Mitogenomic Insights into Phylogeny, Biogeography and Adaptive Evolution of the Genus Typhlomys (Rodentia: Platacanthomyidae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Extraction
2.2. Sequencing and Assembly
2.3. Mitochondrial Genome Annotation
2.4. Comparative Analysis
2.5. Phylogenetic Analysis
2.6. Divergence Time Estimation
2.7. Ancestral Distribution Analysis
2.8. Selection Pressure Analysis
3. Results
3.1. Genome Structure and Organization
3.2. Protein-Coding Genes (PCGs)
3.3. Ribosomal RNA and Transfer RNA
3.4. Overlapping and Intergenic Spacer Regions
3.5. A+T Rich Region
3.6. Phylogenetic Analyses
3.7. Ancestral Distributions
3.8. Selection Pressure in Mitochondrial Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Musser, G.G.; Carleton, M.D. Superfamily Muroidea. In Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd ed.; Wilson, D.E., Reeder, D.M., Eds.; Johns Hopkins University Press: Baltimore, ML, USA, 2005; Volume 5, pp. 894–1531. [Google Scholar]
- Panyutina, A.A.; Kuznetsov, A.N.; Volodin, I.A.; Abramov, A.V.; Soldatova, I.B. A blind climber: The first evidence of ultrasonic echolocation in arboreal mammals. Integr. Zool. 2017, 12, 172–184. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Liu, Q.; Xu, D.M.; Qi, F.Y.; Bai, J.; He, S.W.; Chen, P.; Zhou, X.; Cai, W.Z.; Chen, Z.Z.; et al. Echolocation in soft-furred tree mice. Science 2021, 372, eaay1513. [Google Scholar] [CrossRef]
- Cheng, F.; He, K.; Chen, Z.Z.; Zhang, B.; Wan, T.; Li, J.T.; Zhang, B.W.; Jiang, X.L. Phylogeny and systematic revision of the genus Typhlomys (Rodentia, Platacanthomyidae), with description of a new species. J. Mammal. 2017, 98, 731–743. [Google Scholar] [CrossRef]
- Smith, A.T.; Xie, Y. A Guide to the Mammals of China; Princeton University Press: Princeton, NJ, USA, 2013; pp. 208–209. [Google Scholar]
- Abramov, A.; Balakirev, A.; Rozhnov, V. An enigmatic pygmy dormouse: Molecular and morphological evidence for the species taxonomic status of Typhlomys chapensis (Rodentia: Platacanthomyidae). Zool. Stud. 2014, 53, 34. [Google Scholar] [CrossRef]
- Balakirev, A.E.; Phuong, B.X.; Rozhnov, V.V. Typhlomys (Rodentia, Platacanthomyidae): New species of the genus from northern Vietnam with notes on conservation status and distribution. Biodivers. Data. J. 2024, 12, e133363. [Google Scholar] [CrossRef]
- Su, W.T.; Chen, Z.Z.; Wan, T.; Wang, X.; Zhou, H.Y.; Hu, Y.; Wang, J.H.; Jiang, X.L.; Nie, W.H.; He, K. Taxonomy and distribution of the genus Typhlomys in China based on karyotypic and phylogenetic analyses. Acta Theriol. Sin. 2020, 40, 239–248. [Google Scholar]
- Pu, Y.T.; Wan, T.; Fan, R.H.; Fu, C.K.; Tang, K.Y.; Jiang, X.L.; Zhang, B.W.; Hu, T.L.; Chen, S.D.; Liu, S.Y. A new species of the genus Typhlomys Milne-Edwards, 1877 (Rodentia: Platacanthomyidae) from Chongqing, China. Zool. Res. 2022, 43, 413. [Google Scholar] [CrossRef]
- Wu, J.Y. The Discovery of Chinese Pygmy Dormouse (Typhlomys cinereus) in Qinling mountain. Zool. Res. 1990, 2, 108–126. [Google Scholar]
- Milne-Edwards, A. Sur quelques mammifères et crustacés nouveaux. Bull. Soc Philomath. Paris 1877, 12, 8–10. [Google Scholar]
- Hu, T.L.; Cheng, F.; Xu, Z.; Chen, Z.Z.; Zhang, B.W. Molecular and morphological evidence for a new species of the genus Typhlomys (rodentia: Platacanthomyidae). Zool. Res. 2021, 42, 100. [Google Scholar] [CrossRef] [PubMed]
- Ballard, J.W.; Whitlock, M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004, 13, 729–744. [Google Scholar] [CrossRef]
- Zhou, Z.X.; Liu, B.; Gong, J.; Bai, Y.L.; Yang, J.Y.; Xu, P. Phylogeny and population genetics of species in Takifugu genus based on mitochondrial genome. J. Fish. China 2020, 44, 1792–1803. [Google Scholar]
- Zhang, W.P.; Qiu, B.Y.; Zhang, D.S. Adaptive Evolution Analysis of Mitochondrial Genomes in Anseriform Birds with Various Feeding Habits. Curr. Biotechnol. 2023, 13, 748. [Google Scholar]
- Hu, T.W.; Wang, S.H.; Zhang, D.S. Adaptive Evolution of Mitochondrial Genome in Red-blooded Antarctic Fish and White-blooded Antarctic Fish. Genom. Appl. Biolo. 2021, 40, 3448–3457. [Google Scholar]
- Lv, X.; Cong, H.; Kong, L.; Motokawa, M.; Harada, M.; Wu, Y.; Li, Y.C. The nearly complete mitochondrial genome of chinese pygmy dormouse Typhlomys cinereus (rodentia: Platacanthomyidae). Mitochondrial DNA Resour. 2016, 1, 605–606. [Google Scholar] [CrossRef]
- Chen, X.; Ni, G.; He, K.; Ding, Z.L.; Zhang, Y.P. Capture hybridization of long-range dna fragments for high-throughput sequencing. Methods. Mol. Biol. 2018, 1754, 29–44. [Google Scholar] [PubMed]
- Yang, M.; Song, L.; Zhou, L.; Shi, Y.; Song, N.; Zhang, Y. Mitochondrial genomes of four satyrine butterflies and phylogenetic relationships of the family Nymphalidae (Lepidoptera: Papilionoidea). Int. J. Biol. Macromol. 2020, 145, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, D.; Patrick, M.; Guillaume, S. Novoplasty: De novo assembly of organelle genomes from whole genome data. Nucleic. Acids. Res. 2017, 45, e18. [Google Scholar]
- Bernt, M.; Donath, A.; Juehling, F.; Stadler, P. Mitos: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2012, 69, 313–319. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Hassanin, A.; Leger, N.; Deutsch, J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetic inferences. Syst. Biol. 2005, 54, 277–298. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.L.; JakovliĆ, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandse, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. Figtree ver 1.4.4. Institute of Evolutionary Biology; University of Edinburgh: Edinburgh, UK, 2018. [Google Scholar]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef]
- Beard, K.C.; Qi, T.; Dawson, M.R.; Wang, B.; Li, C. A diverse new primate fauna from middle eocene fissure-fillings in southeastern china. Nature 1994, 368, 604–609. [Google Scholar] [CrossRef]
- Andrew, R.; Drummond, A.J.; Dong, X.; Guy, B.; Marc, A.S. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Harris, A.J.; Blair, C.; He, X. RASP (Reconstruct Ancestral State in Phylogenies): A tool for historical biogeography. Mol. Biol. Evol. 2015, 87, 46–49. [Google Scholar] [CrossRef]
- Ree, R.H.; Smith, S.A. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 2008, 57, 4–14. [Google Scholar] [CrossRef]
- Ronquist, F. Dispersal-vicariance analysis: A new approach to the quantification of historical biogeography. Syst. Biol. 1997, 46, 195–203. [Google Scholar] [CrossRef]
- Landis, M.J.; Matzke, N.J.; Moore, B.R.; Huelsenbeck, J.P. Bayesian analysis of biogeography when the number of areas is large. Syst. Biol. 2013, 62, 789–804. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Kitching, I.; Xu, Z.B.; Huang, Y.X. First mitogenome of subfamily langiinae (lepidoptera: Sphingidae) with its phylogenetic implications. Gene 2021, 789, 145667. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; Haeseler, A.V. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Da Fonseca, R.R.; Johnson, W.E.; O’Brien, S.J.; Ramos, M.J.; Antunes, A. The adaptive evolution of the mammalian mitochondrial genome. BMC Genom. 2008, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.X.; Szymura, J.M.; Hewitt, G.M. Evolution and structural conservation of the control region of insect mitochondrial DNA. Mol. Evol. 1995, 40, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Árnadóttir, E.R.; Moore, K.H.S.; Guðmundsdóttir, V.B.; Ebenesersdóttir, S.S.; Guity, K.; Jónsson, H.; Helgason, A. The rate and nature of mitochondrial DNA mutations in human pedigrees. Cell 2024, 187, 3904–3918. [Google Scholar] [CrossRef]
- Galtier, N.; Roux, C.; Rousselle, M.; Romiguier, J.; Figuet, E.; Glémin, S.; Bierne, N.; Duret, L. Codon Usage Bias in Animals: Disentangling the Effects of Natural Selection, Effective Population Size, and GC-Biased Gene Conversion. Mol. Biol. Evol. 2018, 35, 1092–1103. [Google Scholar] [CrossRef]
- Huang, Y.X.; Xing, Z.P.; Zhang, H.; Xu, Z.B.; Tao, L.L.; Hu, H.Y.; Kitching, I.J.; Wang, X. Characterization of the Complete Mitochondrial Genome of Eight Diurnal Hawkmoths (Lepidoptera: Sphingidae): New Insights into the Origin and Evolution of Diurnalism in Sphingids. Insects 2022, 13, 887. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, K.; Csorba, G.; Hughes, A.C.; Jin, L.; Xiao, Y.; Feng, J. Complete mitochondrial genomes reveal robust phylogenetic signals and evidence of positive selection in horseshoe bats. BMC. Ecol. Evol. 2021, 21, 199. [Google Scholar] [CrossRef] [PubMed]
- Wirth, C.; Brandt, U.; Hunte, C.; Zickermann, V. Structure and function of mitochondrial complex I. Biochim. Biophy. Acta (BBA)-Bioenerg. 2016, 1857, 902–914. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Pan, Z.Q.; Chen, K.K.; Tao, R.S.; Su, C.Y.; Hao, J.S.; Yang, Q. Genetic differentiation and phylogeography of the alpine butterfly Parnassius glacialis (Papilionidae: Parnassinae) in China: Evidence from mitogenomic AT-rich region. Acta Entomol. Sin. 2019, 62, 475–488. [Google Scholar]


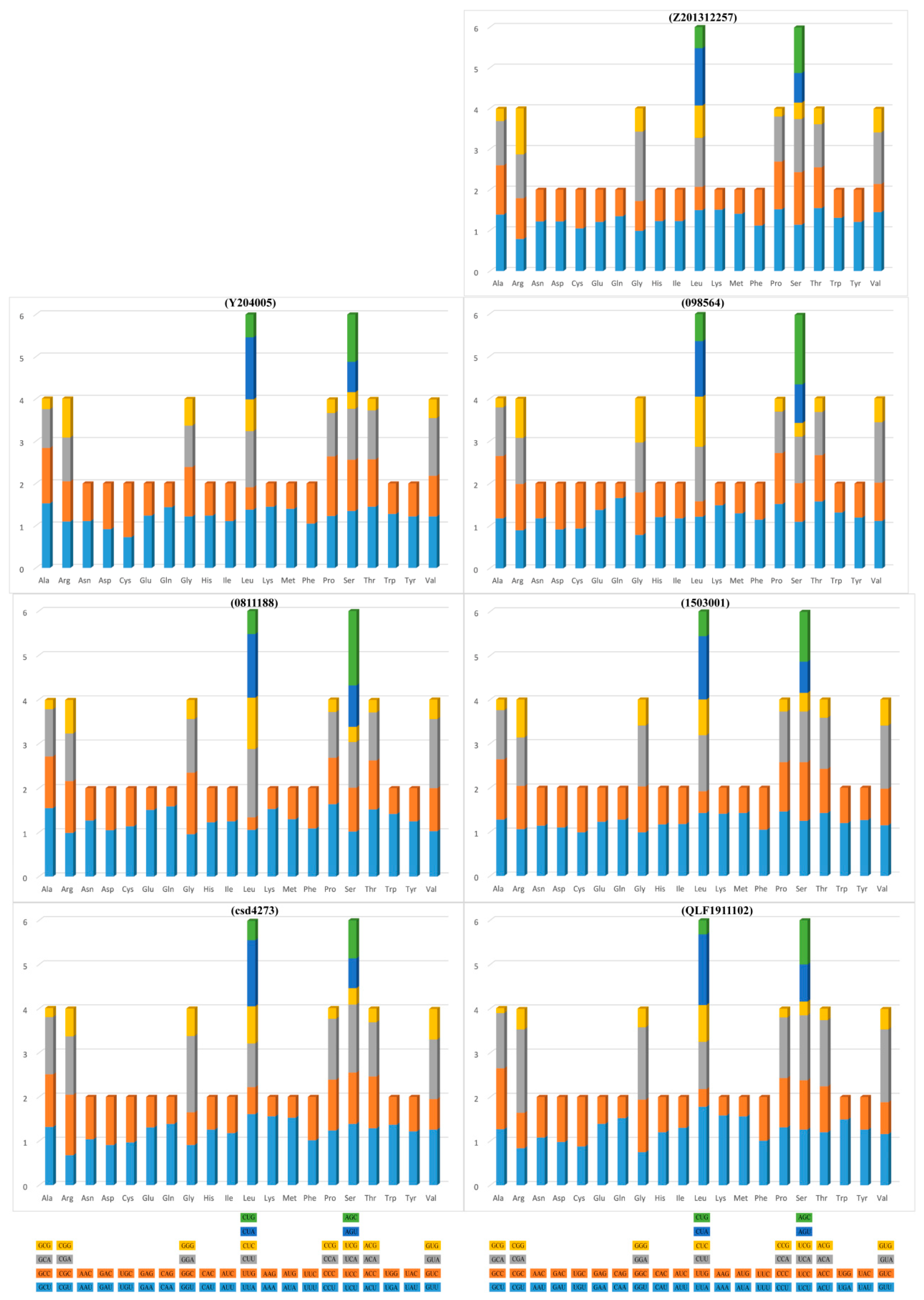
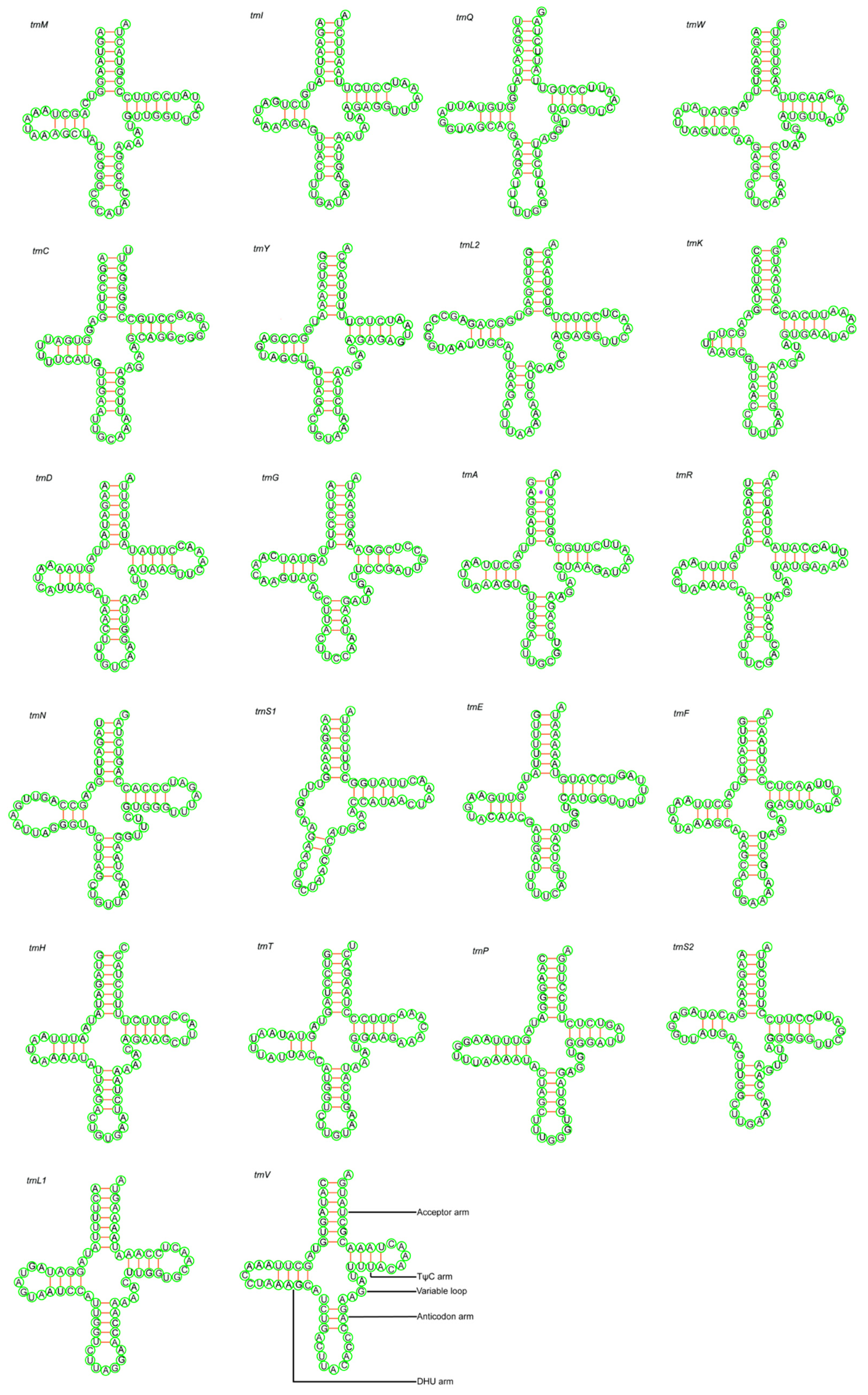
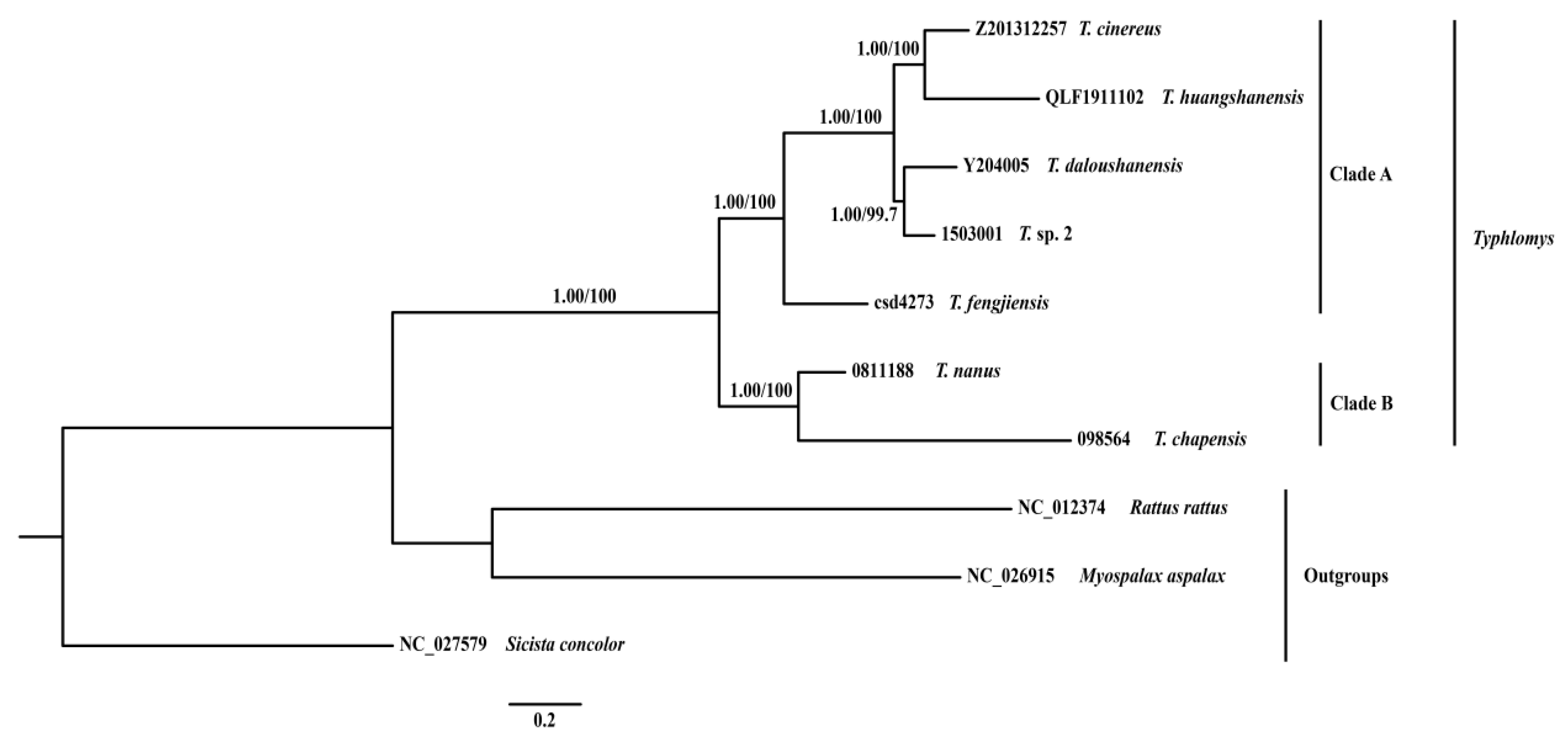



| Species | Field Number | GenBank Accession No. | Location | Latitude (N) | Longitude (E) | Elevation |
|---|---|---|---|---|---|---|
| Typhlomys chapensis | 098564 | PQ825594 | Mt. Huanglian, Yunnan | 22.87 | 103.24 | 2232 |
| Typhlomys cinereus | Z201312257 | PQ825595 | Mt. Wuyi, Fujian | 27.64 | 117.90 | 400 |
| Typhlomys daloushanensis | Y204005 | PQ825596 | Mt. Dalou, Chongqing | 29.02 | 107.19 | 2105 |
| Typhlomys nanus | 0811188 | PQ825597 | Mt. Jiaozi, Yunnan | 26.07 | 102.83 | 3204 |
| Typhlomys huangshanensis | QLF1911102 | PQ825598 | Mt. Qingliangfeng, Anhui | 30.13 | 118.85 | 1303 |
| Typhlomys fengjiensis | CSD4273 | PQ825599 | Fenjie, Chongqing, China | 30.66 | 109.52 | 1883 |
| Typhlomys sp. 2 | 1503001 | PQ825600 | Mt. Dawei, Yunnan | 23.30 | 103.95 | 2043 |
| Rattus rattus | NC_012374 | |||||
| Myosplax aspalax | NC_026915 | |||||
| Sicista concolor | NC_027579 |
| Taxa | Length (bp) | A% | C% | G% | T% | G+C% | A+T% |
|---|---|---|---|---|---|---|---|
| T. chapensis | 17,380 | 33.5 | 26.8 | 12.6 | 27.2 | 39.4 | 60.6 |
| T. cinereus | 16,776 | 34.7 | 24.1 | 11.3 | 29.9 | 35.4 | 64.6 |
| T. daloushanensis | 16,984 | 34.2 | 25.3 | 11.8 | 28.7 | 37.1 | 62.9 |
| T. nanus | 16,665 | 33.9 | 25.9 | 12.3 | 28.0 | 38.2 | 61.8 |
| T. huangshanensis | 16,497 | 34.3 | 23.9 | 11.6 | 30.2 | 35.5 | 64.5 |
| T.fengjiensis | 16,487 | 34.5 | 24.5 | 11.7 | 29.3 | 36.2 | 63.8 |
| T. sp. 2 | 16,490 | 34.3 | 24.7 | 11.8 | 29.9 | 36.5 | 63.5 |
| Position | Strand | Length | Intergenic Nucleotides | Codons | |||
|---|---|---|---|---|---|---|---|
| From | To | Start | Stop | ||||
| trnF | 1 | 69 | + | 69 | 2 | ||
| rrnS | 72 | 1026 | + | 955 | −1 | ||
| trnV | 1026 | 1092 | + | 67 | 0 | ||
| rrnL | 1093 | 2667 | + | 1575 | 0 | ||
| trnL2 | 2668 | 2742 | + | 75 | 1 | ||
| ND1 | 2744 | 3700 | + | 957 | −2 | GTG | TAG |
| trnI | 3699 | 3767 | + | 69 | −3 | ||
| trnQ | 3765 | 3836 | − | 72 | −1 | ||
| trnM | 3836 | 3905 | + | 70 | 0 | ||
| ND2 | 3906 | 4949 | + | 1044 | −2 | ATT | TAG |
| trnW | 4948 | 5015 | + | 68 | 5 | ||
| trnA | 5021 | 5089 | − | 69 | 0 | ||
| trnN | 5090 | 5163 | − | 74 | 2 | ||
| trnC | 5196 | 5263 | − | 68 | 0 | ||
| trnY | 5264 | 5327 | − | 64 | 1 | ||
| COX1 | 5329 | 6873 | + | 1545 | −3 | ATG | TAA |
| trnS2 | 6871 | 6939 | − | 69 | 3 | ||
| trnD | 6943 | 7011 | + | 69 | 1 | ||
| COX2 | 7013 | 7696 | + | 684 | 2 | ATG | TAA |
| trnK | 7699 | 7763 | + | 65 | 1 | ||
| atp8 | 7765 | 7968 | + | 204 | −43 | ATG | TAA |
| atp6 | 7926 | 8606 | + | 681 | −1 | ATG | TAA |
| COX3 | 8606 | 9390 | + | 785 | −1 | ATG | TTA |
| trnG | 9390 | 9457 | + | 68 | 0 | ||
| ND3 | 9458 | 9805 | + | 348 | 0 | ATT | TAA |
| trnR | 9806 | 9874 | + | 69 | 0 | ||
| ND4L | 9875 | 10,171 | + | 297 | −7 | ATG | TAA |
| ND4 | 10,165 | 11,542 | + | 1383 | 0 | ATG | ACT |
| trnH | 11,543 | 11,610 | + | 68 | 0 | ||
| trnS1 | 11,611 | 11,669 | + | 59 | 0 | ||
| trnL1 | 11,670 | 11,739 | + | 70 | 0 | ||
| ND5 | 11,740 | 13,551 | + | 1812 | 1 | ATT | TAA |
| ND6 | 13,553 | 14,077 | − | 525 | 1 | TCT | CAT |
| trnE | 14,079 | 14,148 | − | 70 | 5 | ||
| Cytb | 14,154 | 15,293 | + | 1140 | 2 | ATG | AGA |
| trnT | 15,296 | 15,365 | + | 70 | 1 | ||
| trnP | 15,367 | 15,433 | − | 67 | 321 | ||
| Regions | Size (bp) | A (%) | C (%) | G (%) | T (%) | A+T (%) | C+G (%) | AT-Skew | GC-Skew |
|---|---|---|---|---|---|---|---|---|---|
| Mitochondrial genome | 16,497 | 34.3 | 23.9 | 11.6 | 30.2 | 64.5 | 35.5 | 6.36 | −34.7 |
| PCGs | 11,405 | 32.8 | 25.8 | 10.5 | 31.0 | 63.8 | 36.2 | 2.82 | −42.3 |
| PCGs(J) | 10,880 | 32.4 | 25.6 | 10.6 | 31.3 | 63.8 | 36.2 | 1.72 | −41.4 |
| PCGs(N) | 525 | 40.8 | 29.7 | 6.7 | 22.9 | 63.6 | 36.4 | 28.1 | −63.2 |
| tRNAs | 1509 | 36.4 | 20.1 | 13.9 | 29.6 | 66.0 | 34.0 | 10.3 | −18.2 |
| tRNAs(J) | 956 | 37.4 | 17.4 | 14.4 | 30.8 | 68.2 | 31.8 | 9.68 | −9.43 |
| tRNAs(N) | 553 | 34.7 | 24.8 | 13.0 | 27.5 | 62.2 | 37.8 | 11.6 | −31.2 |
| rRNAs | 2530 | 37.9 | 19.4 | 16.0 | 26.7 | 64.6 | 35.4 | 17.3 | −9.6 |
| A+T | 1064 | 38.3 | 20.8 | 9.7 | 31.3 | 69.5 | 30.5 | / | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Na, C.; Wang, X.; Cheng, Y.; Huang, Y.; He, S.; Khanal, L.; Chen, S.; Jiang, X.; Chen, Z. Mitogenomic Insights into Phylogeny, Biogeography and Adaptive Evolution of the Genus Typhlomys (Rodentia: Platacanthomyidae). Animals 2025, 15, 2823. https://doi.org/10.3390/ani15192823
Na C, Wang X, Cheng Y, Huang Y, He S, Khanal L, Chen S, Jiang X, Chen Z. Mitogenomic Insights into Phylogeny, Biogeography and Adaptive Evolution of the Genus Typhlomys (Rodentia: Platacanthomyidae). Animals. 2025; 15(19):2823. https://doi.org/10.3390/ani15192823
Chicago/Turabian StyleNa, Chao, Xiaohan Wang, Yaxin Cheng, Yixin Huang, Shuiwang He, Laxman Khanal, Shunde Chen, Xuelong Jiang, and Zhongzheng Chen. 2025. "Mitogenomic Insights into Phylogeny, Biogeography and Adaptive Evolution of the Genus Typhlomys (Rodentia: Platacanthomyidae)" Animals 15, no. 19: 2823. https://doi.org/10.3390/ani15192823
APA StyleNa, C., Wang, X., Cheng, Y., Huang, Y., He, S., Khanal, L., Chen, S., Jiang, X., & Chen, Z. (2025). Mitogenomic Insights into Phylogeny, Biogeography and Adaptive Evolution of the Genus Typhlomys (Rodentia: Platacanthomyidae). Animals, 15(19), 2823. https://doi.org/10.3390/ani15192823






