2021, 138, 291–299. [Google Scholar] [CrossRef]
- Xiang, T.; Li, T.; Li, J.; Li, X.; Wang, J. Using machine learning to realize genetic site screening and genomic prediction of productive traits in pigs. FASEB J. 2023, 37, e22961. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD ‘16), San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3149–3157. [Google Scholar]
- Liu, B.; Liu, H.; Tu, J.; Xiao, J.; Yang, J.; He, X.; Zhang, H. An investigation of machine learning methods applied to genomic prediction in yellow-feathered broilers. Poult. Sci. 2025, 104, 104489. [Google Scholar] [CrossRef]
- Wang, X.; Shi, S.; Wang, G.; Luo, W.; Wei, X.; Qiu, A.; Luo, F.; Ding, X. Using machine learning to improve the accuracy of genomic prediction of reproduction traits in pigs. J. Anim. Sci. Biotechnol. 2022, 13, 60. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, J.; Hao, W.; Zong, W.; Liang, M.; Zhao, F.; Zhang, L.; Wang, L.; Gao, H.; Wang, L. An interpretable integrated machine learning framework for genomic selection. Smart Agric. Technol. 2025, 101138, 2772–3755. [Google Scholar] [CrossRef]
- Wang, J.; Chai, J.; Chen, L.; Zhang, T.; Long, X.; Diao, S.; Chen, D.; Guo, Z.; Tang, G.; Wu, P. Enhancing Genomic Prediction Accuracy of Reproduction Traits in Rongchang Pigs Through Machine Learning. Animals 2025, 15, 525. [Google Scholar] [CrossRef]
- Azodi, C.B.; Bolger, E.; McCarren, A.; Roantree, M.; de Los Campos, G.; Shiu, S.H. Benchmarking Parametric and Machine Learning Models for Genomic Prediction of Complex Traits. G3 Genes Genomes Genet. 2019, 9, 3691–3702. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Bao, S.; Zhao, X.; Bai, Y.; Lv, Y.; Gao, P.; Li, F.; Zhang, W. Genome-Wide Association Study and Phenotype Prediction of Reproductive Traits in Large White Pigs. Animals 2024, 14, 3348. [Google Scholar] [CrossRef]
- Chen, D.; Wang, S.J.; Zhao, Z.J.; Ji, X.; Shen, Q.; Yu, Y.; Cui, S.D.; Wang, J.G.; Chen, Z.Y.; Wang, J.Y.; et al. Genomic prediction of pig growth traits based on machine learning. Yi Chuan 2023, 45, 922–932. [Google Scholar] [PubMed]
- Zhang, Y.; Zhang, M.; Ye, J.; Xu, Q.; Feng, Y.; Xu, S.; Hu, D.; Wei, X.; Hu, P.; Yang, Y. Integrating genome-wide association study into genomic selection for the prediction of agronomic traits in rice (Oryza sativa L.). Mol. Breed. 2023, 43, 81. [Google Scholar] [CrossRef]
- Izquierdo, P.; Wright, E.M.; Cichy, K. GWAS-assisted and multitrait genomic prediction for improvement of seed yield and canning quality traits in a black bean breeding panel. G3 Genes Genomes Genet. 2025, 15, jkaf007. [Google Scholar] [CrossRef]



| Trait | Number of Record | Mean | SD | Coefficient of Variation |
|---|---|---|---|---|
| FL/cm 1 | 9610 | 18.90 | 4.90 | 25.91% |
| CD/µm 2 | 5356 | 15.23 1 | 0.81 | 5.31% |
| CP/g 3 | 9040 | 740.32 | 215.19 | 29.07% |
| Method | Optimal Hyperparameters |
|---|---|
| RF | max_depth = 18, n_estimators = 250 |
| XGBoost | n_estimators = 100, learning_rate = 0.1, max_depth = 5 |
| LightGBM | learning_rate = 0.05, max_depth = 5, num_leaves = 20 |
| GBDT | n_estimators = 300, learning_rate = 0.1, max_depth = 5 |
| Hyperparameters | Method | FL 1 | CP 2 | CD 3 |
|---|---|---|---|---|
| Tuning 4 | GBLUP | 0.543 ± 0.0315 | 0.327 ± 0.0478 | 0.396 ± 0.0361 |
| RF | 0.546 ± 0.0699 | 0.352 ± 0.0397 | 0.401 ± 0.0397 | |
| XGB | 0.541 ± 0.0475 | 0.309 ± 0.0794 | 0.387 ± 0.0325 | |
| LightGBM | 0.564 ± 0.0129 | 0.336 ± 0.0799 | 0.402 ± 0.0359 | |
| GBDT | 0.557 ± 0.0423 | 0.320 ± 0.0631 | 0.404 ± 0.0310 | |
| Default 5 | RF | 0.540 ± 0.0509 | 0.336 ± 0.0699 | 0.396 ± 0.0354 |
| XGB | 0.510 ± 0.0443 | 0.299 ± 0.0738 | 0.362 ± 0.0347 | |
| LightGBM | 0.533 ± 0.0445 | 0.314 ± 0.0755 | 0.381 ± 0.0330 | |
| GBDT | 0.475 ± 0.0478 | 0.262 ± 0.0678 | 0.340 ± 0.0388 |
| Hyperparameters | Method | FL 1 | CP 2 | CD 3 | |||
|---|---|---|---|---|---|---|---|
| MSE 4/cm2 | MAE 5/cm2 | MSE/g2 | MAE/g2 | MSE/µm2 | MAE/µm2 | ||
| GBLUP | 16.716 | 3.267 | 0.050 | 0.149 | 0.565 | 0.576 | |
| Tuning | RF | 17.508 | 3.343 | 0.049 | 0.150 | 0.543 | 0.574 |
| XGBoost | 17.419 | 3.340 | 0.053 | 0.154 | 0.578 | 0.591 | |
| LightGBM | 16.359 | 3.239 | 0.049 | 0.149 | 0.537 | 0.570 | |
| GBDT | 16.384 | 3.252 | 0.050 | 0.150 | 0.533 | 0.569 | |
| Default | RF | 17.814 | 3.370 | 0.050 | 0.152 | 0.549 | 0.578 |
| XGBoost | 20.503 | 3.610 | 0.063 | 0.170 | 0.682 | 0.637 | |
| LightGBM | 18.306 | 3.426 | 0.057 | 0.160 | 0.613 | 0.605 | |
| GBDT | 22.502 | 3.769 | 0.068 | 0.179 | 0.736 | 0.662 | |
| Hyper-Parameters | Method | FL 1 | CP 2 | CD 3 |
|---|---|---|---|---|
| GBLUP | 9748.25 s | 19,351.12 s | 62,875.9 s | |
| Tuning | RF | 131.45 s | 128.14 s | 139.05 s |
| XGB | 421.79 s | 433.66 s | 382.62 s | |
| LightGBM | 199.41 s | 233.12 s | 222.49 s | |
| GBDT | 107.23 s | 95.51 s | 82.97 s | |
| Default | RF | 189.47 s | 135.45 s | 189.47 s |
| XGB | 482.26 s | 462.17 s | 401.31 s | |
| LightGBM | 247.33 s | 282.89 s | 235.86 s | |
| GBDT | 200.77 s | 106.18 s | 86.29 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Yan, X.; Li, W.; Xue, S.-H.; Wang, Z.; Su, R. Genomic Selection for Cashmere Traits in Inner Mongolian Cashmere Goats Using Random Forest, Gradient Boosting Decision Tree, Extreme Gradient Boosting and Light Gradient Boosting Machine Methods. Animals 2025, 15, 2940. https://doi.org/10.3390/ani15202940
Liu J, Yan X, Li W, Xue S-H, Wang Z, Su R. Genomic Selection for Cashmere Traits in Inner Mongolian Cashmere Goats Using Random Forest, Gradient Boosting Decision Tree, Extreme Gradient Boosting and Light Gradient Boosting Machine Methods. Animals. 2025; 15(20):2940. https://doi.org/10.3390/ani15202940
Chicago/Turabian StyleLiu, Jiaqi, Xiaochun Yan, Wenze Li, Shan-Hui Xue, Zhiying Wang, and Rui Su. 2025. "Genomic Selection for Cashmere Traits in Inner Mongolian Cashmere Goats Using Random Forest, Gradient Boosting Decision Tree, Extreme Gradient Boosting and Light Gradient Boosting Machine Methods" Animals 15, no. 20: 2940. https://doi.org/10.3390/ani15202940
APA StyleLiu, J., Yan, X., Li, W., Xue, S.-H., Wang, Z., & Su, R. (2025). Genomic Selection for Cashmere Traits in Inner Mongolian Cashmere Goats Using Random Forest, Gradient Boosting Decision Tree, Extreme Gradient Boosting and Light Gradient Boosting Machine Methods. Animals, 15(20), 2940. https://doi.org/10.3390/ani15202940





