The Genetic Structure and Diversity of Different Pigeon Breeds Based on a 5 K Single Nucleotide Polymorphism Chip
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Screening of the Pigeon 5 K Genetic Markers
2.3. Design of Liquid-Phase Probes
2.4. Targeted Sequencing and Genotyping of Pigeon DNA Samples
2.5. Basic Statistics of Genetic Diversity
2.6. Population Analysis
3. Results
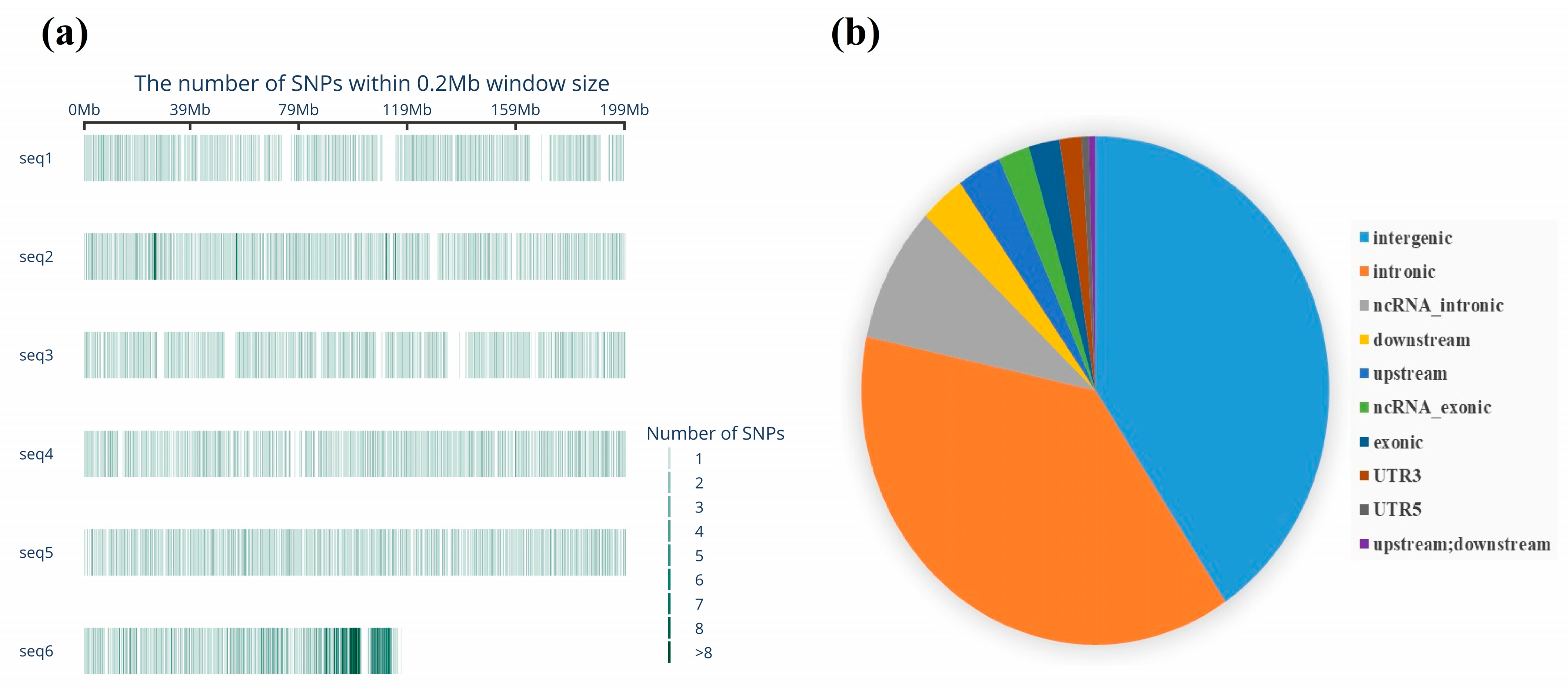
3.1. Basic Statistics of Genetic Diversity
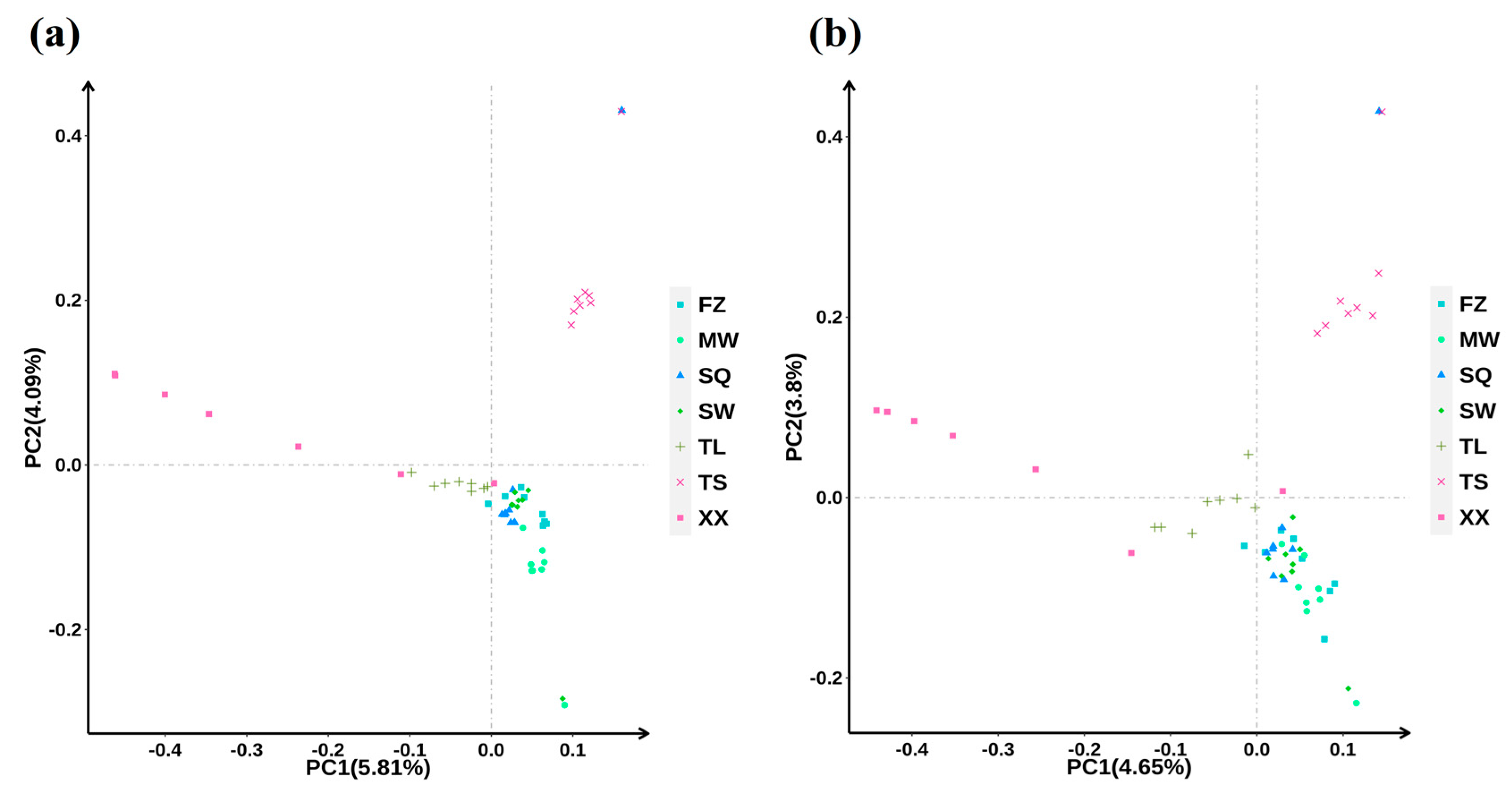
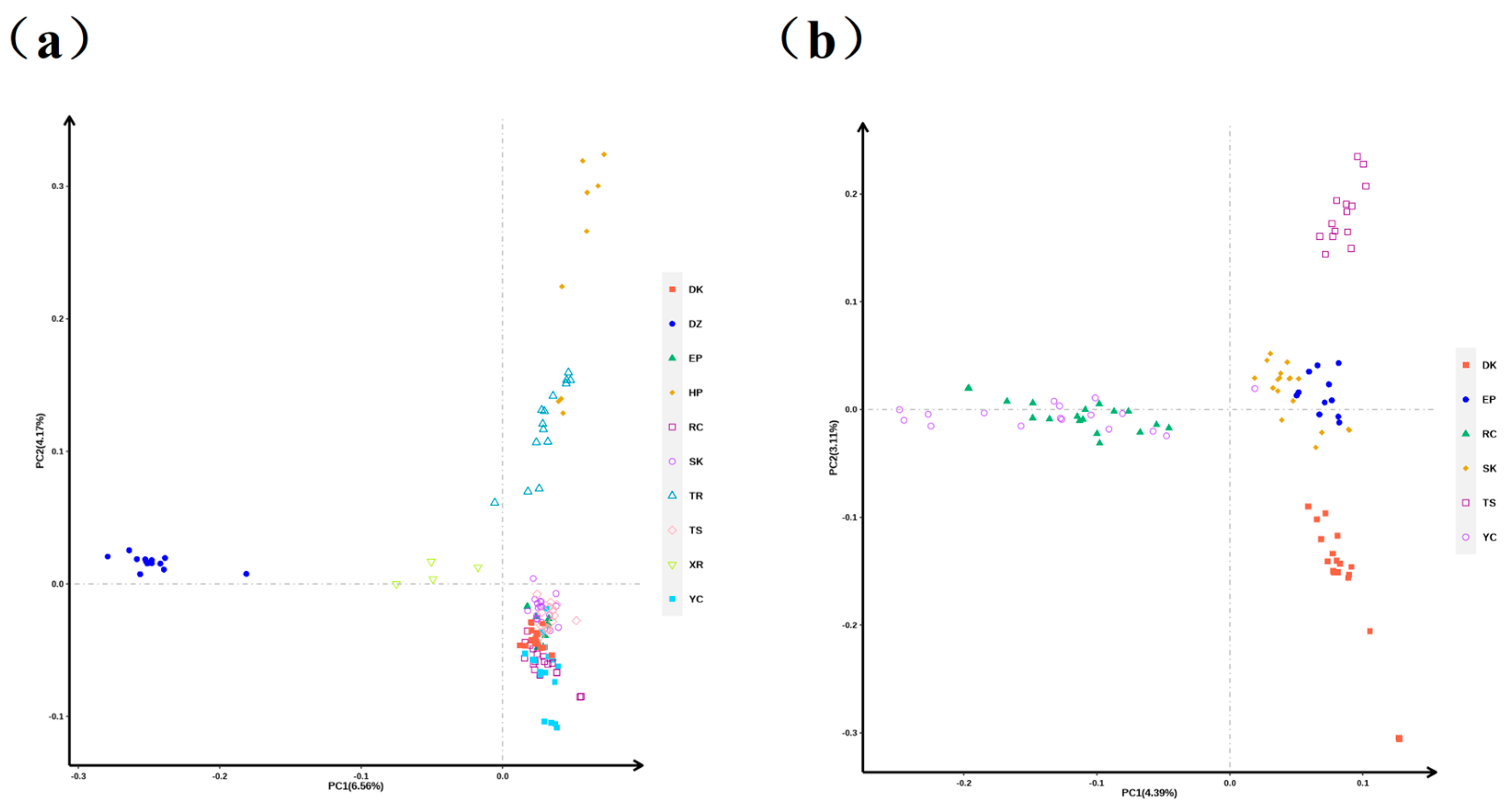
3.2. PCA
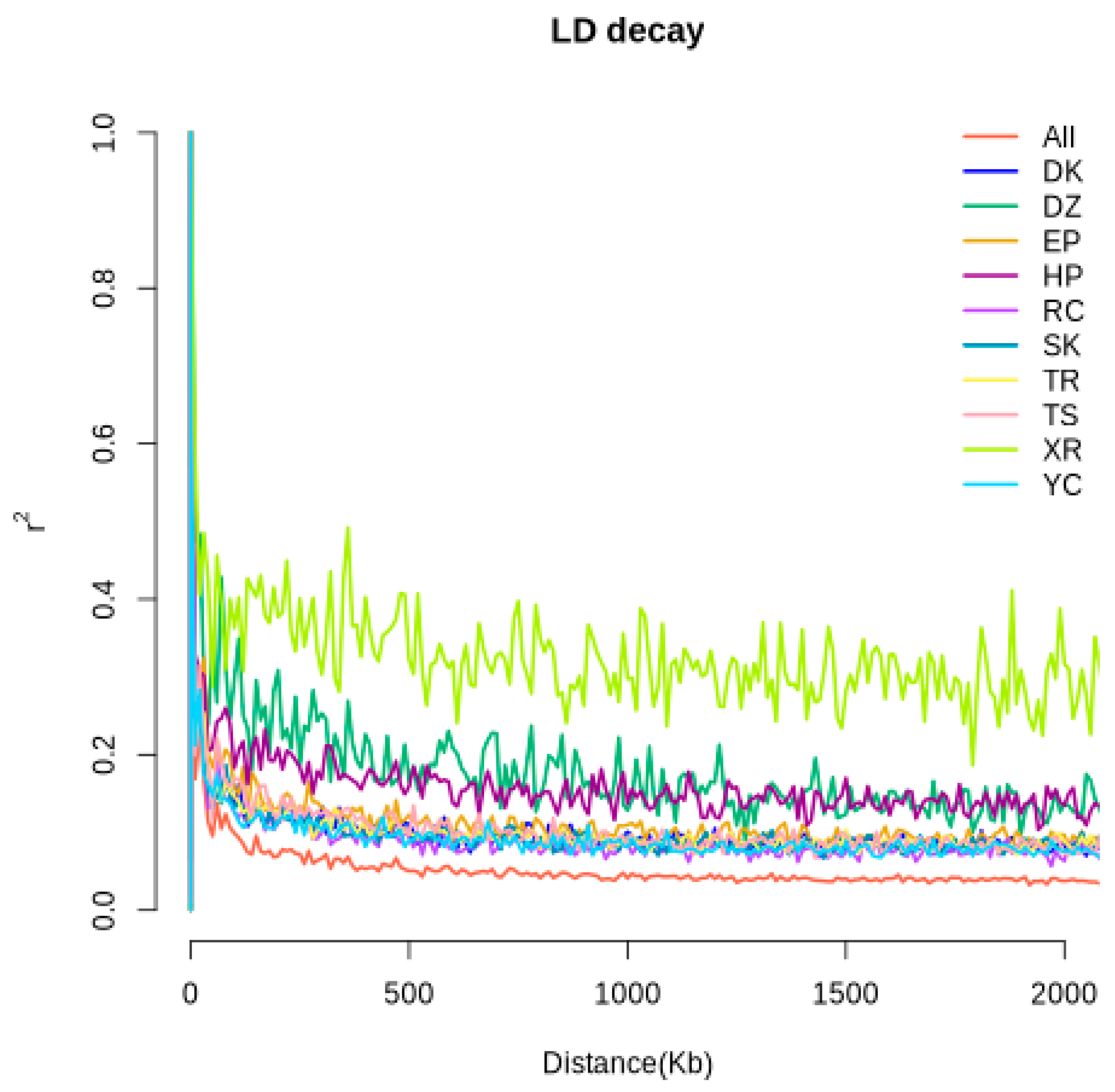
3.3. Population Structure Analysis
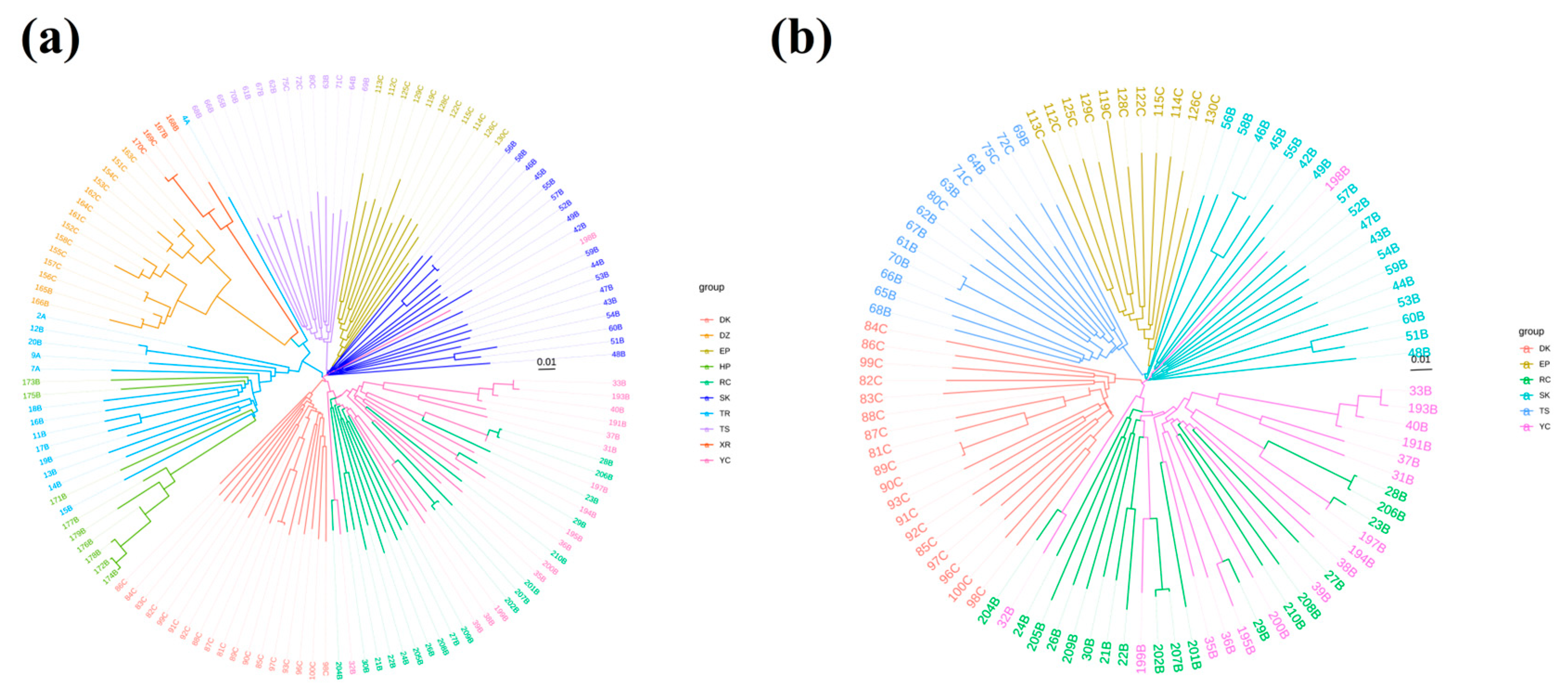
3.4. Phylogenetic Tree Construction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pacheco, G.; van Grouw, H.; Shapiro, M.D.; Gilbert, M.T.P.; Vieira, F.G. Darwin’s Fancy Revised: An Updated Understanding of the Genomic Constitution of Pigeon Breeds. Genome Biol. Evol. 2020, 12, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.A.; Arif, I.A. COI barcodes and phylogeny of doves (Columbidae family). Mitochondrial DNA 2013, 24, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Balog, K.; Wadday, A.S.; Al-Hasan, B.A.; Wanjala, G.; Kusza, S.; Fehér, P.; Stéger, V.; Bagi, Z. MtDNA genetic diversity and phylogeographic insights into giant domestic pigeon (Columba livia domestica) breeds: Connections between Central Europe and the Middle East. Poult. Sci. 2024, 103, 104310. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, M.D.; Kronenberg, Z.; Li, C.; Domyan, E.T.; Pan, H.; Campbell, M.; Tan, H.; Huff, C.D.; Hu, H.; Vickrey, A.I.; et al. Genomic diversity and evolution of the head crest in the rock pigeon. Science 2013, 339, 1063–1067. [Google Scholar] [CrossRef] [PubMed]
- Stringham, S.A.; Mulroy, E.E.; Xing, J.; Record, D.; Guernsey, M.W.; Aldenhoven, J.T.; Osborne, E.J.; Shapiro, M.D. Divergence, convergence, and the ancestry of feral populations in the domestic rock pigeon. Curr. Biol. CB 2012, 22, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Wang, X.; Ding, W.; Xiao, C.; Cai, X.; Lv, W.; Tu, Y.; Zhao, W.; Yao, J.; Yang, C. Whole-genome sequencing reveals the artificial selection and local environmental adaptability of pigeons (Columba livia). Evol. Appl. 2022, 15, 603–617. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; Li, W.; Jin, H.; Zhang, L.; Liu, G. Development and Validation of a 54K Genome-Wide Liquid SNP Chip Panel by Target Sequencing for Dairy Goat. Genes 2023, 14, 1122. [Google Scholar] [CrossRef] [PubMed]
- Podbielska, A.; Radko, A. Genetic Structure of Racing Pigeons (Columba livia) Kept in Poland Based on Microsatellite Markers. Genes 2022, 13, 1175. [Google Scholar] [CrossRef] [PubMed]
- Balog, K.; Mizeranschi, A.E.; Wanjala, G.; Sipos, B.; Kusza, S.; Bagi, Z. Application potential of chicken DNA chip in domestic pigeon species—Preliminary results. Saudi J. Biol. Sci. 2023, 30, 103594. [Google Scholar] [CrossRef] [PubMed]
- Stefaniuk-Szmukier, M.; Andres, K.; Piórkowska, K.; Ropka-Molik, K. Low diversity of mitochondrial DNA in fancy pigeons (Columba livia) revealed by partial D-loop sequencing. Anim. Genet. 2021, 52, 382. [Google Scholar] [CrossRef] [PubMed]
- Speak, S.A.; Birley, T.; Bortoluzzi, C.; Clark, M.D.; Percival-Alwyn, L.; Morales, H.E.; van Oosterhout, C. Genomics-informed captive breeding can reduce inbreeding depression and the genetic load in zoo populations. Mol. Ecol. Resour. 2024, 24, e13967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Mu, C.; Chang, L.; Shen, X.; Bu, Z.; Yang, M.; Fu, S.; Tang, Q.; Liu, P.; Yang, X. Whole-Genome Sequencing for Identifying Candidate Genes Related to the Special Phenotypes of the Taihu Dianzi Pigeon. Animals 2024, 14, 1047. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zhou, H.; Huang, J.; Zhu, W.; Hou, H.; Li, H.; Zhao, L.; Zhang, J.; Liu, J.; Qin, C.; et al. Near telomere-to-telomere assembly of the Tarim pigeon (Columba livia) genome. Sci. Data 2024, 11, 1455. [Google Scholar] [CrossRef] [PubMed]
- Domyan, E.T.; Guernsey, M.W.; Kronenberg, Z.; Krishnan, S.; Boissy, R.E.; Vickrey, A.I.; Rodgers, C.; Cassidy, P.; Leachman, S.A.; Fondon, J.W., 3rd; et al. Epistatic and combinatorial effects of pigmentary gene mutations in the domestic pigeon. Curr. Biol. CB 2014, 24, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Entrikin, R.K.; Erway, L.C. A genetic investigation of roller and tumbler pigeons. J. Hered. 1972, 63, 351–354. [Google Scholar] [CrossRef] [PubMed]

| Group | MAF | PIC | Ao | Ae | Ho | He |
|---|---|---|---|---|---|---|
| All | 0.2574 | 0.2965 | 2.0034 | 1.6084 | 0.3448 | 0.3676 |
| DK | 0.2434 | 0.2718 | 1.9854 | 1.5624 | 0.3622 | 0.3368 |
| DZ | 0.1605 | 0.1724 | 1.6246 | 1.3672 | 0.2494 | 0.2152 |
| EP | 0.2421 | 0.2653 | 1.9568 | 1.5558 | 0.3354 | 0.3297 |
| HP | 0.2251 | 0.2418 | 1.8834 | 1.5129 | 0.3256 | 0.3014 |
| RC | 0.2522 | 0.2771 | 1.9886 | 1.5816 | 0.3534 | 0.3448 |
| SK | 0.2607 | 0.2870 | 1.9943 | 1.6039 | 0.3794 | 0.3576 |
| TR | 0.2600 | 0.2808 | 1.9819 | 1.5974 | 0.3677 | 0.3507 |
| TS | 0.2422 | 0.2672 | 1.9701 | 1.5575 | 0.3498 | 0.3319 |
| XR | 0.1770 | 0.1854 | 1.6118 | 1.4028 | 0.3239 | 0.2329 |
| YC | 0.2518 | 0.2749 | 1.9846 | 1.5790 | 0.3577 | 0.3423 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, H.; Li, X.; Wang, X.; Cai, X.; Tu, Y.; Lv, W.; Shen, X.; Yang, C.; Yao, J. The Genetic Structure and Diversity of Different Pigeon Breeds Based on a 5 K Single Nucleotide Polymorphism Chip. Animals 2025, 15, 2864. https://doi.org/10.3390/ani15192864
Hou H, Li X, Wang X, Cai X, Tu Y, Lv W, Shen X, Yang C, Yao J. The Genetic Structure and Diversity of Different Pigeon Breeds Based on a 5 K Single Nucleotide Polymorphism Chip. Animals. 2025; 15(19):2864. https://doi.org/10.3390/ani15192864
Chicago/Turabian StyleHou, Haobin, Xin Li, Xiaoliang Wang, Xia Cai, Yingying Tu, Wenwei Lv, Xiaohui Shen, Changsuo Yang, and Junfeng Yao. 2025. "The Genetic Structure and Diversity of Different Pigeon Breeds Based on a 5 K Single Nucleotide Polymorphism Chip" Animals 15, no. 19: 2864. https://doi.org/10.3390/ani15192864
APA StyleHou, H., Li, X., Wang, X., Cai, X., Tu, Y., Lv, W., Shen, X., Yang, C., & Yao, J. (2025). The Genetic Structure and Diversity of Different Pigeon Breeds Based on a 5 K Single Nucleotide Polymorphism Chip. Animals, 15(19), 2864. https://doi.org/10.3390/ani15192864





